Understanding the molecular and cellular mechanisms underlying normal development and pathological conditions is an important step towards prevention and developing effective therapeutic approaches for cardiovascular diseases. In recent years, the zebrafish model has rapidly emerged as a powerful genetic system to study cardiac development and function, and some zebrafish mutants have been used as animal models to dissect the molecular causes of cardiovascular disease. Here, we discuss current advancements in zebrafish cardiovascular studies with a focus on the genetic mechanisms regulating cardiac function and the potential of utilizing zebrafish for drug discovery.
Zebrafish heart development
Although the morphological differences between the zebrafish and human heart may seem astounding, the fish heart and mammalian heart undergo similar morphogenetic processes. Cardiac cells of the zebrafish begin as two lateral populations, migrating to the midline and forming the primitive heart tube. As the embryo develops, the heart tube undergoes cardiac looping and further differentiates into distinct cardiac chambers separated by valve leaflets at the atrioventricular (AV) junction [1,2]. Recent optical imaging studies define four distinct physiological developmental stages of zebrafish conduction system that correspond to the cellular and anatomical changes of the heart [3]. Spontaneous, uncoordinated contraction of cardiomyocytes is noted soon after the fusion of bilateral cardiac precursors. By 24 hours post fertilization (hpf), the conduction system begins as a linear wave that travels through the heart from the sinus venosus to the outflow tract, coinciding with the observed peristaltic contraction pattern of the primitive heart tube. By 48 hpf, cardiomyocytes at the AV ring differentiate into cells with slow conducting properties, creating a significant conduction delay at the AV boundary and transitioning the heart from peristaltic contraction to coordinated contraction of the chambers [3,4]. As the ventricle continues to thicken and develop trabeculae, a fast conduction network forms within the ventricle, where a complex conduction pattern proceeding from the outer curvature of the ventricle to its base is observed. Finally, upon maturation of the fast conduction network, the conduction wave transforms to an apex-to-base model across the ventricular myocardium [3,5].
Zebrafish as a genetic model for cardiovascular research
The zebrafish model offers several advantages in examining the genetic mechanisms behind cardiovascular development and function. First, zebrafish produce a large number of easily accessible and transparent embryos, facilitating phenotypic driven genetic screens. Second, the embryonic zebrafish heart is placed at a prominent position at the ventral side of the embryo allowing visual inspection of the patterning and function of the developing heart in live embryos. Furthermore, the zebrafish embryo obtains sufficient oxygenation by passive diffusion making circulation nonessential for the first few days of development providing a unique opportunity to inspect the progression of cardiovascular phenotypes.
A large number of mutations affecting the contractility and/or rhythmicity of the developing zebrafish hearts have been identified [6,7]. Many of these mutants exhibit phenotypic characteristics resembling human heart diseases. Shown in Table 1 are selected zebrafish mutants whose molecular lesions have been determined. Many of these loci encode zebrafish homologues of genes associated with human cardiac diseases. For instance, mutations in essential myosin light chain, regulatory myosin light chain, titin, cardiac troponin T and beta myosin heavy chain have been associated with cardiomyopathy in humans [8–11]. The hearts of zebrafish mutants defective in these genes also show reduced contractility, myofibril disarray and dysmorphic cardiac chambers [12–15], supporting the notion that cellular and molecular mechanisms underlying the contractile machinery are conserved among vertebrates.
Table 1.
