Abstract
MicroRNAs (miRNAs) are endogenous noncoding RNAs that down-regulate gene expression by promoting cleavage or translational arrest of target mRNAs. While most miRNAs are transcribed from their own dedicated genes, some map to introns of ‘host’ transcripts, the biological significance of which remains unknown. Here, we show that prostate cells are naturally devoid of EGF-like domain 7 (Egfl7) transcripts and hence also deficient in a miRNA, miR-126*, generated from splicing and processing of its ninth intron. Use of recombinant and synthetic miRNAs or a specific antagomir established a role of miR-126* in silencing prostein in non-endothelial cells. We mapped two miR-126*-binding sites in the 3′UTR of the prostein mRNA required for translational repression. Transfection of synthetic miR-126* into prostate cancer LNCaP cells strongly reduced the translation of prostein. Interestingly, loss of prostein correlated with reduction of LNCaP cell migration and invasion. Thus, the robust expression of prostein protein in the prostate cells results from a combination of transcriptional activation of the prostein gene and absence of intronic miRNA-126* due to the prostate-specific repression of the Egfl7 gene. We conclude that intronic miRNAs from tissue-specific transcripts, or their natural absence, make cardinal contributions to cellular gene expression and phenotype. These findings also open the door to tissue-specific miRNA therapy.
Keywords: Intronic microRNA, miR-126*, Prostate cancer, Prostein, VE-statin, EGFL7
Introduction
RNA interference (RNAi) is a cellular pathway in which ~22-nt long double-stranded noncoding RNAs, called short interfering RNA (siRNA) or microRNA (miRNA), silence specific target RNAs [1]. In RNAi, the antisense (guide) strand of the siRNA/miRNA is recruited into a ribonucleo-protein complex known as RNA-induced silencing complex (RISC), which engages the target RNA and either degrades the target or inhibits its translation [2, 3]. The miRNA guide strand exhibits imperfect complementarity with specific sequences in the 3′-untranslated region (3′UTR) of the target mRNA, and the resultant RISC represses translation [3–5]. The rules of miRNA-target recognition are not well-defined, although a perfect or near-perfect match with nucleotides 2–8 of the miRNA 5′-end (the ‘seed’ or ‘core’ region) is considered the most important requirement [6–9]. Thus, a given 3′UTR may contain binding sites for multiple miRNA species, and reciprocally, a given miRNA may have target sites on multiple mRNA 3′UTRσ. There are a few hundred experimentally sequenced and predicted human miRNAs [10–13], and together, they hold the potential to regulate thousands of genes impacting a large variety of biological processes. Expression of many miRNAs is regulated in a tissue-specific and temporal manner, the full biological relevance of which remains to be unraveled [13–16].
The vast majority of miRNAs are produced by their individual genes driven by Pol II promoters [4, 17]. The immediate product of transcription is the pri-miRNA that is first trimmed by the nuclear RNase III-like enzyme, Drosha [18]. The product, pre-miRNA, exits from the nucleus and is further processed by the cytoplasmic RNase III-like enzyme, Dicer, to generate the final miRNA. Interestingly, at least two dozen miRNA sequences (in humans) map within specific introns of protein-coding ‘host’ genes [19–22]. Although many of them appear to be conserved across species, the biogenesis and role of such ‘intron-derived’ (or ‘intronic’) miRNAs remains a mystery. It is also not known whether they are in fact produced from the spliced intron or from an alternate internal promoter. In an early study, a number of intronic miRNAs were found to be co-expressed with their host transcripts [14], suggesting that an alternate transcription unit was unlikely. Regarding their processing, recent experimental studies of selected intronic miRNAs have suggested a potentially novel mechanism in which they are generated by a Drosha-mediated cleavage step between splicing commitment and excision, thereby ensuring both miRNA biogenesis and protein synthesis from a single primary transcript [23]. Regardless of the mechanism of excision, we hypothesize that the regulation of the host gene transcription will proportionately affect the abundance of the corresponding intronic miRNA, which will in turn regulate the downstream target genes. In this paper, we provide proof-of-concept of such a relationship between intronic miR-126* and its host gene, ‘epidermal growth factor-like domain 7 or Egfl7 (NP_057299). Also known as VE-statin (vascular endothelial statin) or Neu1, Egfl7 is the first identified inhibitor of mural cell (vascular smooth muscle cells and pericytes) migration, specifically produced by endothelial cells [24–27]. We show that prostate cells are naturally deficient in miR-126*, supporting recent miRNA tissue profiling [28] and that this is due to poor Egfl7 gene expression. In contrast, prostate cells are famously enriched in a number of prostate-specific antigens that includes the most recently discovered protein, prostein (NM_033102), also called prostate cancer-associated protein 6 (PCANAP6) and solute carrier family 45, member 3 (SLC45A3) [29, 30]. We postulated that the scarcity of miR-126* in the prostate might be causally related to the abundance of prostein in this tissue. In other words, prostein may be a natural target of miR-126*. In support of this, our bioinformatic search of cellular targets found a miRNA response element (MRE) corresponding to miR-126* in the 3′ UTR of the prostein mRNA. Here, we provide multiple lines of evidence to support this hypothesis. We show that the absence of miR126* in the prostate is indeed an essential prerequisite for the ability of prostate cells to synthesize prostein. Surprisingly, although prostein is expressed in both normal and cancerous prostate [30], we found that ectopic expression of miR126* led to a significant reduction in the migration and invasiveness of the prostate cancer LNCaP cells ex vivo, suggesting that the natural lack of mir-126* can be a contributing factor to the invasiveness of prostate cancer in the appropriate genetic environment.
