Abstract
Background
Differentiating Spitz nevus (SN) from Spitzoid malignant melanoma (SMM) is one the most difficult problems in Dermatopathology.
Specific Aim
To identify differences on proteomic level between SN and SMM.
Methods
We performed Imaging Mass Spectrometry (IMS) analysis on formalin-fixed, paraffin embedded tissue samples (FFPE) to identify differences on proteomic level between SN and SMM. The diagnosis of SN and SMM was based on histopathologic criteria, clinical features, and follow up data, which confirmed that none of the lesions diagnosed as SN recurred or metastasized. The melanocytic component (tumor) and tumor microenvironment (dermis) from 114 cases of SN and SMM from the Yale Spitzoid Neoplasm Repository were analyzed. After obtaining mass spectra from each sample, classification models were built using a training set of biopsies from 26 SN and 25 SMM separately for tumor and for dermis. The classification algorithms developed on the training data set were validated on another set of 30 samples from SN and 33 from SMM.
Results
We found proteomic differences between the melanocytic components of SN and SMM and identified five peptides that were differentially expressed in the two groups. From these data, 29 out of 30 SN and 26 out of 29 SMM were recognized correctly based on tumor analysis in the validation set. This method correctly classified SN with 97% sensitivity and 90% specificity in the validation cohort.
Conclusions
IMS analysis can reliably differentiate SN from SMM in FFPE tissue based on proteomic differences.
Keywords: Spitz nevus, Spitzoid melanoma, Imaging Mass Spectrometry, Atypical Spitzoid neoplasm, malignant melanoma, MALDI, Matrix-assisted laser desorption ionization, proteomics
INTRODUCTION
Ever since the first description of SN by Sophie Spitz in 1948 pathologists and dermatopathologists in particular have been struggling with the distinction between SN and SMM. To diagnose a young child with melanoma, which has devastating consequences, and subject the child to surgery and chemotherapy is not a simple matter. Imagine if you were the parent of that child?
Spitzoid neoplasms are melanocytic lesions that include a spectrum ranging from completely benign “typical” SN to malignant melanomas that show “Spitzoid” features –”Spitzoid” malignant melanomas (SMM). The gold standard for diagnosing SN and differentiating it from SMM is histopathologic examination applying well-established criteria. However, there are melanocytic lesions, which show conflicting histopathologic criteria and the distinction between a benign SN and SMM may be extremely difficult. These lesions are referred to as “Atypical SN” or “Atypical Spitzoid tumors/neoplasms”.1–4 There is a great interobserver variability and discordance even among expert dermatopathologists regarding Spitzoid neoplasms.2, 5–8
Ancillary techniques such as Comparative genomic hybridization (CGH) and Fluorescent in situ hybridization (FISH) may be helpful. The majority of SN reveal no DNA copy number changes by CGH.9–11 Approximately 20% of SN show an isolated gain of chromosome11p.9 A subset of SN with 11p copy number increases has HRAS mutation; however, that is extremely uncommon in cutaneous melanoma.12, 13 In contrast, more than 95% of conventional melanomas show multiple chromosomal aberrations including gains and loses by CGH.14, 15 B-RAF mutations have been found only in a small subset of SN whereas the majority of conventional melanomas have B-RAF or N-RAS mutations.16 Furthermore, activating hotspot mutations in the B-RAF, N-RAS, and H-RAS genes were not identified in SMM or SN.17, 18 This data suggests that SMM might be a distinct form of melanoma with unknown genes and/or signaling pathways involved in its development.17, 19, 20
Matrix-assisted laser desorption ionization (MALDI) Imaging Mass Spectrometry (IMS) is a powerful method for analyzing metabolites, peptides and proteins, DNA segments, and lipids directly from tissue sections with spatial fidelity. Although gene expression is useful for distinguishing melanocytic nevi from melanomas, it does not always correlate with protein translation and does not account for post-translational modification (PMT). However, both protein expression level and PTM state have fundamental effect on cellular function or dysfunction, therefore, it is more meaningful to analyze proteins and peptides that are involved in the development and progression of diseases, especially cancer. IMS has the ability to discover molecular signatures of diseases and cancer. These molecular signatures are typically comprised of 5–20 different proteins that together result in robust diagnostic patterns.21, 22 IMS-based studies have been used to elucidate molecular signatures of different tumor types and grades including brain, oral, lung, breast, gastric, pancreatic, renal, ovarian and prostate cancer.23–26
The presence of a grey area, in which it is extremely difficult or utterly impossible to distinguish between SN and SMM, prompted this investigation. Since both SN and SMM are composed of large epithelioid cells with abundant cytoplasm containing ample amount of proteins, we hypothesized that there were proteomic differences, which might be able to differentiate between the two groups. We applied IMS to find specific proteomic markers to aid in the diagnosis of SN and SMM.
