Introduction: Poised at the threshold of a genetic revolution
Studies into the genetics of cardiovascular disease and its risk factors have identified many chromosomal loci that contribute to cardiovascular traits. When combined with findings from basic, hypothesis-driven research, we now have a much clearer outline of the genes that contribute to cardiovascular traits (the “parts list”), including many that were unsuspected based on candidate gene approaches. The discovery of so many disease loci promises to remake our understanding of disease mechanisms and susceptibility. Particularly exciting is the prospect that new biomarkers of cardiovascular risk can be identified that are orthogonal to traditional cardiac risk factors; that is, they confer risk through mechanisms that are largely independent of traditional risk factors. The existence of such orthogonal risk factors is suggested by the observation that 15–20% of patients presenting with coronary disease have no traditional risk factors.1
However, the clinical translation of these discoveries faces several challenges. First, the cellular and pathophysiologic significance of most susceptibility alleles is unknown, especially for those at previously unannotated loci. Furthermore, it is not obvious how to integrate an individual's genotype at multiple disease loci with their environmental exposures to arrive at an assessment of individual disease risk; doing so requires not only clinical studies, but also basic investigations into how disease genes are organized into pathways and networks. In the therapeutic realm, studies are just beginning to examine how genotypic information can inform the choice and dosing of medications. More broadly, generalizable approaches are needed to identify genes and pathways that can best be targeted for therapeutic effect.
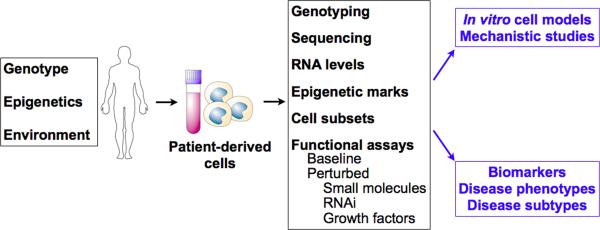
Addressing these translational roadblocks requires patient-focused investigations, as well as experimental systems where the effects of disease alleles can be studied under their native physiologic regulatory mechanisms. Patient-derived cells may be well suited to address these challenges: these cells are increasingly amenable to phenotyping in the context of clinical studies, and are also being used as experimental model systems for mechanistic studies of disease alleles. Because patient cells are intact biological systems subject to physiologic regulation, they are a potentially powerful system for functional and mechanistic studies, including experiments that measure response to specific perturbations (e.g., by small molecule drugs or inhibitory RNAs). In addition, using patient cell samples as a model system permits the study of human disease alleles within their native genetic context, a complementary approach to the study of engineered alleles (overexpression or knockout) in cellular or animal models. Patient-derived cells may yield novel subphenotypes that integrate multiple genetic and environmental influences, help define genotype-phenotype correlations, and provide biomarkers for disease activity and susceptibility. (Figure 1)
Fig. 1.
Phenotypes of patient-derived cells reflect an integration of genetic, epigenetic and environmental influences. Summary of potential utility of patient-derived cells as a source of functional phenotypes, in vitro model systems to study disease biology, and novel biomarkers.
In this review, we discuss the use of patient cells as cellular disease models and a source of novel phenotypes in cardiovascular disease, using current examples such as platelet function assays, endothelial progenitor cell phenotypes, induced pluripotent stem cells (iPSCs), and gene expression profiling of peripheral monocytes. We also discuss emerging technologies that can enable robust and sophisticated cell phenotyping on small numbers of patient-derived cells. A discussion of the use of patient-derived cells for regenerative therapies (such as induced pluripotent stem cells or autologous progenitors) is beyond the scope of this review, but has been reviewed elsewhere.2–6
Platelet reactivity assays: proof-of-concept for relating genetics, functional assays and therapy in individuals
Platelets, cell fragments derived from megakaryocytes, are critical pathogenic factors in acute coronary syndromes and the targets of several drugs. “Resistance” to anti-platelet therapy, or persistent platelet aggregation despite anti-platelet therapy, has emerged as a potential risk factor for secondary acute coronary events.7 A recent meta-analysis of 20 prospective studies including 2930 patients with cardiovascular disease estimated the prevalence of aspirin “resistance” at 28%, and linked aspirin-resistance with an approximately four-fold increased risk of death and major adverse cardiovascular events.8 Clopidogrel resistance has been reported to have a prevalence of 4–30%.7
Platelet aggregation and aspirin responsiveness are strongly heritable, with heritability estimates of 0.48–0.62 and 0.266–0.762, respectively.9, 10 However, several genetic, physiologic, and environmental factors can influence response to anti-platelet therapy, such as metabolizing enzymes, genetic polymorphisms of platelet targets, drug bioavailability, high platelet turnover, drug-drug interactions, and medication non-adherence.
Functional assessments of platelet activity are attractive as a means of integrating these genetic and environmental variables at the level of the individual patient. Available platelet function assays provide quantitative measurements of platelet activation, adhesion, and aggregation (reviewed in11). Several assays measure glycoprotein IIb/IIIa-dependent platelet aggregation induced by an agonist such as ADP (e.g., light transmittance and impedance aggregometry: VerifyNow (Accumetrics), or Plateletworks (Helena Laboratories)). Others measure shear-induced platelet adhesion and aggregation (e.g., Impact (DiaMed) and PFA-100 (Siemens)). Flow cytometry-based methods measure a variety of species associated with platelet activation, such as phosphorylated vasodilator-stimulated phosphoprotein (VASP assay (BioCytex)), activated GP IIb/IIIa, and P-selectin.
These assays have been used in clinical trials to demonstrate a correlation between elevated on-treatment, or residual platelet reactivity (RPR), and adverse cardiovascular outcomes.12, 13 For instance, Breet et al. compared five platelet function assays in 1069 consecutive patients undergoing elective coronary stenting and treated with aspirin and clopidogrel for at least one year. Elevated RPR as determined by three aggregation assays (light transmittance aggregometry, VerifyNow, and Plateletworks) was significantly, but modestly, associated with the primary composite endpoint of all-cause mortality, nonfatal myocardial infarction (MI), stent thrombosis, and ischemic stroke.14 The results are consistent with several smaller studies linking elevated RPR with increased risk of drug-eluting stent (DES) thrombosis,15, 16 major adverse cardiac events following coronary intervention for acute coronary syndromes (ACS),17, 18 or major adverse cardiac events following DES placement for unprotected left main disease.19 Collectively these data suggest that platelet function assays can inform risk assessment during anti-platelet therapy.
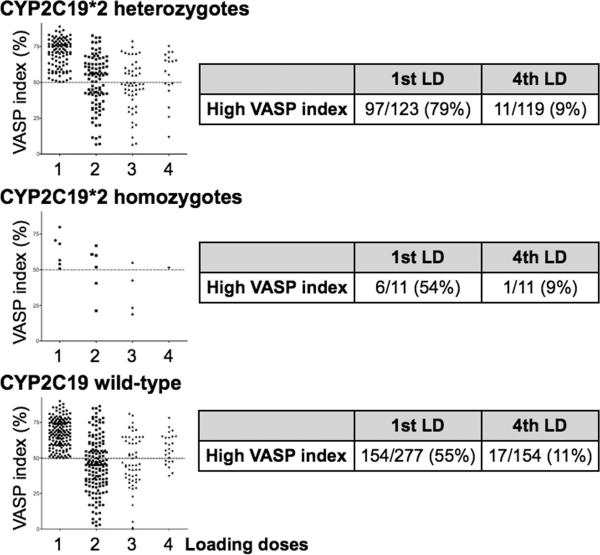
An actively studied question is whether functional platelet assays can guide individual treatment decisions, particularly when genotypic data are available. In a study of 411 patients with non-ST elevation ACS, additional clopidogrel loading doses guided by the VASP platelet function assay achieved effective platelet inhibition in the vast majority of subjects. This held true even in patients bearing the CYP2C19*2 polymorphism, which confers reduced clopidogrel metabolism and lower levels of the active metabolite. Patients with VASP index > 50% received an additional clopidogrel dose followed by a repeat VASP assay, up to a total of 4 clopidogrel loading doses. Using this protocol, 88% of patients bearing at least one copy of the CYP2C19*2 polymorphism achieved a VASP index < 50%.20 (Figure 2)
Fig. 2.