Selected zebrafish mutants with cardiac contractility and/or rhythmic defects
| Zebrafish mutant | Gene | Cardiac Phenotype | Ref |
|---|---|---|---|
| Contractility | |||
| tell tale heart | cardiac myosin light chain 2 | Weak or no contractility; loss of myofibril assembly; extended AV canal region | 8 |
| pickwick | titin | Poor contractility; disrupted sarcomeres | 11 |
| silent heart | cardiac troponin T | Non-contractile heart; disorganized sarcomeres | 12 |
| weak atrium | atrial myosin heavy chain | Weak atrial contractility; disorganized myofibrils; thickening of ventricular wall | 13 |
| T2EGEZ8 | tropomyosin4 | Non-contractile heart; disorganized sarcomeres | 50 |
| cardiofunk | actin, alpha, cardiac muscle 1 | Chamber dilation; blood regurgitation | 51 |
| main squeeze/lost contact | integrin-like kinase | Deterioration contractility; blood vessel dilation | 24,25 |
| dead beat | phospholipase C γ1 | Deterioration ventricular contractility; lacks lumenized vasculature | 52 |
| Rhythmicity | |||
| island beat | L-type calcium channel subunit α1C | Uncoordinated contraction | 15 |
| tremblor | Na/Ca exchanger 1 | Disorganized myofibrils; uncoordinated contraction | 16,17 |
| heart-and-mind | Na/K ATPase α1B1 | Dysmorphic heart tube; uncoordinated contraction | 18 |
| breakdance | kcnh2 | 2:1 rhythm (loss-of-function) | 19,20,42 |
| reggae | kcnh2 | Intermittent cardiac arrest (overactivation) | 21 |
| futka | connexin 36.7/early cardiac connexin | Defective sarcomeres; arrhythmia | 53 |
| santa | cerebral cavernous malformation 1 | Loss of concentric growth of ventricle; absence of AV conduction delay | 3,54 |
| valentine | cerebral cavernous malformation 2 | Loss of concentric growth of ventricle; absence of AV conduction delay | 3,54 |
| slipjig | foxn4 | Failure to loop; lacks AV conduction delay | 26 |
| hobgoblin | tcf2 | AV block | 3 |
In addition to the contractile machinery, mechanisms regulating cardiac rhythmicity appear to be conserved from fish to mammals. As in the mammalian heart, genes regulating ion flux are required for establishing and maintaining coordinated cardiac contraction. Mutations in human cacna1c have been associated with Timothy Syndrome, a severe arrhythmic disorder [17]. The heart of zebrafish embryos mutated in L-type calcium channel subunit α1C (cacna1c) never initiate coordinated contraction, rather individual cardiomyocytes contract spontaneously, which is consistent with the role of L-type calcium channels in calcium influx [16]. In addition, the zebrafish tremblor locus encodes the zebrafish homologue of a cardiac-specific Na/Ca exchanger 1 (NCX1h, also known as slc8a1a) gene. Loss of function of NCX1h disrupts normal calcium transients and leads to a chaotic movement of the heart resembling cardiac fibrillation in humans [18,19]. The Ca2+ extrusion activity of NCX1h is modulated by the cellular concentration of Na+ regulated by the sodium pump. Interestingly, zebrafish embryos deficient in Na,K-ATPase α1b1 also develop cardiac fibrillation [20], further indicating the critical role of calcium homeostasis in regulating cardiac rhythmicity. Furthermore, multiple mutations in the ether-a-go-go-related gene, KCNH2, were identified in zebrafish. Zebrafish embryos carrying loss-of-function mutations of KCNH2 have a prolonged ventricular depolarization whereas cardiac repolarization is accelerated in those embryos homozygous for an activated form of KCNH2 [21–23]. These phenotypes are consistent with the finding that loss of KCNH2 activity causes long QT syndrome, and overactivation of KCNH2 causes short QT syndrome in humans [24,25].
The conserved molecular mechanisms underlying cardiac contraction and rhythm and the phenotypic similarity between zebrafish cardiac mutants and human diseases suggest that zebrafish could serve as a genetic model for discovering genes associated with cardiac diseases. This notion is supported by recent studies on integrin-linked kinase (ILK). ILK is a key transducer of biochemical signals initiated at the plasma membrane by cell-matrix interactions including the interaction between laminin and integrin. Zebrafish ilk mutant hearts have severe myocardial dysfunction resembling human dilated cardiomyopathy (DCM) [26,27]. Morpholino knockdown of laminin α4 (lama4) also results in cardiac dysfunction similar to that observed in ilk mutants, suggesting that ILK mediated cell-matrix signaling has an essential role in regulating cardiac growth and contractility. These findings led Knoll et al. to screen patients with severe DCM and found two mutations in LAMA4 and a mutation in the coding region of ILK [27]. While the causative relationship between mutations in the ILK/laminin signaling pathway and DCM warrant further investigation, these findings suggest that zebrafish may serve as a tool for discovering genetic basis of human cardiac diseases. Recently, molecular cloning of zebrafish mutants with atrioventricular block revealed the roles of transcription factors such as foxn4 and tcf2 in cardiac conduction [28,3]. Whether mutations in these genes also cause similar cardiac defects in mammalian hearts is currently unknown. Future studies investigating the role of genes identified from zebrafish mutants or morphants in mammals will provide further insight into the similarity or differences in mechanisms regulating cardiac function in divergent species and may help discover causes of human cardiovascular development or disease.