MATERIALS AND METHODS
Tumor Specimens
We collected archival FFPE tissue samples of SN and SMM from the Yale Spitzoid Neoplasm Repository. The Institutional Review Board at Yale University approved the study. The histopathology of each case was reviewed by the dermatopathologist (RL) to confirm the diagnosis. The study included histologically unequivocal SN and primary cutaneous SMM, which were diagnosed initially by a board-certified dermatopathologist. The majority of these cases were seen by multiple dermatopathologists at consensus conference at the time of the initial diagnosis. In addition, all cases chosen for the study underwent confirmatory review by at least four other dermatopathologists from Yale Dermatopathology Laboratory. Only cases with an unequivocal consensus diagnosis of SN or melanoma with Spitzoid features (SMM) were included in the study. Histologically ambiguous Spitzoid neoplasms or SN with atypical features were excluded from the study. We selected SN and primary invasive SMM, which had an extensive and densely cellular dermal component with areas, in which tumors cells compiled almost pure populations of melanocytes, which allowed the collection of mostly melanocytes (>95%) for the analyzed samples. Included in the category of SN were melanocytic lesions, which qualified as SN based on diagnostic criteria that have been described previously.27, 28 Compound or predominantly intradermal SN with large nests and/or areas containing large groups of melanocytes without intervening dermis or epithelium were chosen. Excluded from the study were predominantly junctional SN and desmoplastic or purely intradermal SN. The study group of SMM consisted of malignant melanomas with Spitzoid features based on previously described criteria.6, 27, 29, 30 Invasive SMM with large nests of melanocytes and/or areas containing large groups of melanocytes without intervening dermis, epithelium, or inflammatory cells were included in the study. Excluded from the study were superficially invasive SMM with insufficient areas containing pure melanocytic population.
A total of 114 specimens, 56 SN and 58 SMM, were analyzed for tumor and tumor microenvironment (TME - dermis). The samples were randomly divided into two cohorts: a training set and a validation set. The training set consisted of 26 SN and 25 SMM. The validation set consisted of 30 SN and 33 SMM. Five of the 30 cases of SN in the validation set and 2 cases of SMM did not contain sufficient surrounding dermis for analysis. Furthermore, 4 cases of SMM from the validation set contained only dermis and had insufficient melanocytic component for analysis.
Mass Spectrometry Analysis
Serial sections, 5 μm thick, were cut from FFPE tissue blocks using a microtome. One section per sample was mounted onto a conductive glass slide whereas the consecutive serial section was mounted onto a regular glass slide and stained with Hematoxylin and Eosin (H&E), which served as a reference section. Unstained sections were subjected to paraffin removal with xylene and graded ethanol washes. After air drying antigen retrieval was performed by heating the sections in a tris buffer. Antigen retrieved sections were stored in a desiccator at room temperature until matrix deposition for no longer than two days.
Mass spectral profiles were acquired in duplicates from the tissue using a histology-directed profiling approach as follows.31 Digital images were acquired of the histology slide using a Mirax Scan digital microscope slide scanner (Mirax, Budapest, Hungary) at a pixel resolution of 0.23 μm. The dermatopathologist (RL) marked 300 μm diameter color-coded areas of interest (i.e. tumor and TME) on the digital image of the H&E stained section. The goal was to choose pure melanocytic populations for analysis of the tumor and without interference of other cell types such as endothelial cells from blood vessels, red blood cells, inflammatory cells, or epithelium. Samples from dermis in the area underlying the melanocytic lesion were also analyzed as TME.
Using image-processing software (Photoshop), the histology-marked image was merged to an image of the unstained section and the coordinates of annotations were determined. Using fiducial points visible in the image and on the MALDI target plate, an affine transform was used to transfer coordinates into an acoustic robotic microspotter (Labcyte, Sunnyvale, CA). A detailed description of the device and its operational conditions were described elsewhere.32 The spotter is capable of depositing 170 pL volume drops of matrix at very precise locations (spotting error <60 μm) at a rate of 30 Hz. On-tissue digestion was carried out by spotting trypsin solution onto the tissue section at the designated locations (spot diameter ~175 μm). Trypsin was spotted over a series of 40 iterations (one drop each) with drying time (~2 min) between iterations. After trypsin deposition, matrix (10 mg/mL CHCA in a mixture of 50:50:0.1 acetonitrile/H2O/trifluoroacetic acid by volume) was spotted directly onto tryptic spots over 72 iterations (8 passes of 9 drops each). The spots were slightly smaller than the diameter of the annotations placed by the pathologist, allowing for slight placement error. Matrix spot placement accuracy was evaluated before coordinates were transferred to the mass spectrometer. Mass spectra were acquired in reflectron mode using and AutoFlex (Bruker Daltonics, Billerica, MA) equipped with a 355 nm Nd:YAG laser operating at a 1000 Hz repetition. Typically, 15–20 distinct spectra were collected from each tumor or TME area from each section. Peptide markers of interest were later identified directly from the unstained sections using MS/MS sequence analysis using an AutoFlex TOF/TOF (Bruker Daltonics, Billerica, MA). The MS/MS spectra were processed using FlexAnalysis. This included baseline correction, Savitzky-Golay smoothing, and monoisotopic peak picking. The spectra were submitted into a MASCOT (Matrix Science, Boston, MA) database search engine to match tryptic peptide sequences to their respective intact proteins. The MS/MS spectrum search was performed with a peptide tolerance of +/− 0.5 Da and a fragment tolerance of +/− 0.5 Da. The search criteria also included one missed cleavage and variable modifications including lysine acetylation, N-terminus acetylation, C-terminus amidation, and methionine oxidation.