Clopidogrel dosing guided by platelet function assays helps achieve platelet inhibition in carriers of the CYP2C19*2 allele. Graphs show individuals who received additional clopidogrel loading doses (LD) if their post-clopidogrel platelet function remained elevated (according to the VASP assay), up to a total of 4 loading doses. Charts summarize the percentage of patients with elevated VASP index after the 1st and 4th loading doses (if needed). Adapted from20.
In the GRAVITAS trial, 2,214 patients with elevated RPR based on the VerifyNow assay (measured 12–24 hours after DES placement) were randomized to high-dose clopidogrel (600mg followed by 150mg daily) vs. standard dosing (no additional loading followed by 75 mg daily) for 6 months.21 There was no difference in the primary composite end point of cardiovascular death, nonfatal MI, or stent thrombosis at 6 months in high-dose vs. standard clopidogrel dosing. The null result may be due to several factors. First, the overall event rate for the primary composite end point (2.3% in both groups) was relatively low. Second, the arm randomized to high-dose clopidogrel only showed a modest decrease in the incidence of high RPR at 30 days, raising the possibility that more intensive clopidogrel adjustments or an alternative thienopyridine such as prasugrel may have fared better. Subsequent studies may clarify several questions raised by this trial: whether dosing changes triggered by lower RPR thresholds or repeated platelet function assays improve outcomes; whether individuals who experience an adverse event have higher RPR than those who do not; whether RPR could guide the selection of an alternative thienopyridine, such as prasugrel; and whether RPR-guided therapy can improve outcomes in selected high-risk patients. Genetic substudies of GRAVITAS are also pending.22
Overall, platelet function assays have gained traction as tools with both investigational and clinical significance. Several assays are available as point-of-care tests, though caution is still warranted with regard to their widespread use, due to the lack of universal standards for sample handling and defining thresholds for elevated RPR. Nonetheless, clinical data convincingly link functional phenotypes (i.e., elevated RPR) to adverse clinical outcomes. Detailed outcomes trials are underway to determine whether platelet function assays can yield clinically actionable information, particularly in the presence of CYP2C19*2 and other genetic variants that affect clopidogrel activation.
The potential of patient cell-based phenotypes
The promising results of platelet function assays raise the possibility that functional cell-based assays may have broader relevance to the study and treatment of cardiovascular disease. By analogy, oncology has benefited tremendously from routine sampling of malignant cells in the course of usual clinical care. Ready access to cancer samples has resulted in large collections of cancer cell lines that are widely studied across institutions (e.g., the NCI-60 cell lines); primary biopsy specimens are also intensively studied for genetic or phenotypic markers. Patient-derived cancer samples have led to the discovery of novel oncogenes associated with common cancers, the identification of novel disease sub-types (including those with very different prognoses), and the development of new therapies. At several institutions, patient tumor samples are genetically analyzed to identify individuals who are candidates for new molecularly targeted therapies. In sum, studies of patient-derived cancer samples have enabled profound advances in the understanding of disease mechanisms, patient diagnosis and prognosis, development of new therapies, and individualized selection of therapy.
Despite these successes in oncology, however, the broad adoption of patient cell-based studies to cardiovascular disease is not straightforward. To a first approximation, certain carcinogenic mechanisms are driven by somatic mutations that lead to dysregulated cellular proliferation in a cell-autonomous manner. As a result, studying the primary tumor can yield insights into causative genetic aberrations and critical disease mechanisms, even as other approaches are needed to define the role of host mechanisms, germline genotype, and tumor microenvironment. In contrast, cardiovascular traits commonly involve interactions between multiple cell types and organs, communicating through hormones and circulating cells. Study of any individual cell-type may thus convey only a partial view of disease biology. The effects of individual disease alleles are also likely to be more modest.
In addition, access to primary patient samples of many disease-relevant cells, such as endothelial, vascular smooth muscle, or myocardial cells, is minimal during usual clinical care. While samples may be obtained in association with invasive procedures (e.g., percutaneous vascular intervention, carotid endarterectomy, heart explantation, LVAD implantation, and coronary artery bypass grafting), specimens from end-stage disease may be less informative for earlier pathogenic processes. While a limited number of relevant cell lines are used, much research uses primary isolates that are physiologic but difficult to obtain and have short life-spans in culture.
Fortunately, certain cell types are both accessible from patients and clearly involved in disease pathogenesis, such as monocytes and lymphocytes. Furthermore, accessible cells, even if not conventionally regarded as key participants in disease, may share signaling pathways with canonical disease tissues, and thus be useful as surrogate model systems. In perhaps the most prominent historical example of the use of accessible, surrogate cell models, Brown and Goldstein demonstrated regulation of HMG-CoA-reductase by LDL not in liver cells, but in primary skin fibroblasts.23 They also demonstrated LDL-receptor activity in vivo for the first time using circulating blood lymphocytes.24
In the following sections, we highlight recent research that utilizes patient-derived cells to study disease mechanisms or patient populations, and may illustrate paradigms for how these cells could be powerful tools for translational studies. The cells discussed are accessible in the context of patient care or clinical research cohorts, and are appealing for several reasons: they are accessible by routine phlebotomy or skin biopsy (in the case of induced pluripotent stem cells (iPS)); they include inflammatory and immune cells such as monocytes, or iPS-derived cells that directly contribute to cardiovascular pathogenesis; certain cell populations can be purified based on surface marker expression; and peripherally circulating cells are in direct contact with the endothelium, cytokines, and other hormones, thereby providing an integrated readout of cellular responses to environmental factors.
Peripheral Blood Mononuclear Cell Phenotypes
Circulating inflammatory and immune cells, such as peripheral blood mononuclear cells (PBMCs) and T-cells, play important roles in atherosclerosis, remodeling after myocardial infarction, heart failure, and transplant rejection.25–27 In an early example of a cell-based biomarker, the white blood cell count was long ago established as a predictor of myocardial infarction.28 More recently, leukocytosis has been shown to predict CAD burden and adverse outcome after MI.29, 30 Subsequent research has correlated increasing monocyte count with adverse risk or outcomes in cardiovascular disease, particularly acute MI,31 in-stent restenosis,32 subclinical carotid atherosclerosis,33 and post-MI LV dysfunction and aneurysm.34
Several early efforts to use PBMCs as surrogate cell models employed genome-wide gene expression analysis. Differentially regulated genes, many involved in macrophage activation and differentiation, were identified in multiple forms of cardiovascular disease, including atherosclerosis,35, 36 carotid disease,37 ischemic stroke,38 and diabetes.39
In the most clinically advanced application to date of PBMC analysis, gene expression profiling has been used to detect rejection following heart transplantation. In transplant patients undergoing endomyocardial biopsy, Cappola and colleagues identified an expression signature in whole blood that differentiated control patients without rejection from those with stage 3A rejection or higher.40 Recently, Pham et al. randomized cardiac transplant patients to rejection monitoring by routine endomyocardial biopsy vs. a PBMC gene expression signature score, previously shown to have a 99.6% negative predictive value for histologic rejection.41, 42 Over a median follow-up period of 19 months, there was no difference using a non-inferiority comparison between the two groups in the primary composite endpoint of first occurrence of rejection with hemodynamic compromise, graft dysfunction due to other causes, death, or retransplantation. Overall death at two years was also comparable. Notably, gene expression monitoring significantly decreased the number of biopsies performed (0.5 vs. 3.0, P < 0.001). This result awaits further confirmation in larger populations, but constitutes an exciting example of a cell-based phenotype that conveys diagnostic information and can impact patient management.