Chemical genetic screens in zebrafish
Genetic screens in zebrafish have generated useful resources to dissect genes and pathways critical for heart development. However, if a gene or pathway is involved in multiple biological processes, its cardiovascular role may not be easily evaluated in the mutants due to early developmental defects. One complementary approach to overcome this problem is small molecule-based chemical genetic screens. The zebrafish embryos are fertilized and developed ex utero. Chemical inhibitors or agonists can be applied to embryos at desired developmental stages to evaluate their effects on biological processes of interest. In fact, small molecule screens conducted in zebrafish have already discovered compounds that specifically affect the development of the heart, nervous system, hematopoietic cells and pigment cells (for review see [29]). In addition, screening compounds that suppress or enhance phenotypes in a specific mutant background may identify modifiers and/or signaling pathways critical to the biological processes of interest. This approach has been successfully applied in zebrafish. For example the gridlock (grl) mutant carries a mutation in the zebrafish hey2 homologue and blocks aortic blood flow resembling aortic coarctation in humans [30,31]. Multiple compounds were identified in a chemical suppressor screen by their abilities to restore blood flow to the tail of grl mutants [32,33]. Mechanisms of action studies on these compounds lead to the discovery that grd/hey2 regulates vessel formation through the VEGF signaling pathway and that PI3K and ERK mediating opposing effects in artery/vein specification downstream of VEGF [33]. Our laboratory also identified a novel small molecule that suppresses cardiac fibrillation in tre/slc8a1a mutant embryos using a similar strategy (Huang, Kwon, and Chen, unpublished data), attesting to the feasibility of this approach in zebrafish. Whether these compounds could be developed into new therapeutic agents for cardiovascular disorders remain to be seen, yet the zebrafish system affords a potent means to investigate the genetic and molecular pathways responsible for cardiac function.
The success of chemical genetic screens in zebrafish raises interests of using the zebrafish model to discover pharmaceutical agents regulating cardiac physiology. Milan et al. screened a collection of 100 biologically active small molecules for their effects on zebrafish heart rate and found 36 compounds caused bradycardia [34]. Interestingly, eighteen of these compounds were known to cause QT prolongation and torsades de pointes in humans, supporting the feasibility of using zebrafish as a model organism to evaluate drug effects on the cardiovascular system in vivo. From an independent screen of 238 biologically active small molecules, we identified fifteen compounds that reduce the contractility or interfere with the rhythmic contraction of the developing zebrafish heart. Among these, seven are known modulators of ion channels or downstream signaling pathways in mammals (bepridil, amiodarone, niguldipine, pimozide, penitrem A, fluspirilene, and KN-93) [35–41] consistent with the notion that ion flux is critical in regulating cardiac function. Furthermore, these results offer additional verification that the fish heart responds to pharmacological agents in a similar manner as mammalian models.