Data Analysis
Statistical analyses of MS profiles were carried out using ClinPro Tools 2.0 (Bruker Daltonics). Classes of spectra were loaded into the software and baseline correction was achieved using a top hat algorithm with a 10% minimal baseline width. ClinPro Tools automatically normalizes all spectra to their own TIC (Total Ion Current). Thus, for each spectrum the TIC is determined as the sum of intensities from all data points in the spectrum. Peaks in the spectra were selected manually and the maximum intensity within each of the defined peak integration areas was used as the comparative value. The classification model used in this analysis was built in ClinProTools using a genetic algorithm (GA)33 to determine the peak combination that separates best between SN and SMM. Maximum number of peaks was set to 15, maximum number of generations was 50, mutation rate was 0.2 and cross-over rate was 0.5. The number of neighbors for the K-NN classification parameter in the GA settings was set to 3.
For the differentially expressed features identified in the two comparisons of tumor in SN versus tumor in SMM and TME in SN versus TME in SMM, a GA classifier was used to assess the class prediction ability of each individual feature. The prediction accuracy was estimated using a leave-N-out-cross-validation algorithm in which 20% of the data were randomly left out in each of 10 iterations.34 The classification model for SN and SMM were built using the GA, which then classified spectra in the validation cohort based on the supervised learning from the training set.
RESULTS
Characterization of the Study Sample
Our cohort of SN came from patients, who ranged from 1 to 48 years of age (mean=13; SD=10.2), 30 males and 25 females. The lesions were distributed on the head and neck (17), leg (14), back (10), arm (9), buttock (2), abdomen (2), and chest (1). None of the lesions recurred or metastasized and all patients are alive with a follow up ranging from 2 to 20 years (mean=10.7). The SMM cohort comprised patients from 29 to 89 years old (mean=62; SD=14), 32 males and 26 females. The distribution was as follows: leg (23), back (13), arm (12), scalp (4), chest (2), ear (2), and face (2). The depth of the SMM ranged from 0.75–9.0 mm (mean=3.2). The follow up ranged from 1 to 21 years (mean=5). Representative histopathologic features of one patient with SN (case 1) and one with SMM (case 33) are illustrated in Figure 1. A summary of clinical and histopathologic characteristics of patients with SMM is shown in Table 1.
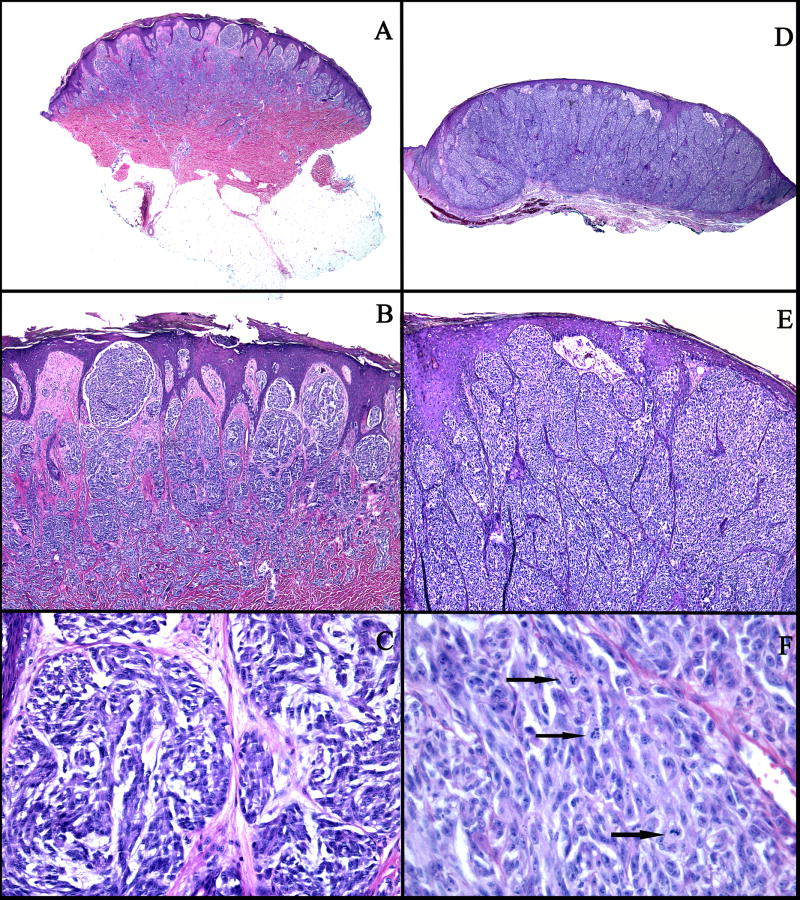
Figure 1.
A–C: An example of a compound SN (case 1): A: Low power view showing a symmetric, well-circumscribed, and wedge-shaped proliferation of melanocytes in the epidermis and in the dermis. B: There are large vertical nests of melanocytes with clefts between the nests and the surrounding hyperplastic epidermis. C: The melanocytes are large, slightly pleomorphic, with vesicular nuclei, prominent nucleoli, and abundant pale cytoplasm. D–F: An example of a SMM (case 33): D: Asymmetric and poorly circumscribed proliferation of melanocytes in the epidermis and dermis. E: Large confluent nests of melanocytes, focally forming sheets in the dermis, and irregular nests and single melanocytes in the epidermis. F: Markedly pleomorphic melanocytes with vesicular nuclei, prominent nucleoli, eosinophilic cytoplasm, and numerous mitotic figures (arrows).
Table 1.