Materials and methods
Plasmid construction, transfection, and RNAi assays
Synthetic siRNAs and 2′-O-methyl antagomirs [31] were obtained from Ambion. miRIDIAN miRNA Mimics were from Dharmacon; these are double-stranded RNA oligonucleotides, chemically enhanced with proprietary design for preferential incorporation of the active strand into RISC and exclusion of the passenger strand. The miRNA-126* reporter firefly luciferase plasmid was constructed by cloning the 1,397-bp 3′UTR of the prostein mRNA (NM_033102) between the SacI and HindIII sites of pMIR-Report vector using real-time reverse-transcription-polymerase chain reaction (RT-PCR; Ambion). Mutation of the miR-126* sites 1 and 2 (Fig. 1) were performed with the QuikChange site-directed mutagenesis kit (Stratagene). All clones were verified by DNA sequencing.
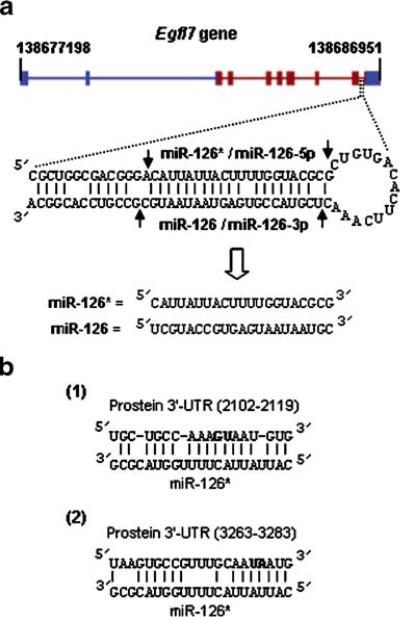
Fig. 1.
a Structure of the Egfl7 gene and host intron. The line diagram on top shows the coding (red) and the untranslated regions (blue) of the human Egfl7 gene on chromosome 9 (base numbers on top). The pre-miRNA, the Dicer cutting sites (arrow) and the final miRNA sequences are shown below. b Two putative sites in the 3′UTR of prostein that recruit miR-126* as shown. The numbers were counted after the termination codon. The bases in bold were mutated in the reporter clones as described in the Results section
The 822-nt long Egfl7 coding sequence (cDNA; NM_016215) was amplified by standard reverse transcription-PCR using primers with EcoRI and BamHI sequences and cloned into the EcoRI and BglII sites of the mammalian expression vector, pCAGGS [32]. This plasmid was called pCAGGS-Egfl7. The clone containing intron-9 inside the coding sequence was made as follows, taking advantage of a unique DraIII site in the preceding exon and a unique EcoNI site in the following exon. In brief, the sequence between the DraIII and the EcoNI site was removed from the pCAGGS-Egfl7 plasmid and was replaced by the corresponding sequence from the genome, amplified by PCR.
Cells were grown in Dulbecco's MEM (Mediatech) supplemented with L-glutamine (2 mM) and 10% heat-inactivated calf serum, and maintained using standard procedures. All plasmids for transfection were purified by the Midiprep kit (Qiagen). Transfections were done to monolayer cells in 96-well plates for luciferase assay or scaled up to 12-well plates for RNA and protein analysis using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For 96-well plates, 0.2 μg of pMIR-based reporter plasmid (expressing firefly luciferase; Ambion) and 0.1 μg of internal control pRL-Con plasmid (expressing Renilla luciferase) were co-transfected, and cells were harvested 44 h later for luciferase assay. For RNAi studies, siRNAs or miRNA-mimics were also transfected similarly, and protein or RNA was measured 34 h later, as described below. A range of siRNA/miRNA-mimic concentrations was tested, equivalent to 10–30 pmol per well of a 12-well plate, and optimal inhibition results are presented. siRNAs were designed by the Whitehead Institute web site (http://jura.wi.mit.edu/bioc/siRNAext/) and then visually selected to satisfy the asymmetry rule [33–35]. All transfection assays were done in triplicate and repeated at least twice.
RNA and protein measurements
For RNA measurements, quantitative RT (qRT)-PCR was performed essentially as described before [36]. In brief, total RNA was collected from adherent tissue culture cells using Trizol (Invitrogen) according to the manufacturer's instructions. RNA was quantified (A260) using a SmartSpec 3000 spectrophotometer (Bio Rad). The RNA was subjected to standard oligo(dT)-cellulose affinity chromatography, and 0.2–0.5 μg of the poly(A)-selected RNA was used for each sample in a 10-μl reverse transcription reaction with 0.25 μl of SuperScript II RT (Invitrogen) and the appropriate reverse (antisense) primer. One-tenth volume of the RT reaction was then used in a 25 μL PCR reaction containing 30 pmol of both forward and reverse primers. Primer sequences are listed in Table 1. The miRNA levels were measured using the mirVana kit (Ambion) and the manufacturer's procedure based on solution hybridization with labeled antisense RNA, followed by RNase digestion, gel electrophoresis and autoradiography.
Table 1.
PCR primer and siRNA sequences
| Gene (NCBI number) | PCR primers (5′ à 3′) |
|---|---|
| Egfl7 (NP_057299) | Forward: GTTACTGGTGCCAGTGTTGG |
| Reverse: TTGCACTGTCCACTCCTGTC | |
| Prostein (NM_033102) | Forward: GATCCTGCCCTACACACTGG |
| Reverse: TCATCAGGCTGTCCTCACTG | |
| siRNA | |
| Egfl7 | 5′ CAGGAGTGGACAGTGCAATdTdT 3′ |
| 3′ dTdTGTCCTCACCTGTCACGTTA 5′ | |
| Prostein (siRNA#1) | 5′ GCAGGTGTTCCTGCCCAAAdTdT 3′ |
| 3′ dTdTCGTCCACAAGGACGGGTTT 5′ | |
| Prostein (siRNA#2) | 5′ GCAGTGAGGACAGCCTGATdTdT 3′ |
| 3′ dTdTCGTCACTCCTGTCGGACTA 5′ |
Protein was measured by immunoblot (Western blot) using the horseradish peroxidase-conjugated secondary antibody and the ECL detection system (Pierce) as described previously [39]. Rabbit antihuman prostein antibody and goat antihuman Egfl7 antibody were from Abcam and R&D Systems, respectively.