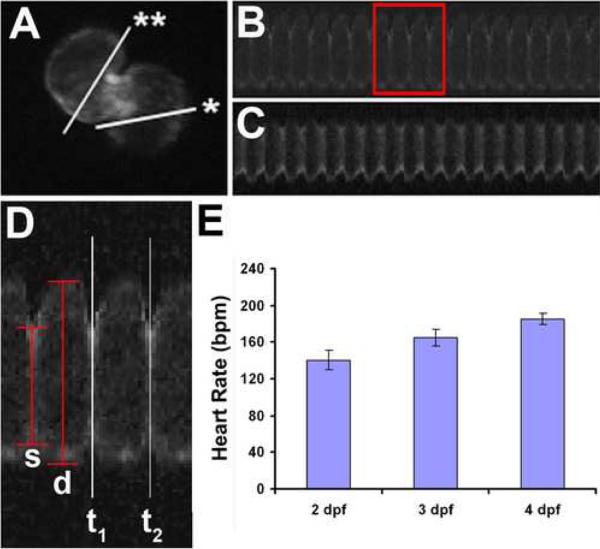
In order to further utilize the zebrafish model to discover pharmaceutical targets regulating cardiac physiology, easily accessible, high-throughput quantitative tools are required to characterize cardiac abnormalities at functional and physiological levels. Over the years, many programs designed for quantifying cardiac contraction have been developed [42–49], including a recent effort to monitor heart rate using an automated system [50]. In addition, we developed a line-scan based methodology allowing a low-cost, high-throughput quantitative means to measure cardiac function parameters [51]. Hereafter, we will refer to this system as zCAS for zebrafish cardiac analysis system. Similar to 2D-echocardiography performed on patients, zCAS produces two-dimensional images of each cardiac chamber from which cardiac function can be assessed. In this system, we designed a custom algorithm to evaluate footage of heartbeats obtained from live embryos and to extract cardiac parameters for physiological analysis. Embryos used for zCAS analysis carry the cmlc2:GFP transgene that drives the expression of the green fluorescent protein (GFP) specifically in cardiomyocytes. As shown in Fig.1, the cardiac chambers of Tg(cmlc2:GFP) embryos are marked by GFP expression. Once a region of the heart is defined, the zCAS program generates a contraction profile to facilitate the quantitative measurement of heart rate and diastolic and systolic widths of the atrium and ventricle (Figure 1C,D). Cardiac parameters such as shortening fraction (SF), ejection fraction, stroke volume and cardiac output are then derived from these values to assess cardiac function. A steady increase in the heart rate of 2-, 3-, and 4-day embryos using zCAS is consistent with measurements previously reported, verifying the validity of zCAS (Figure 1E) [50,52].
Figure 1.
zCAS analysis for embryonic fish cardiac function. (A) Tg(cmlc2:GFP) heart of 2 dpf zebrafish embryo. Asterisk and double asterisk mark regions of the atrium and ventricle scanned. (B) Representative atrial cardiac contraction profile. (C) Representative ventricular cardiac contraction profile. (D) Magnified view of atrial contraction from (B). Heart rate determined by measuring time between two systolic phases, t1 and t2. D: end-diastolic diameter, s: end-systolic diameter. (E) Heart rate measured by the zCAS program during fish development. dpf: days post fertilization.
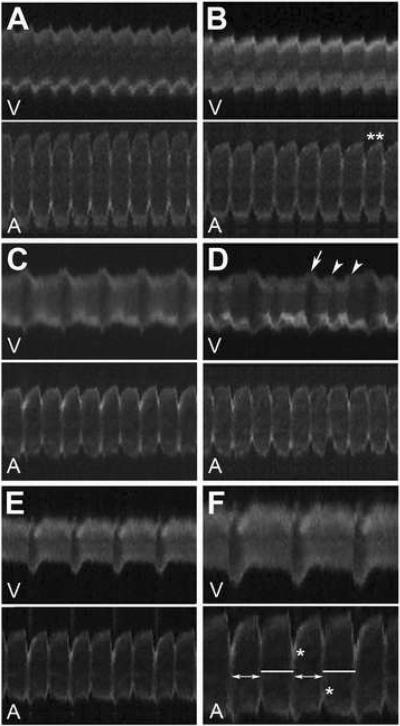
As described above, we have found that treatments of amiodarone, fluspirilene, niguldipine, and pimozide disrupt normal cardiac contraction. We used zCAS to further evaluate the impacts of these compounds on the developing zebrafish heart. We found that embryos treated with pimozide and niguldipine displayed dilated atria and nearly silent ventricles two days after fertilization, and the hearts became silent by three days of development (n= 72/75 and 78/80, respectively; Fig. 2A–B). The SF of the atria of these embryos are not significantly different from untreated siblings, whereas the ventricular SF of embryos treated with pimozide or niguldipine are reduced to 5.6% and 10% of the levels of the untreated embryos, respectively, indicating that pimozide and niguldipine have more profound effects on ventricular contractility.