Histopathologic and clinical characteristics of Spitzoid Malignant Melanoma cases
| Case No. | Age | Gender | Site | Depth (mm) | Clark’s Level | Ulceration/Regression | Mitoses /mm2 | Follow-up (years) | Adverse event(s) | MS Results (tumor) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 72 | F | Leg, lower | 4 | IV | No/Yes | <1 | 1 | DOD; satellitosis, 0/1 SLN | NA |
| 2 | 75 | M | Arm, upper | 1.3 | IV | No/Yes | 2 | 5 | Alive-NED; 0/5 SLN | NA |
| 3 | 54 | M | Back | 1.2 | IV | No/Yes | 2 | 5 | Alive-NED; 0/6 SLN | NA |
| 4 | 64 | F | Knee | 3.8 | IV | No/No | <1 | 3 | Alive-NED; 0/3 SLN | NA |
| 5 | 52 | F | Leg | 1.2 | IV | No/No | 2 | 5 | Alive-NFA | NA |
| 6 | 78 | F | Back | 3.5 | IV | Yes/No | 17 | 4 | Alive-NED | NA |
| 7 | 74 | M | Back | 3 | IV | Yes/No | 6 | 4 | Alive-NED; 0/1 SLN | NA |
| 8 | 48 | F | Leg, lower | 1.3 | IV | No/No | 1 | 4 | Alive-NED;0/2 SLN | NA |
| 9 | 38 | F | Thigh | 1.9 | IV | Yes/No | 26 | 3 | Alive-NED; 1/2 SLN;0/8 CLD | NA |
| 10 | 72 | M | Arm | 5.5 | IV | No/no | 5 | 3 | Alive w/d-multiple brain mets; 0/1 SLN | NA |
| 11 | 76 | F | Leg | 3.1 | IV | Yes/No | 2 | 3 | Alive-NED; 0/2 SLN | NA |
| 12 | 86 | M | Chest | 4.8 | IV | Yes/No | 2 | 3 | Alive-NED; 0/6 SLN | NA |
| 13 | 60 | M | Arm | 5 | IV | No/No | 4 | 2 | Alive w/d-multiple liver, brain, lung mets; 0/19 CLD | NA |
| 14 | 67 | M | Leg | 8 | V | Yes/No | 8 | 1 | DOD; brain and lung mets | NA |
| 15 | 56 | M | Ear | 2.3 | IV | No/No | 2 | 2 | Alive-NED; 0/3 SLN | NA |
| 16 | 88 | F | Leg | 2.5 | IV | No/No | 3 | 4 | NFA | NA |
| 17 | 78 | F | Scalp | 2.9 | IV | No/No | 5 | 2 | Alive-NED; 0/4 SLN | NA |
| 18 | 70 | M | Back | 3.4 | IV | No/No | 2 | 3 | Alive-NED; 1/4 SLN; 0/37 CLD | NA |
| 19 | 50 | M | Back | 3.2 | IV | No/No | 12 | 2 | Alive-NED; 0/2 SLN | NA |
| 20 | 57 | M | Arm | 3.4 | IV | No/No | 3 | 2 | Alive-NED; 0/2 SLN | NA |
| 21 | 53 | M | Leg | 1.9 | IV | No/No | 8 | 2 | Alive-NED; 0/2 SLN | NA |
| 22 | 62 | M | Leg, lower | 2.3 | IV | No/No | 2 | 2 | Alive-NED; 0/3 SLN | NA |
| 23 | 58 | M | Thigh | 2.1 | IV | No/No | 3 | 2 | Alive-NED; 1/3 SLN; 0/11 CLD | NA |
| 24 | 45 | F | Back | 2 | IV | No/No | <1 | 2 | Alive-NED; 1/6 SLN;0/14 CLD | NA |
| 25 | 88 | M | Ear | 3 | IV | No/No | 2 | 2 | Alive-NED | NA |
| 26 | 64 | M | Leg | 7.5 | IV | No/No | 2 | 1 | Alive-NED; 1/4 SLN | + |
| 27 | 43 | M | Temple | 3.8 | IV | Yes/No | 10 | 14 | Alive-NED; 0/2 SLN; 1/29 CLD | + |
| 28 | 60 | M | Arm | 3.5 | IV | No/No | <1 | 4 | Alive-NED; 0/1 SLN | + |
| 29 | 44 | F | Leg | 0.75 | IV | No/No | 1 | 3 | Alive-NFA | + |
| 30 | 52 | F | Arm | 2.2 | IV | Yes/No | 4 | 3 | Alive-NED | + |
| 31 | 65 | F | Back | 1.1 | IV | No/No | <1 | 2 | Alive-NED | + |
| 32 | 51 | F | Leg | 0.9 | IV | No/No | <1 | 2 | Alive-NED; 0/4 SLN | + |
| 33 | 89 | F | Arm | 3 | IV | No/No | 4 | 2 | NFA | + |
| 34 | 58 | M | Back | 1.1 | III | No/No | 5 | 2 | Alive-NED; 0/2 SLN | + |
| 35 | 57 | F | Arm | 0.95 | IV | Yes/Yes | 2 | 4 | Alive-NED; 0/2 SLN | + |
| 36 | 46 | F | Arm | 1.3 | IV | No/No | 2 | 3 | Alive-NED | + |
| 37 | 52 | F | Abdomen | 13 | IV | Yes/No | 6 | 2 | Alive w/d - local recurrence, multiple brain and subcutaneous mets; 0/6 SLN; 1/19 CLD | + |