A growing body of work has identified monocyte subpopulations with distinct functions in normal and pathological states, particularly in atherosclerosis and healing after MI (reviewed in43, 44). In mice, monocytes expressing high levels of the lymphocyte antigen complex Ly6-C (Ly6-Chi) secrete numerous inflammatory and proteolytic mediators, increase in response to hypercholesterolemia, home to sites of injury, and differentiate into foam cells.45 Ly6-Clo monocytes, in contrast, have an immunomodulatory role, and mediate granulation and wound-healing. Moreover, LV healing after myocardial infarction involves sequential mobilization of Ly6-Chi monocytes followed by Ly6-Clo monocytes.46
Although humans lack direct equivalents of the murine Ly6-Chi and Ly6-Clo monocytes, functional monocyte subsets also contribute to human cardiovascular disease and post-MI LV healing.43, 44, 47 Increased CD14+CD16+ monocytes are associated with coronary artery disease and carotid intima-media thickness.48, 49 After anterior STEMI, CD14+CD16− and CD14+CD16+ monocytes sequentially accumulate in infarcted tissue, analogous to the murine observations; the magnitude of the early CD14+CD16− monocyte peak was negatively correlated with myocardial salvage.50, 51 These studies demonstrate strong biologic links between monocyte subset dynamics and cardiovascular disease. Further clinical studies and functional assays for monocyte subsets (that complement FACS-based counts of cell number) may illuminate the role of these subsets in disease, and lead to the use of monocyte phenotypes as biomarkers of CAD risk or adverse LV remodeling after MI.
Endothelial Progenitor Cells
Cells termed “endothelial progenitor cells” (EPCs) have been intensively studied as a potential biomarker and source of cells for cardiovascular regenerative therapy. EPCs, broadly defined, refer to circulating bone-marrow derived cells that exhibit endothelial properties in vitro, and promote neovascularization and endothelial repair in vivo.52 Asahara et al. first described cells with these properties, and identified them as circulating cells expressing CD34 -- as well as varying degrees of Flk1 (VEGF receptor 2), CD45, and CD31.53
Several methods have been used in the literature to isolate EPCs from the PBMC fraction, including FACS and in vitro culture assays. FACS-based definitions typically include combinations of CD34, VEGF receptor 2 (VEGFR-2, or KDR) or the hematopoietic stem cell marker AC133. (These markers may not be required for EPC activity, however; a murine subpopulation of lin−/c-Kit−/Sca-1− cells has been reported that differentiates into endothelial-like cells in vitro with high replicative potential.54) Culture-based approaches include a colony-forming-unit (CFU) assay,55 and an early outgrowth EPC assay for uptake of acetylated LDL (AcLDL) and Ulex europaeus agglutinin I.56
In the absence of a specific marker for EPCs, none of these assays isolates a homogeneous cell population, and the different assays identify distinct functional cell subsets, each of which likely correlates with different biological activities. Furthermore, many lines of evidence suggest that (a) vasculogenic properties of EPCs likely arise from paracrine effects as opposed to true progenitor cell activity, and (b) commonly used EPC definitions identify cells of hematopoietic and not endothelial origin, including varying proportions of monocytes, lymphocytes, and platelets.57–60 For example, in early outgrowth EPC protocols that lack a pre-plating step, platelet contamination may lead to uptake of platelet microparticles by adherent mononuclear cells.58, 61 Contaminating platelets and platelet microparticles can confer binding to Ulex europaeus agglutinin I and transfer of platelet proteins to the mononuclear cells,62 resulting in apparent endothelial patterns of staining which are in fact due to platelet-derived proteins. Platelet microparticles can also alter the functional phenotype of early outgrowth EPCs, increasing expression of proangiogenic paracrine factors, and enhancing EPC adhesion to injured vascular endothelium with accelerated reendothelialization in vivo.62 In light of the difficulty of isolating a homogeneous cell population that confers EPC-like properties, unbiased approaches such as genome-wide gene expression may provide empiric signatures of circulating cells important for vascular repair and neovascularization.63
Yoder and colleagues have identified an endothelial colony-forming cell (ECFC; also called late outgrowth endothelial cells) that forms in vitro colonies with a typical cobblestone appearance and shows a robust in vitro capacity for proliferation, self-renewal, and tube formation.64, 65 ECFCs are most abundant in umbilical cord blood and are present in peripheral circulation at very low concentrations (~1 colony per 108 mononuclear cells plated). Thus ECFCs could conceivably be isolated from cord blood and used for mechanistic studies and genotype-phenotype correlations. Their scarcity in peripheral blood, however, may render their use in adult clinical studies impractical, barring improved methods for their detection.
Multiple studies in selected patient populations have reported that EPCs are inversely associated with overall cardiovascular risk, clinical outcomes, traditional cardiovascular risk factors, and markers of endothelial function.55, 56, 66–68 In two large community cohorts, EPCs showed only sparse associations with traditional cardiovascular risk factors.69, 70 Despite this, in the community-based Framingham Heart Study, the EPC CFU phenotype was inversely associated with the Framingham risk score and subclinical phenotypes of cardiovascular disease, such as coronary and aortic calcification.71 Furthermore, a genome-wide association study revealed that SNPs at the SLC22A3-LPAL2-LPA locus on chromosome 6, a locus previously associated with myocardial infarction,72 were highly associated with CFU (P=4.9×10−7) and risk of MI (P=1.1×10−4). Specifically, SNPs associated with lower CFU numbers were associated with increased risk of MI, and vice versa.70 These data provide genetic support for the notion that EPCs mediate risk of cardiovascular disease and MI at least in part through mechanisms orthogonal to traditional risk factors. Going forward, patient-derived EPCs and ECFCs may be useful as biomarkers of cardiovascular risk, or model systems to study vascular repair. However, further efforts are needed to clarify which cell subsets contribute to observed EPC phenotypes in vitro, which in vivo biological processes are mediated by EPCs, and whether simply measuring number, rather than function, of EPCs is a faithful surrogate for in vivo activity.
Lymphoblast Cell Lines
Lymphoblast cell lines (LCLs), created by the transformation of human B-lymphocytes by Ebstein-Barr virus, were originally devised as a renewable source of DNA for genetic studies. For instance, with the collection of 270 LCLs genotyped by the International HapMap Project73 (representing individuals of Northern European, African, and Asian ancestry), investigators can leverage publicly available datasets of genome-wide SNP genotyping and gene expression for their studies. LCLs possess certain limitations that must be addressed or mitigated for individual studies, including a restricted repertoire of expressed genes related to their B-lymphocyte lineage, variable growth rates, and the potential for confounding effects of EBV infection.74
Nonetheless, the availability of LCLs in association with many phenotyped patient cohorts, coupled with their capacity for in vitro expansion, make patient-derived LCLs an appealing model system for functional studies. LCLs have been widely used to study the genetic determinants of variation in human gene expression.75 LCLs have also been used successfully as experimental models in the study of congenital hyperinsulinism/hyperammonemia,76 pharmacogenomic studies,77–79 ion channel disorders,80 and Huntington's disease.81 In Huntington's disease, LCLs are much more accessible than primary neuronal tissue and display disease-relevant phenotypes. For instance, the ratio of cellular [ATP]/[ADP] shows an inverse correlation with the length of the unstable triplet repeat in both human LCLs and striatal neurons from mice expressing mutant huntingtin (with 111 triplet repeats).81 This surrogate phenotype led to the discovery of a defect in mitochondrial energy metabolism in Huntington's disease and is also correlated with other Huntington's subphenotypes.82
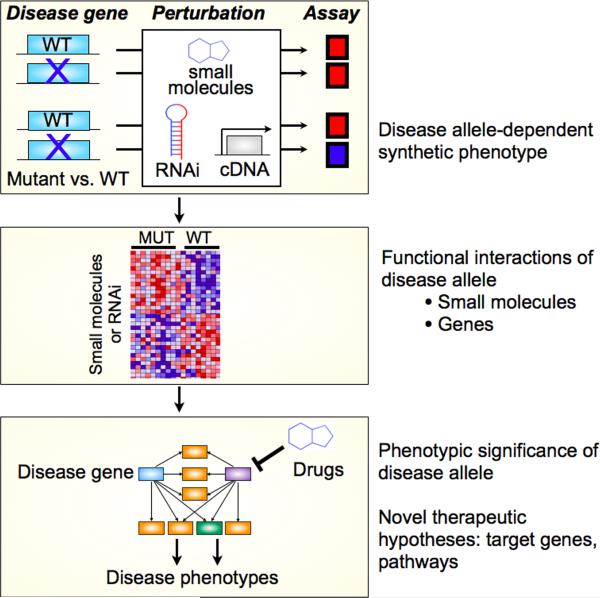
More broadly, the existence of genotyped LCLs from clinical cohorts has opened new avenues for assigning function to disease alleles. LCLs have been used to elucidate the function of susceptibility alleles identified through genome-wide association studies (GWAS) for systemic lupus erythematosus (SLE)83 and childhood asthma.84 Importantly, the capacity of LCLs for in vitro expansion enables their use in high-throughput perturbational screening. For instance, a chemical-genetic interaction screen was performed in LCLs from related individuals with a monogenic form of diabetes (Maturity Onset Diabetes of the Young type 1, or MODY1), caused by mutation in the orphan nuclear hormone receptor HNF4α.85 LCLs were treated with approximately 4,000 marketed drugs and other compounds with characterized mechanisms. By analyzing screening data for LCLs from 10 MODY1 individuals and from 8 wild-type related controls, classes of compounds were identified that induced distinct effects between mutant and wild-type LCLs (i.e., a chemical-gene interaction). Many of these functional interactions were preserved in pancreatic β-cells, a canonical cell model for diabetes. Furthermore, several of these compounds favorably modulated a key disease phenotype, insulin secretion from pancreatic β-cells.85 Thus LCLs can serve as a surrogate cell system for functional studies of disease alleles. The use of LCLs in perturbational screens (by small molecules or inhibitory RNA, for instance) allows the logic of synthetic genetic interaction screens to be applied to human cells, in order to elucidate networks of genes that interact with disease alleles, and identify proteins or pathways that can be modulated for therapeutic effects. (Figure 3)
Fig. 3.