Cell migration and invasion assay
Wounding assay for migration was performed as follows. One day after transfection of the indicated miRNA or siRNA, at which time prostein silencing was maximal (Fig. 3a), the LNCaP cell monolayer was wounded with a p10 tip, incubation was continued, and images were captured at 2 h intervals until 12 h. The rate of migration was measured by quantifying the total distance traveled by cells from the edge of the wound to its center divided by the number of hours.
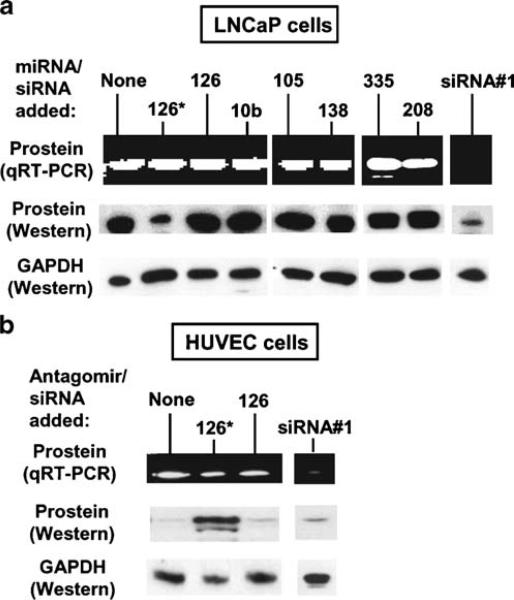
Fig. 3.
Silencing of prostein by miR-126*. The LNCaP cells (a) were transfected with anti-prostein siRNA or the indicated miRNA-mimics, and the HUVEC (b) were transfected with anti-prostein siRNA or antagomiR-126*. The RNA and protein were measured as described. Representative qRT-PCR samples after the same number of PCR cycles were analyzed on agarose gel and stained with EtBr. Equal sample loading was ascertained by staining for 28S RNA and Western blotting for GAPDH (from a different gel run) as in Fig. 2. Anti-prostein siRNA no. 1 is described in Table 1; siRNA no. 2 showed essentially similar inhibition (data not shown)
For the modified Boyden chamber assay, LNCaP cells were first grown in monolayers to about 70% confluency and transfected with the desired miRNA/siRNA or mock-transfected. At 24 h post-transfection, cells were trypsinized, and a fixed number of cells (5×105) in serum-free medium was added to the upper chamber of each Matrigel insert in a 24-well plate format (BD BioCoat, 8 μM pore size). The lower chamber contained 15% serum-containing medium. Cells were incubated for 36 h, and invasion was detected by staining the bottom of the membrane with Mayer's hematoxylin solution. Stained cells were visualized under a microscope (magnification ×100), counted in four random fields, and the average number was taken. Each treatment was performed in triplicate wells (n=3). Data were analyzed by one-way ANOVA followed by Bonferroni's test.
Results
Cell type-specific expression of Egfl7 and its intronic miR-126*
Two miRNAs, miR-123 and miR-126, were originally identified in mouse [13], but miR-123 was later found to be processed from the 5′-half of the same precursor (pre-miRNA) as miR-126 and was hence renamed miR-126* (also known as miR-126-5p; Fig. 1a). The corresponding human orthologs, miR-126 and miR-126*, were predicted from homology to the mouse counterparts, but their expression was not verified. An interesting feature of the miR-126 locus is that in all vertebrates tested to date, including human and mice, its sequence maps in a specific intron of the Egfl7 gene (Fig. 1a). The human Egfl7 gene on chromosome 9, for example, contains nine introns, and the miR-126/126* precursor locates within intron-9 (Fig. 1a).
We first determined the expression of Egfl7 gene and miR-126/126* in human cells representative of four different lineages: A549 (alveolar epithelial), HeLa (cervical fibroblast), HUVEC (umbilical vein endothelial), and LNCaP (fibroblastoid prostate cancer). Our analysis clearly revealed that only the HUVEC cells made appreciable amounts of Egfl7 mRNA and miR-126/126* (Fig. 2). The Egfl7 protein level showed a parallel pattern (Fig. 2), suggesting that the mRNA is functional.
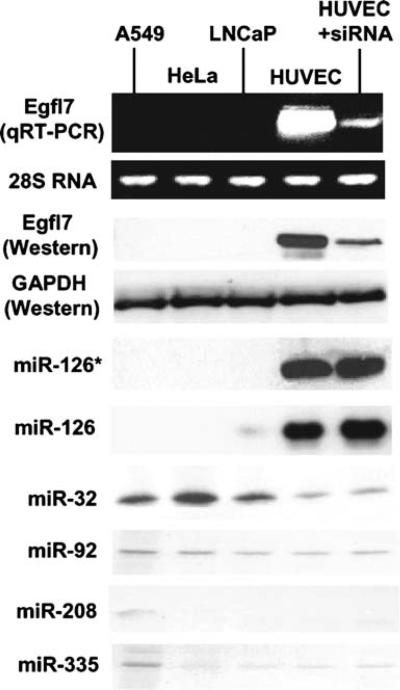
Fig. 2.