| 38 | 75 | M | Scalp | 2.1 | IV | No/No | 2 | 3 | DOD-satellitosis, multiple mets; 0/1 SLN | + |
| 39 | 59 | M | Leg | 2 | IV | No/No | <1 | 7 | Alive-NED; 0/7 SLN | + |
| 40 | 62 | M | Arm | 4.5 | IV | No/No | <1 | 7 | Alive-NED; 1/4 SLN and 0/5 CLD | + |
| 41 | 86 | M | Leg, lower | 5 | IV | No/No | 4 | 4 | DOD-local recurrence and multiple mets | + |
| 42 | 58 | F | Knee | 6 | IV | Yes/No | 6 | 2 | Alive-NED | + |
| 43 | 29 | M | Back | 2.5 | IV | Yes/No | 2 | 1 | Alive-NED; 0/2 SLN; 5/10 CLD | + |
| 44 | 50 | F | Thigh | 4.8 | IV | No/No | <1 | 1 | Alive-NED; 1/3 SLN; 0/4 CLD | − |
| 45 | 76 | M | Back | 2.1 | IV | Yes/No | 6 | 1 | Alive-NED; 0/4 SLN | + |
| 46 | 57 | M | Back | 4.8 | IV | Yes/No | 10 | 1 | NFA | + |
| 47 | 87 | F | Arm | 2.3 | IV | Yes/No | 18 | 1 | NFA | + |
| 48 | 38 | M | Chest | 8 | IV | Yes/No | 1 | 21 | Alive; R axilla-1/18 CLD; L axilla-4/15 CLD | + |
| 49 | 61 | M | Leg, lower | 2 | IV | Yes/No | 5 | 20 | Alive w/d; satellitosis, multiple mets; 2/25 CLD | − |
| 50 | 75 | M | Scalp | 2.1 | IV | No/Yes | <1 | 3 | DOD-satellitosis, local recurrence, multiple mets | − |
| 51 | 59 | M | Back | 2.9 | IV | No/No | <1 | 14 | Alive w/d; soft tissue mets; 0/12 CLD | + |
| 52 | 79 | F | Cheek | 9 | V | Yes/No | 2 | 2 | Alive-NFA | + |
| 53 | 69 | M | Back | 1.8 | IV | No/No | 17 | 2 | Alive-NED; 1/2 SLN in R axilla; 3/10 SLN in L axilla; 0/19 CLD | + |
| 54 | 51 | M | Knee | 1 | III | No/No | <1 | 2 | Alive-NFA | + |
| 55 | 53 | F | Leg | 1 | IV | No/No | 1 | 2 | Alive-NED; 0/2 SLN | ++ |
| 56 | 48 | F | Thigh | 0.8 | III | Yes/No | <1 | 8 | DOD; multiple mets | ++ |
| 57 | 62 | F | Scalp | 3.5 | IV | No/No | <1 | 2 | DOD-multiple mets; 1/11 CLD | ++ |
| 58 | 63 | F | Leg | 5.5 | IV | Yes/No | 5 | 2 | Alive-NED; 0/3 SLN | ++ |
DOD-died of disease; NED-no evidence of disease; SLN-sentinel lymph node(s); CLD-completion lymphadenectomy; mets-metastases; w/d – with disease; LN-lymph nodes; NA-Not applicable; NFA-no follow up available; ++ - correct recognition based on dermis only (no tumor available); Cases 1–25 are part of the teaching set, cases 26–58 are part of the validation set.
Mass Spectrometry Analysis
Mass spectra from each dotted area (Figure 2) on each sample from the training set were obtained in duplicates. Data was analyzed using GA, as described above, and classification models were built using a training set of biopsies from 26 SN and 25 SMM separately for tumor and for TME. Each peak in the mass spectra corresponds to a peptide at a specific m/z ratio. The spectra generated at each spot on the digested tissue typically contain many hundreds of distinct peaks with a S/N>3. For the areas containing tumor, 5 peaks with the following m/z ratios, which were able to best discriminate between SN and SMM, were identified: 976.49, 1060.18, 1336.72, 1410.74, and 1428.77 (Figure 3). 12 peaks were found discriminatory and were used to build a classification model for the TME. Their m/z values were: 713.19, 1251.75, 1287.70, 1365.81, 1428.81, 1685.92, 2519.26, 2632.31, 2773.26, 3224.46, 3287.51, and 3411.84.
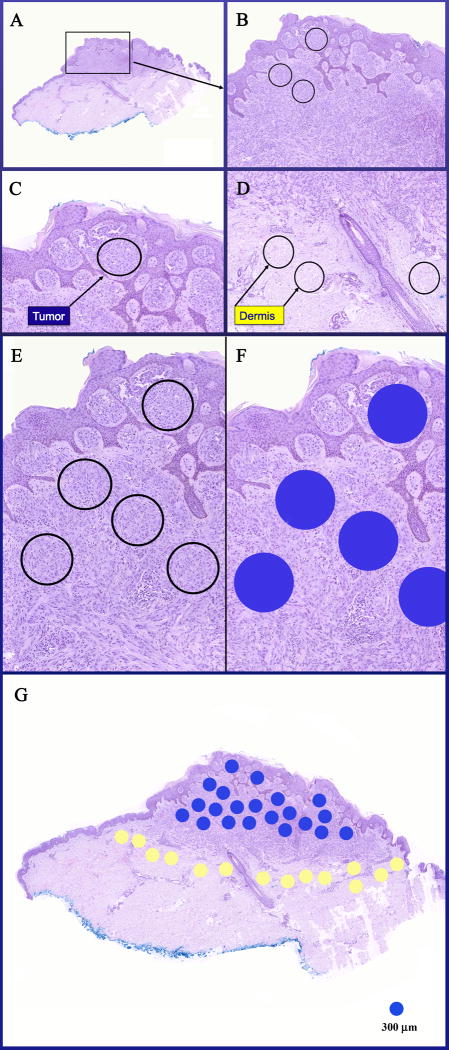
Figure 2.