Figure 2.
Cardiac contraction profiles of chemically-treated Tg(cmlc2:GFP) zebrafish embryos at 2 dpf. (A) Niguldipine-treated embryos; (B) Pimozide-treated embryos, double asterisk (**) mark anomaly in interval between contractions; (C, D) Amiodarone-treated, white arrow and arrowheads mark phases of major and minor relaxation, respectively; (E, F) Fluspirilene-treated, lines and double headed arrows denote relaxation periods alternating between 0.63 and 0.525 s, respectively. Maximum atrial wall contraction is marked by an asterisk (*). V: ventricle line scan; A: atrial line scan.
Treatment of fluspirilene and amiodarone on the other hand, caused cardiac arrhythmia. Most of amiodarone- and fluspirilene-treated embryos (n = 39/78 and 43/59, respectively) displayed a 2:1 AV block, a phenotype clearly noted by visual inspection. Interestingly, cardiac contraction profiles produced by zCAS uncovered other subtle defects. The atria of fluspirilene treated embryos displayed an alternating beat-by-beat rate (between 0.525 seconds/beat and 0.63 seconds/beat) resembling alternans in mammals (Fig. 2E–F). In addition, the atria of amiodarone- and fluspirilene-treated embryos exhibited an alternating contracting force of the left and right walls, where a contraction of the more forceful left atrial wall is followed by a contraction of a more forceful right wall (Fig. 2C–D, F). Furthermore, zCAS analysis revealed minor relaxation periods between each ventricular beat in amiodarone-treated embryos using zCAS (Figure 2D). These observations demonstrate that zCAS is a sensitive and reliable high throughput tool, which can assist in evaluating the physiological impact of genetic mutations and small molecules on the embryonic heart.
Conclusion
Cardiovascular studies in zebrafish have primarily focused on the embryonic heart. Studies using zebrafish cardiac mutants and morphants have revealed insights into cellular and molecular mechanisms regulating embryonic cardiac contractility and rhythmicity. The ease of conducting chemical genetics screens in zebrafish, together with the advances in analytical technology such as zCAS, provide a cost-effective and reliable means for determining the effects of small molecules on the cardiovascular system. Success in combining chemical genetics with zebrafish mutants and/or morphants facilitates modifier screens for specific cardiovascular phenotypes and demonstrates the feasibility of using this approach to elucidate molecular pathways critical for regulating cardiovascular functions as well as to discover new therapeutic agents. One remaining question is whether the fish model could be utilized for investigating adult cardiac disorders. Recent studies showed that adult fish heterozygous for the KCNH2 mutation have prolonged QT interval similar to the dominant trait observed in humans [21], suggesting that the study of adult heterozygous fish may afford new insight into the progression of cardiac disease. Developing technologies required for imaging and physiological studies in adult fish will greatly facilitate these lines of study.