The direct relationship between Egfl7 transcript and its intronic miRNAs. Analyses were done in the three cell lines indicated above without treatment or after 34 h of treatment with anti-Egfl7 siRNA (Table 1) in HUVEC, as detailed in the Materials and methods section. Egfl7 mRNA was estimated by qRT-PCR, and representative samples at the same number of PCR cycles were analyzed on agarose gel followed by ethidium bromide (EtBr) staining for a visual representation. Western blot for the proteins and RNase-protection assay for miRNAs have been described. Equal RNA and protein amounts in the same samples were documented by the EtBr-stained 28S RNA on agarose gel and Western for the GAPDH control
As discussed earlier, the exact mechanism of biogenesis of intronic miRNAs remains unknown. However, if it requires the splicing machinery, it should happen in the nucleus, and therefore, it should be possible to knock down the processed mRNA in the cytoplasm without inhibiting the upstream generation and processing of the intronic miRNA. This was tested by transfecting the HUVEC with siRNA against the Egfl7 mRNA, which indeed caused considerable degradation of the mRNA (Fig. 2) with no effect on the pre-excised miRNA-126/126*. Together, these results suggest that miR-126/126* are truly intronic miRNAs from the Egfl7 transcript and that the prostate cells are naturally deficient in both.
Prostein is a specific target of ectopic miR-126*
We initially performed a computational search to identify the possible mRNAs targets of the Egfl7 intronic miRNAs. Prostein was one of the targets of miR-126* predicted by PicTar [40] but not by a number of others, including DIANA [9] or Miranda [11]. The two predicted sites for miR-126* were both in the 3′UTR of prostein mRNA (Fig. 1b), and none was found for miR-126. The minimum free energy (MFE) values of the two binding sites were −19.9 and −17.9 kcal/mol, respectively, suggestive of a moderate strength of miR-126*-binding. Equivalent sites were also found in the 3′UTR of the prostein orthologs in a number of other species, including a few mammals and chicken and fish. As prostein is a prostate-specific gene with little or no expression in other tissues, we hypothesized that miR-126* is a bona fide silencer of prostein and that the expression of protein in prostate capitalize on the absence of miR-126* in this tissue.
We, therefore, conducted a series of experiments to test this hypothesis. First, we measured protein expression levels in the four cell lines mentioned above. We used qRT-PCR and Western to confirm that both prostein mRNA and protein are abundant in LNCaP cells (Fig. 3a) but undetectable in other cell types (representative HUVEC is shown in Fig. 3b [29, 30]). We then showed that introduction of exogenous synthetic miR-126*-mimic in LNCaP cells silenced prostein protein expression, while the mRNA level remained unaffected (Fig. 3a). Prostate is also naturally deficient in a few other miRNAs, notably miR-10b, 105, -138, -144, -181c, -299, and -301, but none of them had any predicted site in the prostein mRNA [9]. To ascertain that the effect of miR-126* on the protein level was not due to a nonspecific stimulation of RNAi, we performed transfection with three of these miRNA-mimics (miR-10b, -105, -138) and found no effect on prostein (Fig. 3a), demonstrating specificity of miR-126*. To rule out that the inability of the miRNAs to degrade prostein mRNA is not because of a lack of specific components of the RNAi pathway, we transfected LNCaP cells with synthetic siRNAs designed against prostein mRNA. This resulted in mRNA degradation and loss of protein (Fig. 3a). A general inhibition of translation was also ruled out by the unchanged levels of GAPDH control.
In addition to miR-126*, four other miRNAs were predicted by PicTar and/or TargetScan to have target sites on prostein mRNA. They were: miR-32, -92, -208, and 335. Our results (Fig. 2) show that while miR-32 is expressed modestly in all cells (slightly lower in HUVEC), the others are expressed very poorly (miR-92, -335) or not at all (miR-208). To test whether ectopic introduction of any of these miRNAs would reduce prostein levels, we performed similar transfection experiments in LNCaP cells with the corresponding miRNA-mimics and found that they had no discernible effect. We present the negative results with miR-335 and miR-208 (Fig. 3a) as representative of this set.
Taken together, these results show that ectopic introduction of synthetic miR-126* is sufficient to significantly lower prostein protein levels. Later, we demonstrate this with intronically generated miR-126* as well.
Relationship between prostein and miR-126* in non-prostate cells
It is to be recalled that vascular HUVEC cells are naturally low in prostein transcripts, suggesting lack of transcription. However, these cells are also high in miR-126* due to Egfl7 gene expression. As shown (Fig. 3b), the HUVEC cells were indeed found to make small but detectable amounts of prostein transcript. In contrast, the protein level was practically undetectable, suggesting a translational silencing in addition to transcriptional regulation. We reasoned that the translation silencing is due to high levels of miR-126*. To determine the contribution of miR-126*-mediated silencing in the backdrop of already poor transcription, we tested if reduction of miRNA-126* by antisense can elevate the prostein protein level in these cells. In fact, when transfected with an antagomir [31] against miR-126*, HUVEC cells produced substantially larger amounts of prostein protein (Fig. 3b). Control antagomir-126 had no effect, showing specificity. The siRNA against prostein reduced both transcript and protein levels, confirming the existence of a functional RNAi pathway. These results show that the lack of prostein in vascular cells is a combined result of low transcription of the gene and translational suppression by miR-126*.
Two miR-126* target sites in the 3′UTR of prostein mRNA
To map the functional miR-126* sites on the prostein mRNA, we constructed firefly luciferase reporter plasmids containing CMV promoter and the 1,397-nucleotide long prostein 3′UTR. We transfected LNCaP and HUVEC cells with the construct, and luciferase activities were measured. Note that these two cells are naturally contrasting of miR-126* levels. All assays were normalized by co-transfection with a constitutive Renilla luciferase plasmid. Results (Table 2) show that the luciferase activity was much lower in HUVEC cells that are naturally rich in miR-126*. Transfection with miR-126*-mimic drastically reduced the luciferase activity in LNCaP cells, essentially to the level in HUVEC cells, showing that the 3′UTR segment contains sequences needed for miR-126*-dependent translational repression. Transfection of control miRNA-mimics of miR-126 or -138 had no effect.