A case of SMM (case 53) with circled foci marking the locations of the tumor (melanocytic) and dermal (TME) components to be studied by IMS. A&B: Low power views of an asymmetric melanocytic proliferation in the dermis. C, E, &F: Densely cellular areas of tumor containing pure population of melanocytes without intervening epithelial component, vessels, and collagen are selected for IMS analysis and marked blue for tumor. D: Circled areas in the dermis representing TME to be studied by IMS. G: An entire section with multiple foci marked blue for tumor and yellow for dermis to be subjected to IMS analysis. Each dot is with a diameter of 300 μm.
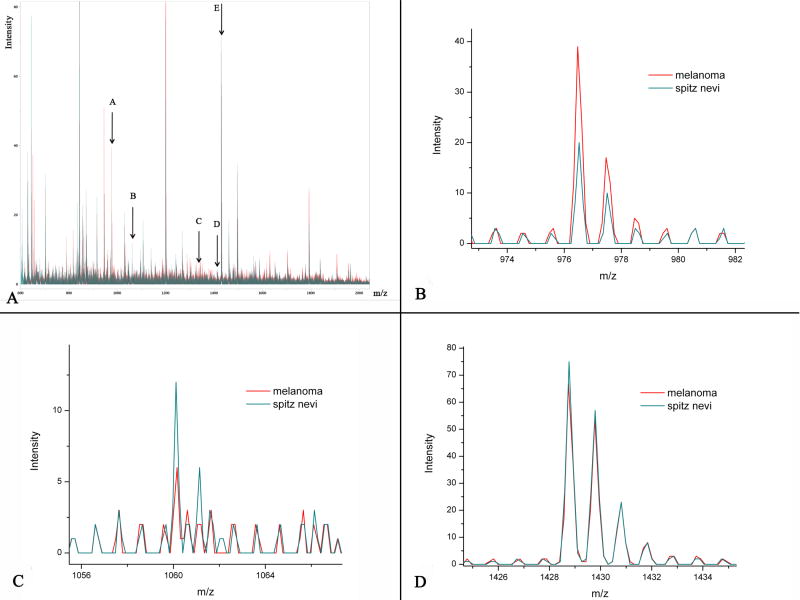
Figure 3.
A: Overlay of the average spectra for SN and SMM in the training set. Arrows labeled A, B, C, D, and E mark the 5 peptides of interest discriminating between SN and SMM. B: Peak with m/z ratio=976.49 showing a higher intensity in SMM than in SN. This peak corresponds to Actin. C: Tryptic peptide with m/z ratio=1060.18 show a higher intensity in SN. D: Tryptic peptide with m/z ratio=1428.77 corresponding to Vimentin.
The GA model was run against all spectra in the dataset and a summary table of the results was generated. The number of spectra in favor of SN and the number of spectra in favor of SMM were calculated. If the proportion of all spectra in favor of SN was higher, the case was classified as SN. Otherwise it was classified as SMM.
After a molecular signature for both tumor and dermis was determined based on data from the training set it was then tested on a validation cohort of 30 SN (30 cases with tumor, of which 25 also contained sufficient amount of dermis for analysis) and 33 SMM (29 cases with tumor of which 2 did not have sufficient amount of dermis, as well as 4 additional cases containing only dermis and not enough tumor component). The method was able to correctly recognize 29 out of 30 SN (97%) in the validation cohort. There was only one SN, which was incorrectly classified. In this particular case only 6 spectra were obtained due to very little tissue left in the block, whereas in the remaining cases of SN the average number of tumor spectra obtained was 16. If this case were excluded from the analysis the recognition would be 100% for SN. Twenty-six out of 29 SMM (90%) were also recognized correctly based on tumor proteomic differences. Thus this algorithm showed a sensitivity of 97% and specificity of 90% in correctly identifying SN based on tumor analysis in the validation set.
The dermis in 25 cases from SN and 31 cases of SMM was analyzed using the algorithm developed from the training set based on samples from TME. 12 peptides within the TME of SN and SMM showed differences in their expression. The method correctly classified 28 out of 31 (90%) SMM and 16 out of 25 SN (64%) and showed a sensitivity of 64% and specificity of 90% for TME in identifying SN.
Protein Identification
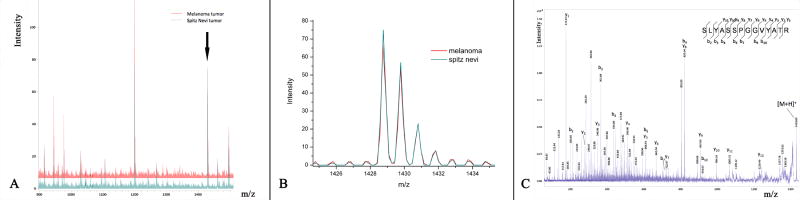
We identified two of the proteins differentially expressed by melanocytes in SN and SMM as Actin (m/z = 976.49) and Vimentin (m/z = 1428.61). Vimentin was identified against MASCOT database search with a probability based Mowse score of 108 and a P-value of 1.1e-008 (Figure 4).