Perturbational screens on patient-derived cells can clarify the functional significance of disease alleles and lead to new therapeutic hypotheses. Using the logic of synthetic genetic interactions, patient cells can be screened for small molecules, RNAi, or cDNA that cause different phenotypes in cells that are mutant or wild-type at a disease gene. This identifies small molecules or genes that functionally interact with a disease gene, identifies pathways that may connect the disease allele to disease phenotypes, and provides clues for new therapeutic targets and hypotheses. Adapted from85.
Induced pluripotent stem cells (iPSCs)
iPSCs have recently emerged as a transformative technology for patient-specific cellular disease models (in addition to their potential as a source of cells for regenerative therapies).86–88 Adult somatic cells, such as skin fibroblasts, are reprogrammed into pluripotent stem cells with defined factors, and may then be differentiated into specific cell lineages. iPS-derived cardiomyocyte models have been developed for long QT syndrome type 1 (caused by mutation in KCNQ1)89 and type 2 (caused by mutation in KCNH2).90 In both cases, cardiomyocytes display altered activity of the expected current (IKs and IKr, respectively), prolonged action potential duration, and enhanced arrhythmogenicity.
iPS-derived cells can also model cardiac manifestations of complex multi-organ syndromes. For instance, iPSC were derived from patients with LEOPARD syndrome bearing mutation in the PTPN11 gene (encoding the SHP2 phosphatase).91 LEOPARD iPS-derived cardiomyocytes show increased size, sarcomeric organization and NFATC4 nuclear localization consistent with hypertrophic cardiomyopathy (commonly observed in LEOPARD patients). In addition, LEOPARD iPSCs recapitulate the expected abnormalities in RAS-MAPK signaling characteristic of the syndrome. iPSC have also been derived from patients with Timothy syndrome (due to mutation in the L-type calcium channel Ca(V)1.2, and associated with a long QT syndrome);92 differentiated ventricle-like cells display abnormal Ca+2 transients, prolonged action potentials, and irregular electrical activity and contraction. In Hutchison Gilford Progeria (HGP, caused by mutation in lamin A, and associated with premature atherosclerosis), iPS-derived vascular smooth muscle and mesenchymal stem cells have decreased viability in response to stress (such as hypoxia); these defects may underlie the impaired ability of HGP iPS-derived mesenchymal stem cells to rescue hind limb ischemia in vivo.93
iPS-derived cellular models may also facilitate the discovery of new therapies. For instance, iPS-derived cardiomyocyte models of long QT syndrome were successfully used to evaluate the effects of established and novel drugs on action potential duration and arrhythmic susceptibility.89, 90 These and other iPS-derived cardiomyocyte models may provide a platform to screen drug candidates for their propensity to prolong the QT interval, an adverse effect of great concern in drug development. More generally, iPS-derived cellular models may facilitate drug discovery by providing a ready source of traditionally non-accessible human cells (e.g., cardiomyocytes) that harbor clinically relevant disease mutations in their native genetic context.
Emerging technology platforms for phenotyping of patient-derived cells
With the exception of LCLs and iPSCs, the patient-derived cells mentioned above have limited in vitro proliferative capacity. As a result, the number of cells available for study is principally determined by the sample size feasibly obtained from subjects, such as the volume of peripheral blood. These limitations can be mitigated by the use of sensitive detection methodologies (often with amplification steps as in gene expression studies) or the use of relatively simple phenotypes, such as cell counts based on FACS measurements or CFU assays. However, functional studies would benefit from technology platforms that enable more detailed phenotyping on limited samples. Ideally, such platforms would facilitate (i) isolation of potentially rare cells from peripheral blood at a reasonable yield and purity, (ii) phenotypic analysis of small numbers (i.e., hundreds to thousands) of cells with minimal subculturing, and (iii) preserved cell viability. Finally, these platforms should be sufficiently inexpensive and robust to be applied to patient samples.
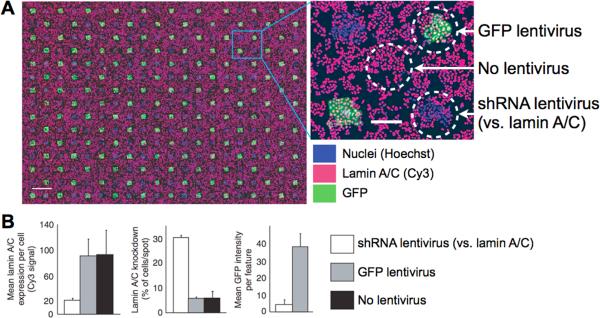
One approach to decreasing cell requirements is to measure cell phenotypes in formats denser than 384-well assay plates. For instance, cellular microarrays consist of small molecules, or lentiviruses encoding cDNA or shRNA, printed at high density on glass slides. When cell suspensions are incubated on the slides, cells attach and are perturbed by the compounds, cDNA, or shRNA.94–96 Cellular phenotypes arising from these perturbations can then be quantified using automated microscopy. This format allows perturbational screens to be performed on ~100 cells per spot (a decrease of approximately one order of magnitude compared to screening in 384-well plates), with approximately 5,000 spots per glass slide.96 (Figure 4)
Fig. 4.
Cell microarray incubated with HeLa cells demonstrates both lentiviral gene knockdown and gene expression on printed spots. Each spot is approximately 250 μm in diameter and contains ~150 cells. A. Cells in contact with a GFP lentivirus spot show effective GFP expression (circle at top right); cells in contact with shRNA against lamin A/C show loss of Cy3 lamin A/C signal (lower right). The circle at center shows HeLa cells attached to the slide but not touching a lentivirus spot. B. Quantitation of lamin A/C knockdown and GFP expression from cell microarrays. Scale bar, 1 mm (left panel) and 200 μm (right). Adapted from96.