Table 2.
Sites in the prostein 3′UTR required for miR-126*-mediated silencing
| Nature of the 3′UTR site and the miRNA added | Luciferase activity in: |
|
|---|---|---|
| LNCaP | HUVEC | |
| Wild type | 100 | 18±5 |
| Wild type + miR-126* | 14±4 | 16±6 |
| Wild type + miR-126 | 94±8 | 20±4 |
| Wild type + miR-138 | 98±6 | 12±8 |
| Mutant 1 | 105±6 | 92±6 |
| Mutant 1 + miR-126* | 98±5 | 90±8 |
| Mutant 2 | 110±10 | 90±8 |
| Mutant 2 + miR-126* | 96±5 | 92±8 |
Transfection and luciferase assays were performed as described in the Materials and methods section. All numbers were normalized against Renilla luciferase activity; the value for wild-type prostein 3′UTR was taken as 100, and others were expressed as its percentage. Mutants 1 and 2 refer to mutations in the miR-126*-binding sites 1 and 2, respectively, as indicated in Fig. 1b.
We then mutated each of the two predicted miR-126*-binding sites (Fig. 1b) in the 3′UTR within the sequence complementary to the seed region of the miRNA. In the first site (2,102–2,119) GU was mutated to CA, and in the second site (3,263–3,283), UA was mutated to AU. The transfection experiment was then repeated as above, and luciferase was assayed. Results (Table 2) clearly show that each mutation led to a substantial relief of translational inhibition by miR-126* that was either exogenously added to LNCaP cells or naturally present in HUVEC cells. We conclude that these are the functional sites of miR-126*-binding in prostein 3′UTR and that both sites are required for optimal and robust inhibition of prostein translation. The luciferase results also show that only miR-126* is necessary and sufficient to repress prostein translation and that the rest of the Egfl7 transcript is unlikely to be needed.
Prostein is important for the motility and invasiveness of LNCaP prostate cancer cells
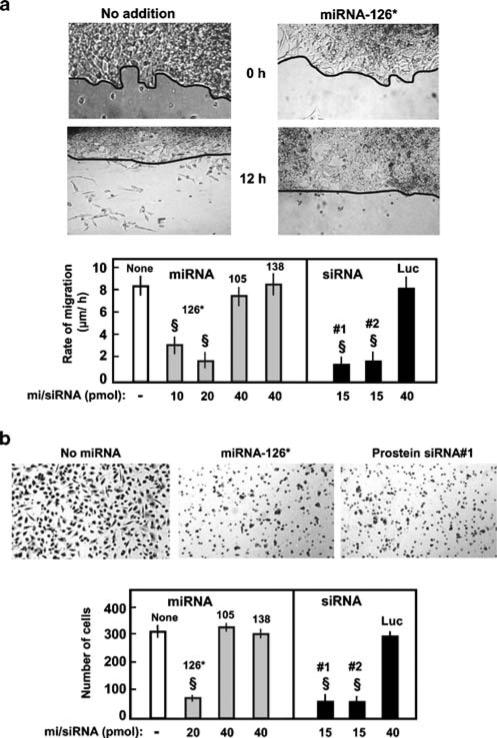
Although the various prostate-specific antigens are diagnostic tools for prostate enlargement and cancer, it has never been addressed whether they have any relation with the cancerous phenotype of the cells. As the LNCaP cells produce abundant amounts of prostein, we were interested to test whether its regulation by miR-126* will have any effect on the growth or invasiveness of the cells. This was tested in a standard monolayer-wounding assay in which the number of motile LNCaP cells beyond the scratch line was counted at various times after wounding. Similar experiments were conducted on a monolayer transfected with miR-126*-mimic. In parallel, a Matrigel invasion assay was also performed to obtain a measure of the migration of these transfected cells. The results (Fig. 4) clearly reveal a significant reduction of both the migration rate and invasiveness in miR-126*-treated cells. To rule out that this is due to an effect of miR-126* on other targets, we repeated the same experiment with two anti-prostein siRNA (Table 1) that had strongly silenced prostein levels before (Fig. 3), which also resulted in inhibition of migration and invasion. Two control miRNA-mimics, namely, miR-105 and miR-138, and a control siRNA against luciferase had no affect on either property. We conclude that prostein is an important determinant of motility and invasiveness of prostate cancer, and thus, the absence of miR-126* in these cells is a clinically relevant factor.
Fig. 4.