Figure 4.
A&B: The peak corresponding to Vimentin with m/z 1428.77. C: Example of an MS/MS spectrum acquired directly from the FFPE tissue section and sequenced as the tryptic peptide Vimentin.
DISCUSSION
Our study shows that SN and SMM can be successfully distinguished using MALDI-IMS analysis based on detection of proteomic differences. MALDI-IMS can profile tryptic peptides in FFPE sections and has been used to identify proteomic patterns to accurately diagnose and classify different tumor types and grades including brain, oral, lung, breast, gastric, pancreatic, renal, ovarian and prostate cancer.23–26 Behavior of genome products is difficult to predict from the gene sequence alone and measurement of gene expression at the protein level is more informative, since protein contains information that collectively indicates the actual rather than the potential functional state.35
We were able to differentiate SN from SMM with 97% sensitivity and 90% specificity in the validation cohort, when the discriminatory criteria obtained from the training cohort for tumor were used. The GA algorithm used in our experiments incorporated 5 peaks for the tumor, determined through statistical comparison of the peaks in the training sets for both SN and SMM. In addition to analysis of the tumor, a proteomic signature for TME (Tumor Micro-Environment) in the dermis in cases of SN and SMM was developed. Interestingly, the method was able to classify correctly 28 out of 31 SMM based on proteomic differences in the dermis alone. It is conceivable that growth factors, cytokines, and other components are being actively secreted in the immediate TME surrounding SMM whereas they are not present in such quantities near SN. Using data from the TMI may be helpful in correctly diagnosing cases with equivocal results based on tumor analysis.
Molecular imaging and signature identification by MS allows us to look beyond classic histology. Statistical analysis of MS data uses computer generated statistical models in the construction of the algorithm and data are analyzed objectively. Proteomic signatures established using MS classification might be used as a supplement to standard histology. Our successful use of FFPE tissue further supports the practicability of combining MS analysis with histopathology in evaluation of Spitzoid melanocytic lesions.
MS analysis could be particularly useful in cases, which are histologically equivocal and a firm diagnosis of SN or SMM cannot be made with absolute certainty. The next step is to explore the Spitzoid lesions in the gray, histologically equivocal area, for which clinical follow up data is available, classify these lesions in either the benign or malignant category based on their proteomic signature, and correlate the results with the clinical information. Our goal is to more reliably differentiate between SN and SMM and significantly narrow the gray zone, in which difficult Spitzoid melanocytic neoplasms with unknown biologic potential cannot be reliably diagnosed microscopically. Future studies would be necessary to confirm our findings and use larger sample sets to achieve better statistical confidence.
This is the first study employing IMS as a novel tool in the evaluation of SN and SMM. Based on our results IMS is an excellent ancillary method to confirm the diagnosis of a SN with 97% accuracy. Our findings demonstrate that this technology may be a valuable adjunct to histopathologic evaluation of Spitzoid lesions. The identification of differences on a protein level within the melanocytes in SN and SMM provides us with a more objective method using molecular and biochemical protein biomarkers to aid in the diagnosis and improve the diagnostic accuracy of conventional histopathologic examination. Furthermore, identification of protein expression profiles, which discriminate between SN and SMM, may translate into tumor biomarkers that can be incorporated into standard diagnostic and treatment strategies.
Acknowledgments
Funding: Developmental Research Project Grant (R.Lazova, PI), Yale SPORE in Skin Cancer, NIH/NCI (P50 CA121974); Developmental Research Project Grant and Department of Defense grant (W81XWH-05-1-0179) (R. Caprioli, PI) and NIH/NIGMS (5R01 GM58008) (R. Caprioli, PI)
The authors would like to thank Jamie Allen as well as Carolyn Truini and Dr. Stephan Ariyan for their help. The authors would also like to thank the group of dermatopathologists from Yale Dermatopathology Laboratory for reviewing the cases.