Microfluidic chips combine nanotechnology with microfluidic large-scale integration, resulting in devices that manipulate nanoliter fluid volumes across thousands of integrated, miniaturized components for detailed cellular processing, functional assays and phenotyping.97–100 The miniaturization of these devices allows significant economies of scale for cells, reagents, and labor (such as pipetting). Devices are typically fabricated from a biocompatible substrate (such as polydimethylsiloxane, or PDMS), with controllable valves that enable sample loading and mixing of fluid streams. Cell subpopulations may be presorted before loading onto the chip, or potentially sorted on-chip. Integrated fluidic circuits can accomplish a wide variety of manipulations, including sample processing, incubation with small molecules, and parallel handling of multiple samples. Phenotyping can be performed on the level of single cells or cell populations using such readouts as fluorescent microscopy or quantitative RT-PCR.101–104
Microfluidic applications have phenotyped circulating cells from patients. For instance, a microfluidic device can capture neutrophils from a drop of whole blood, generate chemoattractant gradients, and monitor chemotaxis at the level of the individual cell.105, 106 Another promising application detects and characterizes circulating tumor cells (CTCs) in patients with epithelial cancers,107 particularly prostate cancer.108 Cells are captured by immobilized antibodies against a cell surface marker (e.g., EpCAM), fluorescently labeled with antibodies against prostate-specific antigen (PSA), and then subjected to semi-automated image capture and analysis. This device allows the monitoring of CTC numbers after primary tumor resection, and extraction of DNA or RNA for analysis. Nanopatterning of the PDMS surface has been used to enhance the efficiency of CTC capture. For instance, three-dimensional nanotexturing increases the surface area for capture,109 and nanochannels etched in a herringbone pattern induce microvortices, chaotic mixing, and increased encounters between flowing CTCs and immobilized capture antibodies.110 In another example, a microfluidic device was combined with a nuclear magnetic resonance nanosensor array and targeted magnetic nanoparticles, allowing highly sensitive detection of cancer cells bearing specific surface markers.111
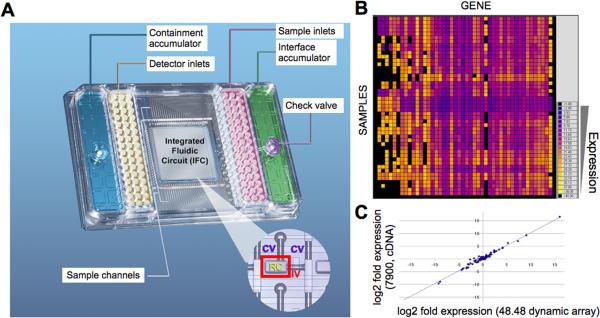
The integration of microfluidics with sophisticated measurements of gene expression, proteins,112 and epigenetic marks113 may yield devices that can interrogate pathway activity in patient-derived cells. For instance, Tay et al. combined fluorescence microscopy and gene expression to characterize transduction of TNF-α signals into NF-κB-dependent transcription at the resolution of single cells.114 Robust microfluidic platforms for quantitative RT-PCR gene expression analyses are now commercially available, and may be useful to study patient samples.115 (Figure 5)
Fig. 5.
Miniaturized quantitative RT-PCR analysis enabled by microfluidic integrated fluidic circuits. A. Image of a 48 × 48 microfluidic chip. The integrated fluidic circuit (at center) contains 2304 reaction chambers (RC, outlined by a red box in inset), bounded by control valves (CV) and interface valves (IV). B. Heatmap of RT-PCR Ct values from individual reaction chambers on a single chip. C. Correlation between expression levels obtained by microfluidic (nanoliter scale) and conventional (microliter scale) quantitative RT-PCR. r = 0.986. Adapted from115.
Conclusions
The notion of cell-based phenotypes in cardiovascular disease is not new to this moment. As mentioned above, patient-derived cell models figured prominently in the work of Brown and Goldstein almost 40 years ago. The advantages of studying patient cells were also recognized in the early days of peripheral blood gene expression analysis for atherosclerosis.116
In light of several recent developments, patient-derived cells increasingly represent a powerful tool for both patient-focused and basic science studies in cardiovascular disease. The great success of human genetic studies has identified dozens of novel genes that contribute to cardiovascular traits. For instance, 95 loci were recently associated with serum lipids, which together explain 10–12% of the observed variation.117 These discoveries created two opportunities: to understand the functional significance of novel disease loci and how they contribute to disease; and to translate these findings into improved clinical care, through novel biomarkers or therapies. The large number of disease loci found to contribute to cardiovascular traits makes both of these goals challenging, and highlights the need for new approaches that can be generalized to many loci and diseases.
Patient-derived cells are uniquely positioned to address these unmet needs, because they can be obtained directly from phenotyped patients, and also serve as model systems for in vitro studies. As clinical phenotypes and outcomes become mapped onto sophisticated cell-based measurements, patient cells will increasingly help draw connections between disease alleles, clinical phenotypes, circulating biomarkers, cellular pathways, and therapeutic hypotheses. An important translational goal is the development of improved biomarkers that can impact clinical decisions; to date, using genetic scores and serum biomarkers to improve cardiovascular risk classification over that conferred by traditional risk factors has proven surprisingly difficult.118–123 Cell-based phenotypes can integrate the effects of multiple genetic and environmental factors, and represent a new dimension for circulating biomarkers. Overall, patient cell phenotyping is likely to become an important tool to study genetically complex cardiovascular traits, providing a complementary approach to genetic, physiologic, proteomic, and metabolomic methods.
Acknowledgments
Funding Sources This work was supported by grant NIH/NHLBI K08 HL077186 (S.Y.S.) and the de Gunzburg Family Foundation at Massachusetts General Hospital (S.Y.S.).
Footnotes
Disclosures None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Khot UN, Khot MB, Bajzer CT, Sapp SK, Ohman EM, Brener SJ, Ellis SG, Lincoff AM, Topol EJ. Prevalence of conventional risk factors in patients with coronary heart disease. JAMA. 2003;290:898–904. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- 2.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 3.Hare JM, Chaparro SV. Cardiac regeneration and stem cell therapy. Curr Opin Organ Transplant. 2008;13:536–542. doi: 10.1097/MOT.0b013e32830fdfc4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle AJ, McNiece IK, Hare JM. Mesenchymal stem cell therapy for cardiac repair. Methods Mol Biol. 2010;660:65–84. doi: 10.1007/978-1-60761-705-1_5. [DOI] [PubMed] [Google Scholar]
- 5.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J Mol Cell Cardiol. 2011;50:327–332. doi: 10.1016/j.yjmcc.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Sweeny JM, Gorog DA, Fuster V. Antiplatelet drug `resistance'. Part 1: mechanisms and clinical measurements. Nat Rev Cardiol. 2009;6:273–282. doi: 10.1038/nrcardio.2009.10. [DOI] [PubMed] [Google Scholar]
- 8.Krasopoulos G, Brister SJ, Beattie WS, Buchanan MR. Aspirin “resistance” and risk of cardiovascular morbidity: systematic review and meta-analysis. BMJ. 2008;336:195–198. doi: 10.1136/bmj.39430.529549.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donnell CJ, Larson MG, Feng D, Sutherland PA, Lindpaintner K, Myers RH, D'Agostino RA, Levy D, Tofler GH. Genetic and environmental contributions to platelet aggregation: the Framingham heart study. Circulation. 2001;103:3051–3056. doi: 10.1161/01.cir.103.25.3051. [DOI] [PubMed] [Google Scholar]