Motility (wounding) and invasion (Boyden chamber) assay of miRNA-treated LNCaP cells. Both assays are detailed in the Materials and methods section. a Motility assay. Top: Representative pictures of 20 pmol miR-126*-treated and control untreated cells at two time points (0, 12 h) are shown (from 12-well plates). Due to the uneven growth of LNCaP monolayers, the wound line is hand-traced for a clear reference. Cells that migrated below the line were counted. Bottom: Migration rates of LNCaP cells in micron per hour were determined from the wounding experiment described above. The various miRNA numbers are indicated, and prostein siRNA no. 1 and siRNA no. 2 are described in Table 1. Control luciferase siRNA sequence has been described [35]. b Modified Boyden chamber assay. Top: Representative photographs (×100) of migrated LNCaP cells transfected with no miRNA (control), miR-126* (20 pmol), and prostein siRNA no. 1. Bottom: Graphical presentation of the number of migrated cells transfected with the indicated mi/siRNAs. Data are mean±SEM; n=3; §P<0.05
Intronic miR-126*, spliced and processed from recombinant Egfl7 gene, is functional
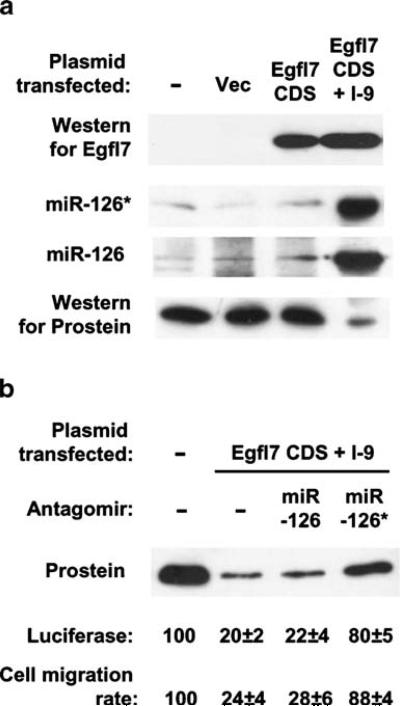
In the previous experiments, we demonstrated that synthetic and ectopic miR-126* and anti-miR-126*, respectively, suppressed and elevated prostein translation. Nonetheless, a direct demonstration of regulation by intracellularly expressed miR-126* was still lacking. To accomplish this, we constructed two plasmids in the pCAGGS vector [32], one containing just the Egfl7 coding sequence (no intron) and the other containing the same coding sequence plus only the ninth intron placed between its natural exons. Transcription in this vector occurs from a strong chicken β-actin promoter aided by a CMV enhancer, and is thus, relatively independent of tissue.
Results (Fig. 5a) show that both plasmids, when transfected into LNCaP cells (that are naturally deficient in Egfl7), expressed substantial amounts of recombinant Egfl7 protein, measured by Western blot. The identical mobility and expression levels suggested that intron-9 was properly spliced out of its natural sequence context. The intron-harboring clone additionally produced miR-126* and miR-126 as shown, providing direct proof that these miRNAs are indeed products of Egfl7 intron-9 and that we produced them ectopically in prostate cells from an actual splicing event, similar to its natural mechanism of synthesis. Interestingly, prostein translation was also strongly suppressed in these cells, whereas cells expressing recombinant Egfl7 alone (without intron-9) produced normal amounts of prostein.
Fig. 5.
Translational silencing by miRNA from a recombinantly spliced intron. a LNCaP monolayers were transfected with (from left to right) no plasmid, pCAGGS vector alone, pCAGGS containing Egfl7 coding sequence (CDS), or pCAGGS containing Eglf7 coding sequence with intron-9 (I-9). Expressions of Eglf7, miR-126*/126 and prostein were measured as before. b In parallel cultures, LNCaP cells were additionally transfected with the indicated antagomirs and either co-transfected with miR-126*-reporter luciferase plasmid or subjected to migration assay as described previously. Numbers were expressed as percentage of values from untransfected cells
Although recombinant intron-9 produced both miR-126* and miR-126, we already know from previous results that only the former inhibits prostein translation. Nevertheless, to formally exclude an effect of miR-126, we transfected these cells with either antagomir-126 or antagomir-126* (Fig. 5b). Antagomir-126 failed to restore prostein levels, whereas antagomir-126* did. The functionality of the intronic miR-126* was further documented by suppression of luciferase from the miR-126*-reporter construct used earlier. Again, antagomir-126 had no effect on luciferase, whereas antagomir-126* restored activity. Lastly, we tested whether intronic miR-126* could reduce LNCaP cell migration in wounding assay, and it, in fact, did (Fig. 5b). Transfection with the intron-9 clone indeed reduced migration, and like the luciferase assay, antagomir-126 had no effect, whereas antagomir-126* restored migration. These results not only established the physiological function of intronic miR-126* spliced in situ, but also ruled out the possibility that silencing of prostein might be due to Egfl7 protein or some other intron or sequence in the Egfl7 transcript.
Discussion
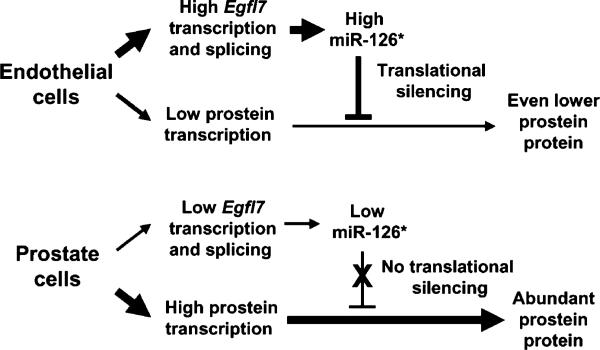
Despite the fact that introns occupy a substantial portion of all eukaryotic genomes, they are generally viewed as sequences that are only to be removed and destroyed [41, 42]. The discovery of intronic miRNA for the first time raised the possibility that some excised introns may in fact have a cellular function. In this paper, we provide the following major findings on the properties and function of miR-126*: (1) It is a product of an intron of the Egfl7 gene that is not expressed in the prostate; (2) It can inhibit the translation of prostein by binding to two sites in the 3′UTR of the prostein mRNA; (3) Its absence is thus essential for the abundant and specific expression of prostein in the prostate cancer cells, which in turn plays an apparently important role in the migratory ability of these cells. Reciprocally, it also appears that in the non-prostate tissues, particularly in endothelial cells, an abundance of Egfl7 transcription and miR-126* may contribute to the overall silencing of prostein expression. Summing up, our results document the inverse relationship between the expression of a miRNA-host gene (Egfl7) and that of its target mRNA (prostein). The regulation is schematically depicted in Fig. 6.
Fig. 6.