References
- 1.Binder SW, Asnong C, Paul E, Cochran AJ. The histology and differential diagnosis of Spitz nevus. Semin Diagn Pathol. 1993;10:36–46. [PubMed] [Google Scholar]
- 2.Barnhill RL, Flotte TJ, Fleischli M, Perez-Atayde A. Cutaneous melanoma and atypical Spitz tumors in childhood. Cancer. 1995;76:1833–45. doi: 10.1002/1097-0142(19951115)76:10<1833::aid-cncr2820761024>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Crotty KA, Scolyer RA, Li L, Palmer AA, Wang L, McCarthy SW. Spitz naevus versus Spitzoid melanoma: when and how can they be distinguished? Pathology. 2002;34:6–12. doi: 10.1080/00313020120111212-1. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara G, Argenziano G, Soyer HP, et al. The spectrum of Spitz nevi: a clinicopathologic study of 83 cases. Arch Dermatol. 2005;141:1381–7. doi: 10.1001/archderm.141.11.1381. [DOI] [PubMed] [Google Scholar]
- 5.Ackerman AB. Discordance among expert pathologists in diagnosis of melanocytic neoplasms. Hum Pathol. 1996;27:1115–6. doi: 10.1016/s0046-8177(96)90301-9. [DOI] [PubMed] [Google Scholar]
- 6.Barnhill RL, Argenyi ZB, From L, et al. Atypical Spitz nevi/tumors: lack of consensus for diagnosis, discrimination from melanoma, and prediction of outcome. Hum Pathol. 1999;30:513–20. doi: 10.1016/s0046-8177(99)90193-4. [DOI] [PubMed] [Google Scholar]
- 7.Farmer ER, Gonin R, Hanna MP. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol. 1996;27:528–31. doi: 10.1016/s0046-8177(96)90157-4. [DOI] [PubMed] [Google Scholar]
- 8.Rapini RP. Spitz nevus or melanoma? Semin Cutan Med Surg. 1999;18:56–63. doi: 10.1016/s1085-5629(99)80009-1. [DOI] [PubMed] [Google Scholar]
- 9.Bastian BC, Wesselmann U, Pinkel D, Leboit PE. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999;113:1065–9. doi: 10.1046/j.1523-1747.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 10.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163:1765–70. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvell JD, Kohler S, Zhu S, Hernandez-Boussard T, Pollack JR, van de Rijn M. High-resolution array-based comparative genomic hybridization for distinguishing paraffin-embedded Spitz nevi and melanomas. Diagn Mol Pathol. 2004;13:22–5. doi: 10.1097/00019606-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000;157:967–72. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Engen-van Grunsven AC, van Dijk MC, Ruiter DJ, Klaasen A, Mooi WJ, Blokx WA. HRAS-mutated Spitz tumors: A subtype of Spitz tumors with distinct features. Am J Surg Pathol. 34:1436–41. doi: 10.1097/PAS.0b013e3181f0a749. [DOI] [PubMed] [Google Scholar]
- 14.Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–5. [PubMed] [Google Scholar]
- 15.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 16.Fullen DR, Poynter JN, Lowe L, et al. BRAF and NRAS mutations in spitzoid melanocytic lesions. Mod Pathol. 2006;19:1324–32. doi: 10.1038/modpathol.3800653. [DOI] [PubMed] [Google Scholar]
- 17.Lee DA, Cohen JA, Twaddell WS, et al. Are all melanomas the same? Spitzoid melanoma is a distinct subtype of melanoma. Cancer. 2006;106:907–13. doi: 10.1002/cncr.21686. [DOI] [PubMed] [Google Scholar]
- 18.Gill M, Cohen J, Renwick N, Mones JM, Silvers DN, Celebi JT. Genetic similarities between Spitz nevus and Spitzoid melanoma in children. Cancer. 2004;101:2636–40. doi: 10.1002/cncr.20680. [DOI] [PubMed] [Google Scholar]
- 19.Gaiser T, Kutzner H, Palmedo G, et al. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol. 23:413–9. doi: 10.1038/modpathol.2009.177. [DOI] [PubMed] [Google Scholar]
- 20.Raskin L, Ludgate M, Iyer RK, et al. Copy number variations and clinical outcome in atypical spitz tumors. Am J Surg Pathol. 2011;35:243–52. doi: 10.1097/PAS.0b013e31820393ee. [DOI] [PubMed] [Google Scholar]
- 21.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman TA, Monroe EB, Tucker KR, Rubakhin SS, Sweedler JV. Chapter 13: Imaging of cells and tissues with mass spectrometry: adding chemical information to imaging. Methods Cell Biol. 2008;89:361–90. doi: 10.1016/S0091-679X(08)00613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groseclose MR, Massion PP, Chaurand P, Caprioli RM. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;8:3715–24. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathan CO, Amirghahri N, Rice C, Abreo FW, Shi R, Stucker FJ. Molecular analysis of surgical margins in head and neck squamous cell carcinoma patients. Laryngoscope. 2002;112:2129–40. doi: 10.1097/00005537-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Oppenheimer SR, Mi D, Sanders ME, Caprioli RM. Molecular analysis of tumor margins by MALDI mass spectrometry in renal carcinoma. J Proteome Res. 9:2182–90. doi: 10.1021/pr900936z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 27.Paniago-Pereira C, Maize JC, Ackerman AB. Nevus of large spindle and/or epithelioid cells (Spitz’s nevus) Arch Dermatol. 1978;114:1811–23. [PubMed] [Google Scholar]
- 28.Mones JM, Ackerman AB. Melanomas in prepubescent children: review comprehensively, critique historically, criteria diagnostically, and course biologically. Am J Dermatopathol. 2003;25:223–38. doi: 10.1097/00000372-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Reed RJ. Atypical spitz nevus/tumor. Hum Pathol. 1999;30:1523–6. doi: 10.1016/s0046-8177(99)90180-6. [DOI] [PubMed] [Google Scholar]
- 30.Spatz A, Barnhill RL. The Spitz tumor 50 years later: revisiting a landmark contribution and unresolved controversy. J Am Acad Dermatol. 1999;40:223–8. doi: 10.1016/s0190-9622(99)70192-1. [DOI] [PubMed] [Google Scholar]
- 31.Cornett DS, Mobley JA, Dias EC, et al. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5:1975–83. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78:827–34. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 33.Holland J. Adaptation in Natural and Artificial Systems. Ann Arbor: University of Michigan Press; 1975. [Google Scholar]
- 34.Harrell F., Jr . Regression Modeling Strategies. New York: Springer; 2001. [Google Scholar]
- 35.Conrad DH, Goyette J, Thomas PS. Proteomics as a method for early detection of cancer: A review of proteomics, exhaled breath condensate, and lung cancer screening. J Intern Med. 2008;23(1 SUPPL):78–84. doi: 10.1007/s11606-007-0411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]