- 10.Faraday N, Yanek LR, Mathias R, Herrera-Galeano JE, Vaidya D, Moy TF, Fallin MD, Wilson AF, Bray PF, Becker LC, Becker DM. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115:2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- 11.Michelson AD. Methods for the measurement of platelet function. Am J Cardiol. 2009;103:20A–26A. doi: 10.1016/j.amjcard.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Gori AM, Marcucci R, Migliorini A, Valenti R, Moschi G, Paniccia R, Buonamici P, Gensini GF, Vergara R, Abbate R, Antoniucci D. Incidence and clinical impact of dual nonresponsiveness to aspirin and clopidogrel in patients with drug-eluting stents. J Am Coll Cardiol. 2008;52:734–739. doi: 10.1016/j.jacc.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 13.Bonello L, Tantry US, Marcucci R, Blindt R, Angiolillo DJ, Becker R, Bhatt DL, Cattaneo M, Collet JP, Cuisset T, Gachet C, Montalescot G, Jennings LK, Kereiakes D, Sibbing D, Trenk D, Van Werkum JW, Paganelli F, Price MJ, Waksman R, Gurbel PA. Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol. 2010;56:919–933. doi: 10.1016/j.jacc.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 14.Breet NJ, van Werkum JW, Bouman HJ, Kelder JC, Ruven HJ, Bal ET, Deneer VH, Harmsze AM, van der Heyden JA, Rensing BJ, Suttorp MJ, Hackeng CM, ten Berg JM. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA. 2010;303:754–762. doi: 10.1001/jama.2010.181. [DOI] [PubMed] [Google Scholar]
- 15.Buonamici P, Marcucci R, Migliorini A, Gensini GF, Santini A, Paniccia R, Moschi G, Gori AM, Abbate R, Antoniucci D. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol. 2007;49:2312–2317. doi: 10.1016/j.jacc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 16.Sibbing D, Braun S, Morath T, Mehilli J, Vogt W, Schomig A, Kastrati A, von Beckerath N. Platelet reactivity after clopidogrel treatment assessed with point-of-care analysis and early drug-eluting stent thrombosis. J Am Coll Cardiol. 2009;53:849–856. doi: 10.1016/j.jacc.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Patti G, Nusca A, Mangiacapra F, Gatto L, D'Ambrosio A, Di Sciascio G. Point-of-care measurement of clopidogrel responsiveness predicts clinical outcome in patients undergoing percutaneous coronary intervention results of the ARMYDA-PRO (Antiplatelet therapy for Reduction of MYocardial Damage during Angioplasty-Platelet Reactivity Predicts Outcome) study. J Am Coll Cardiol. 2008;52:1128–1133. doi: 10.1016/j.jacc.2008.06.038. [DOI] [PubMed] [Google Scholar]
- 18.Marcucci R, Gori AM, Paniccia R, Giusti B, Valente S, Giglioli C, Buonamici P, Antoniucci D, Abbate R, Gensini GF. Cardiovascular death and nonfatal myocardial infarction in acute coronary syndrome patients receiving coronary stenting are predicted by residual platelet reactivity to ADP detected by a point-of-care assay: a 12-month follow-up. Circulation. 2009;119:237–242. doi: 10.1161/CIRCULATIONAHA.108.812636. [DOI] [PubMed] [Google Scholar]
- 19.Migliorini A, Valenti R, Marcucci R, Parodi G, Giuliani G, Buonamici P, Cerisano G, Carrabba N, Gensini GF, Abbate R, Antoniucci D. High residual platelet reactivity after clopidogrel loading and long-term clinical outcome after drug-eluting stenting for unprotected left main coronary disease. Circulation. 2009;120:2214–2221. doi: 10.1161/CIRCULATIONAHA.109.883454. [DOI] [PubMed] [Google Scholar]
- 20.Bonello L, Armero S, Ait Mokhtar O, Mancini J, Aldebert P, Saut N, Bonello N, Barragan P, Arques S, Giacomoni MP, Bonello-Burignat C, Bartholomei MN, Dignat-George F, Camoin-Jau L, Paganelli F. Clopidogrel Loading Dose Adjustment According to Platelet Reactivity Monitoring in Patients Carrying the 2C19 2* Loss of Function Polymorphism. J Am Coll Cardiol. 2010;56:1630–1636. doi: 10.1016/j.jacc.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Price MJ, Berger PB, Teirstein PS, Tanguay JF, Angiolillo DJ, Spriggs D, Puri S, Robbins M, Garratt KN, Bertrand OF, Stillablower ME, Aragon JR, Kandzari DE, Stinis CT, Lee MS, Manoukian SV, Cannon CP, Schork NJ, Topol EJ. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA. 2011;305:1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 22.Price MJ, Berger PB, Angiolillo DJ, Teirstein PS, Tanguay JF, Kandzari DE, Cannon CP, Topol EJ. Evaluation of individualized clopidogrel therapy after drug-eluting stent implantation in patients with high residual platelet reactivity: design and rationale of the GRAVITAS trial. Am Heart J. 2009;157:818–824. doi: 10.1016/j.ahj.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein JL, Brown MS. Familial hypercholesterolemia: identification of a defect in the regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity associated with overproduction of cholesterol. Proc Natl Acad Sci U S A. 1973;70:2804–2808. doi: 10.1073/pnas.70.10.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho YK, Brown S, Bilheimer DW, Goldstein JL. Regulation of low density lipoprotein receptor activity in freshly isolated human lymphocytes. J Clin Invest. 1976;58:1465–1474. doi: 10.1172/JCI108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nature Rev Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 26.Libby P, Ridker PM, Hansson GK. Inflammation in atherosclerosis: from pathophysiology to practice. J Am Coll Cardiol. 2009;54:2129–2138. doi: 10.1016/j.jacc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolakis S, Lip GY, Shantsila E. Monocytes in heart failure: relationship to a deteriorating immune overreaction or a desperate attempt for tissue repair? Cardiovasc Res. 2010;85:649–660. doi: 10.1093/cvr/cvp327. [DOI] [PubMed] [Google Scholar]
- 28.Friedman GD, Klatsky AL, Siegelaub AB. The leukocyte count as a predictor of myocardial infarction. N Engl J Med. 1974;290:1275–1278. doi: 10.1056/NEJM197406062902302. [DOI] [PubMed] [Google Scholar]
- 29.Cannon CP, McCabe CH, Wilcox RG, Bentley JH, Braunwald E. Association of white blood cell count with increased mortality in acute myocardial infarction and unstable angina pectoris. OPUS-TIMI 16 Investigators. Am J Cardiol. 2001;87:636–639. doi: 10.1016/s0002-9149(00)01444-2. [DOI] [PubMed] [Google Scholar]
- 30.Sabatine MS, Morrow DA, Cannon CP, Murphy SA, Demopoulos LA, DiBattiste PM, McCabe CH, Braunwald E, Gibson CM. Relationship between baseline white blood cell count and degree of coronary artery disease and mortality in patients with acute coronary syndromes: a TACTICS-TIMI 18 (Treat Angina with Aggrastat and determine Cost of Therapy with an Invasive or Conservative Strategy- Thrombolysis in Myocardial Infarction 18 trial)substudy. J Am Coll Cardiol. 2002;40:1761–1768. doi: 10.1016/s0735-1097(02)02484-1. [DOI] [PubMed] [Google Scholar]
- 31.Horne BD, Anderson JL, John JM, Weaver A, Bair TL, Jensen KR, Renlund DG, Muhlestein JB. Which white blood cell subtypes predict increased cardiovascular risk? J Am Coll Cardiol. 2005;45:1638–1643. doi: 10.1016/j.jacc.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 32.Fukuda D, Shimada K, Tanaka A, Kawarabayashi T, Yoshiyama M, Yoshikawa J. Circulating monocytes and in-stent neointima after coronary stent implantation. J Am Coll Cardiol. 2004;43:18–23. doi: 10.1016/j.jacc.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Chapman CM, Beilby JP, McQuillan BM, Thompson PL, Hung J. Monocyte count, but not Creactive protein or interleukin-6, is an independent risk marker for subclinical carotid atherosclerosis. Stroke. 2004;35:1619–1624. doi: 10.1161/01.STR.0000130857.19423.ad. [DOI] [PubMed] [Google Scholar]
- 34.Maekawa Y, Anzai T, Yoshikawa T, Asakura Y, Takahashi T, Ishikawa S, Mitamura H, Ogawa S. Prognostic significance of peripheral monocytosis after reperfused acute myocardial infarction:a possible role for left ventricular remodeling. J Am Coll Cardiol. 2002;39:241–246. doi: 10.1016/s0735-1097(01)01721-1. [DOI] [PubMed] [Google Scholar]
- 35.Seo D, Wang T, Dressman H, Herderick EE, Iversen ES, Dong C, Vata K, Milano CA, Rigat F, Pittman J, Nevins JR, West M, Goldschmidt-Clermont PJ. Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- 36.Patino WD, Mian OY, Kang JG, Matoba S, Bartlett LD, Holbrook B, Trout HH, 3rd, Kozloff L, Hwang PM. Circulating transcriptome reveals markers of atherosclerosis. Proc Natl Acad Sci U S A. 2005;102:3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patino WD, Kang JG, Matoba S, Mian OY, Gochuico BR, Hwang PM. Atherosclerotic plaque macrophage transcriptional regulators are expressed in blood and modulated by tristetraprolin. Circ Res. 2006;98:1282–1289. doi: 10.1161/01.RES.0000222284.48288.28. [DOI] [PubMed] [Google Scholar]