Summary model. The reciprocal relationship between Egfl7 mRNA and prostein translation is shown schematically. The thickness of the lines is roughly symbolic of expression levels
Egfl7 is expressed at high levels in the vasculature associated with tissue proliferation and is abundant in such organs as heart, lung, ovary, and uterus [24–27]. In the developing embryo, it is expressed very early during endothelial differentiation and later in all embryonic blood vessels. Although the exact physiological role of Egfl7 in vivo is still a matter of speculation, available evidence indicates that its normal function is to regulate the dynamism of the vasculature by reorganizing the vascular bed in response to angiogenic stimuli and preventing an over-recruitment of smooth muscle cells around the newly established capillaries. Conversely, its suppression, as seen in prostate cancer cells, may facilitate the uncontrolled growth and proliferation of smooth muscle cells and vascular tissues characteristic of a tumor. Regardless of the precise role of Egfl7 in the prostate, we show here that its natural absence leads in turn to the absence of its intronic mRNA, which indirectly promotes the invasiveness of LNCaP prostate cancer cells by allowing the expression of prostein.
As we have shown here, other miRNAs with in silico predicted target sites on prostein mRNA did not silence prostein when experimentally tested, suggesting that miR-126* may be the only miRNA that regulates prostein. In recent years, attempts have been made to define the general features of miRNA-target base-pairing for improved silencing efficiency by combining computational and experimental approaches [7–9]. As mentioned before, the most important requirement is annealing of the core sequence of nt 2 to 8 of the miRNA [7, 8]. Interestingly, both the miR-126* sites of prostein partially deviate from this rule (Fig. 1). The first site has a mismatch at nt-4, but then continuous annealing for the next eight nucleotides. The second site anneals with the first nucleotide of miR-126*, which is considered irrelevant to RISC formation [7]. Both sites have additional consensus features important for optimal silencing [8, 9]. First, they match additional nucleotides of miR-126*. Second, they are sufficiently far from the prostein stop codon, being 102, 1,262 nt away, respectively. Third, both are positioned away from the center of the 1,381-nt long 3′UTR. Finally, multiple miRNA sites on the same target mRNA are known to boost silencing [4, 8, 43], and this is also the case with miR-126*, as both sites were needed for its optimal effect (Table 2).
Are there other miR-126* targets relevant for the prostate? Although we do not have the answer, mRNA sequences of none of the other prostate-specific genes revealed miR-126* sites; these include: PSA (the classic prostate-specific antigen, also called kallikrein 3; NM_001648), PAP (prostatic acid phosphatase; NM_001099), PSGR (prostate-specific G protein-coupled receptor; AF311306), prostate stem cell antigen (PSCA; AJ297436), PSMA (prostate-specific membrane antigen; NM_004476), and AIbZIP (androgen-induced bZIP protein; NM_130898). On the other hand, some of these genes and others that are also up-regulated in prostate cancer have putative target sites for other miRNAs that are also relatively scarce in the prostate as listed earlier. For instance, miR-301 has predicted sites in the 3′UTRσ of the transmembrane prostate androgen-induced protein (TMEPAI), testis-, and prostate-specific protein kinase 2 (TESK2) and p21-activated kinase 6 (PAK6, which is high in testis and prostate). There is a putative site for miR-152 in PSA, and for miR-20 in PAP, but both miRNAs are poorly expressed in all tissues, including prostate.
Prostate cancer, in its various degrees of aggressiveness, is a leading cause of death among men, particularly in the industrialized nations. It is second only to heart disease, and the need for an early and specific diagnosis has been long recognized for better treatment and prognosis. A large set of prostate-specific proteins have been characterized, but their physiological roles remain essentially unknown. We emphasize that in spite of our results, it is too premature to advocate the use of miR-126* as a general therapy for prostate cancer. First, although we have demonstrated an importance of prostein in the migration on LNCaP cells that are poorly invasive and androgen-dependent in nature, two other established prostate cancer cell lines of epithelial origin, namely, DU145 and PC3, which are more invasive and androgen-independent, poorly express prostein [44]. This may suggest that prostein levels inversely correlate with invasiveness in all forms of prostate cancer; however, in a broader survey of other prostatic cell lines and pathological samples, prostein was found in both normal and cancer cells with a variety of invasive scores [30, 44]. Second, Egfl7 transcript and miR-126* levels of all three prostate cell lines were equally low (data not shown). Clearly, prostein and miR-126* cannot be the sole determinants of invasiveness, carcinogenesis or progression of prostate cancer, making it quite likely that other genes and miRNAs are also involved in this multivariate disease that await discovery. Indeed, hybridization-based miRNA-profiling revealed that at least 51 miRNAs were differentially expressed between benign and malignant prostate tumors, of which 37 were down-regulated and 14 up-regulated [28]. Although it remains unknown whether these miRNAs actually regulate prostate cancer, it is interesting to note that hierarchical clustering revealed closeness of miRNA profile between the invasive DU145 and PC3 cells, whereas both were distant from the noninvasive LNCaP cell line. Regardless, prostein does exhibit many features that may be relevant for understanding prostate cancer [30]. First, the prostein gene is located in a region of chromo-some 1 that determines susceptibility to hereditary prostate cancer, suggesting that it may play a role in prostate cancer malignancy. Second, orthologs of prostein are found in many other vertebrates, including dog, rat, mouse, opossum, cow, chicken, zebrafish, pufferfish, sea urchin, frog, and nonhuman primates (data not shown), suggesting that it may have an evolutionarily conserved signaling function, which is characteristic of oncogenes. Third, it also shows significant homology with AIM1 (absent in melanoma-1; NP_057264), a likely regulator of the malignant expression of human melanoma [45, 46]. Finally, like AIM1, prostein belongs to the solute-carrier family and is exported to the cell surface and, therefore, may signal to the neighboring cells in a tissue. It would be interesting to determine the identity and mechanism of the interacting partners of prostein and whether their expression is also regulated by miRNAs, which may eventually pave the way to control specific forms of prostate cancers using the naturally occurring miRNA pathway.