- 38.Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr., Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu H, Goldin E, Baird AE. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- 39.Cipolletta C, Ryan KE, Hanna EV, Trimble ER. Activation of peripheral blood CD14+ monocytes occurs in diabetes. Diabetes. 2005;54:2779–2786. doi: 10.2337/diabetes.54.9.2779. [DOI] [PubMed] [Google Scholar]
- 40.Horwitz PA, Tsai EJ, Putt ME, Gilmore JM, Lepore JJ, Parmacek MS, Kao AC, Desai SS, Goldberg LR, Brozena SC, Jessup ML, Epstein JA, Cappola TP. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation. 2004;110:3815–3821. doi: 10.1161/01.CIR.0000150539.72783.BF. [DOI] [PubMed] [Google Scholar]
- 41.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 42.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Bogaev RC, Elashoff B, Baron H, Yee J, Valantine HA. Gene-Expression Profiling for Rejection Surveillance after Cardiac Transplantation. N Engl J Med. 2010;362:1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 43.Nahrendorf M, Pittet MJ, Swirski FK. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woollard KJ, Geissmann F. Monocytes in atherosclerosis: subsets and functions. Nat Rev Cardiol. 2010;7:77–86. doi: 10.1038/nrcardio.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swirski FK, Weissleder R, Pittet MJ. Heterogeneous in vivo behavior of monocyte subsets in atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:1424–1432. doi: 10.1161/ATVBAHA.108.180521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hristov M, Leyendecker T, Schuhmann C, von Hundelshausen P, Heussen N, Kehmeier E, Krotz F, Sohn HY, Klauss V, Weber C. Circulating monocyte subsets and cardiovascular risk factors in coronary artery disease. Thromb Haemost. 2010;104:412–414. doi: 10.1160/TH10-01-0069. [DOI] [PubMed] [Google Scholar]
- 48.Schlitt A, Heine GH, Blankenberg S, Espinola-Klein C, Dopheide JF, Bickel C, Lackner KJ, Iz M, Meyer J, Darius H, Rupprecht HJ. CD14+CD16+ monocytes in coronary artery disease and their relationship to serum TNF-alpha levels. Thromb Haemost. 2004;92:419–424. doi: 10.1160/TH04-02-0095. [DOI] [PubMed] [Google Scholar]
- 49.Rogacev KS, Ulrich C, Blomer L, Hornof F, Oster K, Ziegelin M, Cremers B, Grenner Y, Geisel J, Schlitt A, Kohler H, Fliser D, Girndt M, Heine GH. Monocyte heterogeneity in obesity and subclinical atherosclerosis. Eur Heart J. 2010;31:369–376. doi: 10.1093/eurheartj/ehp308. [DOI] [PubMed] [Google Scholar]
- 50.Tsujioka H, Imanishi T, Ikejima H, Kuroi A, Takarada S, Tanimoto T, Kitabata H, Okochi K, Arita Y, Ishibashi K, Komukai K, Kataiwa H, Nakamura N, Hirata K, Tanaka A, Akasaka T. Impact of heterogeneity of human peripheral blood monocyte subsets on myocardial salvage in patients with primary acute myocardial infarction. J Am Coll Cardiol. 2009;54:130–138. doi: 10.1016/j.jacc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 51.Ikejima H, Imanishi T, Tsujioka H, Kuroi A, Tanimoto T, Kitabata H, Hirata K, Akasaka T. Effect of human peripheral monocyte subsets on coronary flow reserve in infarct-related artery in patients with primary anterior acute myocardial infarction. Clin Exp Pharmacol Physiol. 2010;37:453–459. doi: 10.1111/j.1440-1681.2009.05324.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoder MC. Is endothelium the origin of endothelial progenitor cells? Arterioscler Thromb Vasc Biol. 2010;30:1094–1103. doi: 10.1161/ATVBAHA.109.191635. [DOI] [PubMed] [Google Scholar]
- 53.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 54.Zhu S, Liu X, Li Y, Goldschmidt-Clermont PJ, Dong C. Aging in the atherosclerosis milieu may accelerate the consumption of bone marrow endothelial progenitor cells. Arterioscler Thromb Vasc Biol. 2007;27:113–119. doi: 10.1161/01.ATV.0000252035.12881.d0. [DOI] [PubMed] [Google Scholar]
- 55.Hill JM, Zalos G, Halcox JP, Schenke WH, Waclawiw MA, Quyyumi AA, Finkel T. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N Engl J Med. 2003;348:593–600. doi: 10.1056/NEJMoa022287. [DOI] [PubMed] [Google Scholar]
- 56.Vasa M, Fichtlscherer S, Aicher A, Adler K, Urbich C, Martin H, Zeiher AM, Dimmeler S. Number and migratory activity of circulating endothelial progenitor cells inversely correlate with risk factors for coronary artery disease. Circ Res. 2001;89:E1–7. doi: 10.1161/hh1301.093953. [DOI] [PubMed] [Google Scholar]
- 57.Medina RJ, O'Neill CL, Sweeney M, Guduric-Fuchs J, Gardiner TA, Simpson DA, Stitt AW. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med Genomics. 2010;3:18. doi: 10.1186/1755-8794-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, Boulanger CM, Westwood N, Urbich C, Willeit J, Steiner M, Breuss J, Xu Q, Kiechl S, Mayr M. Proteomic analysis reveals presence of platelet microparticles in endothelial progenitor cell cultures. Blood. 2009;114:723–732. doi: 10.1182/blood-2009-02-205930. [DOI] [PubMed] [Google Scholar]
- 59.Deb A, Patterson C. Hard luck stories: the reality of endothelial progenitor cells continues to fall short of the promise. Circulation. 2010;121:850–852. doi: 10.1161/CIR.0b013e3181d4c360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson MR, Yoder MC. Endothelial progenitor cells: quo vadis? J Mol Cell Cardiol. 2011;50:266–272. doi: 10.1016/j.yjmcc.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prokopi M, Mayr M. Proteomics: a reality-check for putative stem cells. Circ Res. 2011;108:499–511. doi: 10.1161/CIRCRESAHA.110.226902. [DOI] [PubMed] [Google Scholar]
- 62.Mause SF, Ritzel E, Liehn EA, Hristov M, Bidzhekov K, Muller-Newen G, Soehnlein O, Weber C. Platelet microparticles enhance the vasoregenerative potential of angiogenic early outgrowth cells after vascular injury. Circulation. 2010;122:495–506. doi: 10.1161/CIRCULATIONAHA.109.909473. [DOI] [PubMed] [Google Scholar]
- 63.Karra R, Vemullapalli S, Dong C, Herderick EE, Song X, Slosek K, Nevins JR, West M, Goldschmidt-Clermont PJ, Seo D. Molecular evidence for arterial repair in atherosclerosis. Proc Natl Acad Sci U S A. 2005;102:16789–16794. doi: 10.1073/pnas.0507718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ingram DA, Mead LE, Tanaka H, Meade V, Fenoglio A, Mortell K, Pollok K, Ferkowicz MJ, Gilley D, Yoder MC. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood. 2004;104:2752–2760. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 65.Yoder MC, Mead LE, Prater D, Krier TR, Mroueh KN, Li F, Krasich R, Temm CJ, Prchal JT, Ingram DA. Redefining endothelial progenitor cells via clonal analysis and hematopoietic stem/progenitor cell principals. Blood. 2007;109:1801–1809. doi: 10.1182/blood-2006-08-043471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt-Lucke C, Rossig L, Fichtlscherer S, Vasa M, Britten M, Kamper U, Dimmeler S, Zeiher AM. Reduced number of circulating endothelial progenitor cells predicts future cardiovascular events: proof of concept for the clinical importance of endogenous vascular repair. Circulation. 2005;111:2981–2987. doi: 10.1161/CIRCULATIONAHA.104.504340. [DOI] [PubMed] [Google Scholar]
- 67.Werner N, Kosiol S, Schiegl T, Ahlers P, Walenta K, Link A, Bohm M, Nickenig G. Circulating endothelial progenitor cells and cardiovascular outcomes. N Engl J Med. 2005;353:999–1007. doi: 10.1056/NEJMoa043814. [DOI] [PubMed] [Google Scholar]
- 68.Boilson BA, Kiernan TJ, Harbuzariu A, Nelson RE, Lerman A, Simari RD. Circulating CD34+ cell subsets in patients with coronary endothelial dysfunction. Nat Clin Pract Cardiovasc Med. 2008;5:489–496. doi: 10.1038/ncpcardio1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao Q, Kiechl S, Patel S, Oberhollenzer F, Weger S, Mayr A, Metzler B, Reindl M, Hu Y, Willeit J, Xu Q. Endothelial progenitor cells, cardiovascular risk factors, cytokine levels and atherosclerosis--results from a large population-based study. PLoS One. 2007;2:e975. doi: 10.1371/journal.pone.0000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw SY, Cheng S, Cupples LA, Larson MG, McCabe EL, Ngwa JS, Wang YA, Martin RP, Klein RJ, Hashmi B, Ajijola OA, Lau E, O'Donnell CJ, Vasan RS, Cohen KS, Wang TJ. Genetic and Clinical Correlates of Early Outgrowth Colony Forming Units. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.110.958470. Epub April 14 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng S, Cohen KS, Shaw SY, Larson MG, Hwang SJ, McCabe EL, Martin RP, Klein RJ, Hashmi B, Hoffmann U, Fox CS, Vasan RS, O'Donnell CJ, Wang TJ. Association of colony-forming units with coronary artery and abdominal aortic calcification. Circulation. 2010;122:1176–1182. doi: 10.1161/CIRCULATIONAHA.109.931279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, Grosshennig A, Linsel-Nitschke P, Perret C, DeSuremain M, Meitinger T, Wright BJ, Preuss M, Balmforth AJ, Ball SG, Meisinger C, Germain C, Evans A, Arveiler D, Luc G, Ruidavets JB, Morrison C, van der Harst P, Schreiber S, Neureuther K, Schafer A, Bugert P, El Mokhtari NE, Schrezenmeir J, Stark K, Rubin D, Wichmann HE, Hengstenberg C, Ouwehand W, Ziegler A, Tiret L, Thompson JR, Cambien F, Schunkert H, Samani NJ. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 73.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Yakub I, Birren BW, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, Rivas M, Dermitzakis ET, Cahir-McFarland E, Kieff E, Hafler D, Daly MJ, Altshuler D. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.MacMullen C, Fang J, Hsu BY, Kelly A, de Lonlay-Debeney P, Saudubray JM, Ganguly A, Smith TJ, Stanley CA. Hyperinsulinism/hyperammonemia syndrome in children with regulatory mutations in the inhibitory guanosine triphosphate-binding domain of glutamate dehydrogenase. J Clin Endocrinol Metab. 2001;86:1782–1787. doi: 10.1210/jcem.86.4.7414. [DOI] [PubMed] [Google Scholar]
- 77.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci U S A. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr., Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 79.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Badner JA, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui Y, Su YR, Rutkowski M, Reif M, Menon AG, Pun RY. Loss of protein kinase C inhibition in the beta-T594M variant of the amiloride-sensitive Na+ channel. Proc Natl Acad Sci U S A. 1997;94:9962–9966. doi: 10.1073/pnas.94.18.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seong IS, Ivanova E, Lee JM, Choo YS, Fossale E, Anderson M, Gusella JF, Laramie JM, Myers RH, Lesort M, MacDonald ME. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 82.Reis SA, Thompson MN, Lee JM, Fossale E, Kim HH, Liao JK, Moskowitz MA, Shaw SY, Dong L, Haggarty SJ, Macdonald ME, Seong IS. Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr127. Epub April 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Pons-Estel B, Petri M, Daly M, Gregersen PK, Martin J, Altshuler D, Behrens TW, Alarcon-Riquelme ME. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 84.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 85.Shaw SY, Blodgett DM, Ma MS, Westly EC, Clemons PA, Subramanian A, Schreiber SL. Disease allele-dependent small-molecule sensitivities in blood cells from monogenic diabetes. Proc Natl Acad Sci U S A. 2011;108:492–497. doi: 10.1073/pnas.1016789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narsinh K, Narsinh KH, Wu JC. Derivation of human induced pluripotent stem cells for cardiovascular disease modeling. Circ Res. 2011;108:1146–1156. doi: 10.1161/CIRCRESAHA.111.240374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Unternaehrer JJ, Daley GQ. Induced pluripotent stem cells for modelling human diseases. Philos Trans R Soc Lond B Biol Sci. 2011;366:2274–2285. doi: 10.1098/rstb.2011.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dambrot C, Passier R, Atsma D, Mummery CL. Cardiomyocyte differentiation of pluripotent stem cells and their use as cardiac disease models. Biochem J. 2011;434:25–35. doi: 10.1042/BJ20101707. [DOI] [PubMed] [Google Scholar]
- 89.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 90.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 91.Carvajal-Vergara X, Sevilla A, D'Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF, Stewart CL, Colman A. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 94.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 95.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. Proc Natl Acad Sci U S A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Methods. 2006;3:117–122. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 97.Whitesides GM. The origins and future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 98.Melin J, Quake SR. Microfluidic large-scale integration: the evolution of design rules for biological automation. Annu Rev Biophys Biomol Struct. 2007;36:213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 99.Wang CJ, Levchenko A. Microfluidics technology for systems biology research. Methods Mol Biol. 2009;500:203–219. doi: 10.1007/978-1-59745-525-1_7. [DOI] [PubMed] [Google Scholar]
- 100.Hong J, Edel JB, deMello AJ. Micro- and nanofluidic systems for high-throughput biological screening. Drug Discov Today. 2009;14:134–146. doi: 10.1016/j.drudis.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 101.Wang MM, Tu E, Raymond DE, Yang JM, Zhang H, Hagen N, Dees B, Mercer EM, Forster AH, Kariv I, Marchand PJ, Butler WF. Microfluidic sorting of mammalian cells by optical force switching. Nat Biotechnol. 2005;23:83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 102.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 104.King KR, Wang S, Irimia D, Jayaraman A, Toner M, Yarmush ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip. 2007;7:77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Agrawal N, Toner M, Irimia D. Neutrophil migration assay from a drop of blood. Lab Chip. 2008;8:2054–2061. doi: 10.1039/b813588f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Irimia D, Balazsi G, Agrawal N, Toner M. Adaptive-control model for neutrophil orientation in the direction of chemical gradients. Biophys J. 2009;96:3897–3916. doi: 10.1016/j.bpj.2008.12.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wan Y, Mahmood MA, Li N, Allen PB, Kim YT, Bachoo R, Ellington AD, Iqbal SM. Nanotextured substrates with immobilized aptamers for cancer cell isolation and cytology. Cancer. 2011 doi: 10.1002/cncr.26349. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr., Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haun JB, Castro CM, Wang R, Peterson VM, Marinelli BS, Lee H, Weissleder R. Micro-NMR for Rapid Molecular Analysis of Human Tumor Samples. Sci Transl Med. 2011;3:71ra16. doi: 10.1126/scitranslmed.3002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diercks AH, Ozinsky A, Hansen CL, Spotts JM, Rodriguez DJ, Aderem A. A microfluidic device for multiplexed protein detection in nano-liter volumes. Anal Biochem. 2009;386:30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu AR, Hiatt JB, Lu R, Attema JL, Lobo NA, Weissman IL, Clarke MF, Quake SR. Automated microfluidic chromatin immunoprecipitation from 2,000 cells. Lab Chip. 2009;9:1365–1370. doi: 10.1039/b819648f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kang JG, Patino WD, Matoba S, Hwang PM. Genomic analysis of circulating cells: a window into atherosclerosis. Trends Cardiovasc Med. 2006;16:163–168. doi: 10.1016/j.tcm.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr., Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D'Agostino RB, Vasan RS. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- 119.Ridker PM, Buring JE, Rifai N, Cook NR. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds Risk Score. JAMA. 2007;297:611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- 120.Zethelius B, Berglund L, Sundstrom J, Ingelsson E, Basu S, Larsson A, Venge P, Arnlov J. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 121.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 122.Melander O, Newton-Cheh C, Almgren P, Hedblad B, Berglund G, Engstrom G, Persson M, Smith JG, Magnusson M, Christensson A, Struck J, Morgenthaler NG, Bergmann A, Pencina MJ, Wang TJ. Novel and conventional biomarkers for prediction of incident cardiovascular events in the community. JAMA. 2009;302:49–57. doi: 10.1001/jama.2009.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]