Acknowledgments
This work was supported in part by the US National Institute of Health (NIH) grant AI059267 to S.B. We thank Dr. Ratna Chakrabarti (University of Central Florida) for the LNCaP cells, Dr. Lalita Shevde-Samant (Mitchell Cancer Institute) for the PC3 and DU145 cell lines and advice with the Boyden chamber and wounding assays, Dr. Ramesh Pillai (EMBL Grenoble, France) for the pRL-Con plasmid, and Dr. Lance Ford (Bioo Scientific Corp., Austin, Texas) for helpful discussions on miRNA analysis and assay.
Abbreviations
- bp
base pair
- RNAi
RNA interference
- siRNA
small interfering RNA
- miRNA
microRNA
- RISC
RNAi-induced silencing complex
- RSV
respiratory syncytial virus
- UTR
untranslated region
- (Egfl7)
EGF-like domain 7
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HUVEC
human umbilical vein endothelial cells
Biography
 Alla Musiyenko received her B.Sc. and M.Sc. from Kiev State University, Kiev, Ukraine, and worked as a Research Assistant in the Human Genetic Department, Institute of Molecular Biology and Genetics, National Academy of Science of Ukraine. Currently, she is a Research Technologist III at the University of South Alabama, USA.
Alla Musiyenko received her B.Sc. and M.Sc. from Kiev State University, Kiev, Ukraine, and worked as a Research Assistant in the Human Genetic Department, Institute of Molecular Biology and Genetics, National Academy of Science of Ukraine. Currently, she is a Research Technologist III at the University of South Alabama, USA.
 Sailen Barik received his Ph.D. in Biochemistry from Bose Institute, Calcutta University, India. He is presently a Professor in the Department of Biochemistry and Molecular Biology at the University of South Alabama, Mobile, Alabama, USA. He holds a joint appointment in the Department of Microbiology and Immunology. His research interests include host–pathogen interactions and RNA interference.
Sailen Barik received his Ph.D. in Biochemistry from Bose Institute, Calcutta University, India. He is presently a Professor in the Department of Biochemistry and Molecular Biology at the University of South Alabama, Mobile, Alabama, USA. He holds a joint appointment in the Department of Microbiology and Immunology. His research interests include host–pathogen interactions and RNA interference.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Barik S. Silence of the transcripts: RNA interference in medicine. J Mol Med. 2005;83:764–773. doi: 10.1007/s00109-005-0690-0. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. Erratum in: PLoS Biol (2005) 3:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landgraf P. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying SY, Lin SL. Intron-derived microRNAs—fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 22.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA). J Biomed Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soncin F, Mattot V, Lionneton F, Spruyt N, Lepretre F, Begue A, Stehelin D. VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J. 2003;22:5700–5711. doi: 10.1093/emboj/cdg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, De Sauvage FJ, Ye W. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- 26.Fitch MJ, Campagnolo L, Kuhnert F, Stuhlmann H. EGFL7, a novel epidermal growth factor-domain gene expressed in endothelial cells. Dev Dyn. 2004;230:316–324. doi: 10.1002/dvdy.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campagnolo L, Leahy A, Chitnis S, Koschnick S, Fitch MJ, Fallon JT, Loskutoff D, Taubman MB, Stuhlmann H. EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am J Pathol. 2005;167:275–284. doi: 10.1016/S0002-9440(10)62972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 29.Cunha AC, Weigle B, Kiessling A, Bachmann M, Rieber EP. Tissue-specificity of prostate specific antigens: compara tive analysis of transcript levels in prostate and non-prostatic tissues. Cancer Lett. 2006;236:229–238. doi: 10.1016/j.canlet.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Kalos M, Stolk JA, Zasloff EJ, Zhang X, Houghton RL, Filho AM, Nolasco M, Badaro R, Reed SG. Identification and characterization of prostein, a novel prostate-specific protein. Cancer Res. 2001;61:1563–1568. [PubMed] [Google Scholar]
- 31.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 32.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 34.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. Cell Erratum in:505. [DOI] [PubMed] [Google Scholar]
- 35.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2004;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 36.Bitko V, Garmon NE, Cao T, Estrada B, Oakes JE, Lausch RN, Barik S. Activation of cytokines and NF-kappa B in corneal epithelial cells infected by respiratory syncytial virus: potential relevance in ocular inflammation and respiratory infection. BMC Microbiol. 2004;4:28. doi: 10.1186/1471-2180-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 38.Bitko V, Barik S. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 2001;1:34. doi: 10.1186/1471-2180-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- 40.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 41.Naora H, Deacon NJ. Relationship between the total size of exons and introns in protein-coding genes of higher eukaryotes. Proc Natl Acad Sci USA. 1982;79:6196–6200. doi: 10.1073/pnas.79.20.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clement JQ, Qian L, Kaplinsky N, Wilkinson MF. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5:206–220. doi: 10.1017/s1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalos M, Askaa J, Hylander BL, Repasky EA, Cai F, Vedvick T, Reed SG, Wright GL, Jr., Fanger GR. Prostein expression is highly restricted to normal and malignant prostate tissues. The Prostate. 2004;60:246–256. doi: 10.1002/pros.20043. [DOI] [PubMed] [Google Scholar]
- 45.Trent JM, Stanbridge EJ, McBride HL, Meese EU, Casey G, Araujo DE, Witkowski CM, Nagle RB. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science. 1990;247:568–571. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- 46.Ray ME, Su YA, Meltzer PS, Trent JM. Isolation and characterization of genes associated with chromosome-6 mediated tumor suppression in human malignant melanoma. Oncogene. 1996;12:2527–2533. [PubMed] [Google Scholar]