Acknowledgement
We thank members of the Chen Lab for discussion, Q. Lu and Y. Wang for discussion and developing zCAS algorithm and J. Saxe and J. Huang for small molecule libraries. C.T.N. was supported by a predoctoral fellowship from the Training Grant in Genetic Mechanisms. This work is supported in part by grants from NIH and AHA to J.-N. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yelon D. Cardiac patterning and morphogenesis in zebrafish. Dev. Dyn. 2001;222:552–563. doi: 10.1002/dvdy.1243. [DOI] [PubMed] [Google Scholar]
- 2.Scherz PJ, et al. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–1187. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 3.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:1006–1019. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Milan DJ, et al. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133:1135–1132. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 5.Sedmera D, et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H1152–H1160. doi: 10.1152/ajpheart.00870.2002. [DOI] [PubMed] [Google Scholar]
- 6.Stainer DYR, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 7.Chen JN, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 8.Poetter K, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat. Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 9.Gerull B, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat. Genet. 2003;30:201–204. doi: 10.1038/ng815. [DOI] [PubMed] [Google Scholar]
- 10.Theirfelder L, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 11.Klassan S, et al. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 12.Rottbauer W, et al. Cardiac myosin light chain-2: A novel essential component of thick-myofilament assembly and contractility of the heart. Circ. Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 14.Sehnert AJ, et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 15.Berdougo E, et al. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- 16.Rottbauer W, et al. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel α1 subunit. Dev. Cell. 2001;1:265–275. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 17.Spawlski I, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc. Natl. Acad. Sci. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langenbacher AD, et al. Mutation in sodium-calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc. Natl. Acad. Sci. 2005;102:17699–17704. doi: 10.1073/pnas.0502679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebert AM, et al. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc. Natl. Acad. Sci. 2005;102:17705–17710. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu X, et al. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- 21.Langheinrich U, et al. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol. Appl. Pharmacol. 2003;193:370–382. doi: 10.1016/j.taap.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Arnaout R, et al. Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassel D, et al. Deficient zebrafish ether-a-go-go related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation. 2008;117:866–875. doi: 10.1161/CIRCULATIONAHA.107.752220. [DOI] [PubMed] [Google Scholar]
- 24.Splawski I, et al. Spectrum of mutations in long-QT syndrome genes: KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 25.Schimpf R, et al. Short QT syndrome. Cardiovasc. Res. 2005;67:357–366. doi: 10.1016/j.cardiores.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 26.Bendig G, et al. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes & Dev. 2006;20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoll R, et al. Laminin-α4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 28.Chi NC, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes & Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zon LI, Peterson RI. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 30.Weinstein BM, et al. Gridlock, a localized heritable vascular patterning defect in zebrafish. Nat. Med. 1995;1:1143–1147. doi: 10.1038/nm1195-1143. [DOI] [PubMed] [Google Scholar]
- 31.Zhong TP, et al. Gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 32.Peterson RT, et al. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 33.Hong CC, et al. Artery/vein specification is governed by opposing phosphatidylinositol-3 kinase and MAP kinase/ERK signaling. Curr. Biol. 2006;16:1366–1372. doi: 10.1016/j.cub.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milan DJ, et al. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107:1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 35.Gill A, et al. Pharmacology of bepridil. Am. J. Cardiol. 1992;69:11D–16D. doi: 10.1016/0002-9149(92)90953-v. [DOI] [PubMed] [Google Scholar]
- 36.Kodama I, et al. Amiodarone: ionic and cellular mechanisms of action of the most promising class III agent. Am. J. Cardiol. 1999;84:20R–28R. doi: 10.1016/s0002-9149(99)00698-0. [DOI] [PubMed] [Google Scholar]
- 37.Handrock R, Herzig S. Stereoselectivity of Ca2+ channel block by dihydropyridines: no modulation by the voltage protocol. Eur. J. Pharmacol. 1996;309:317–321. doi: 10.1016/0014-2999(96)00465-7. [DOI] [PubMed] [Google Scholar]
- 38.Enyeart JJ, et al. Antipsychotic pimozide is a potent Ca2+ channel blocker in heart. Mol. Pharmacol. 1990;26:16373–16379. [PubMed] [Google Scholar]
- 39.Knaus H-G, et al. Tremorgenic indole alkaloids potently inhibit smooth muscle high-conductance calcium-activated potassium channels. Biochemistry. 1994;33:5819–5828. doi: 10.1021/bi00185a021. [DOI] [PubMed] [Google Scholar]
- 40.Galizzi J-P, et al. Neuroleptics of the diphenylbutylpiperidine series are potent calcium channel inhibitors. Proc. Natl. Acad. Sci. 1986;83:7513–7517. doi: 10.1073/pnas.83.19.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson ME, et al. KN-93, an inhibitor of multifunctional Ca++/calmodulin-dependent protein kinase, decreases early afterdepolarizations in rabbit heart. J. Pharmacol. Exp. Ther. 1998;287:996–1006. [PubMed] [Google Scholar]
- 42.Ho YL, et al. Assessment of zebrafish cardiac performance using Doppler echocardiography and power angiography. Ultrasound Med. Biol. 2002;28:1137–1143. doi: 10.1016/s0301-5629(02)00564-1. [DOI] [PubMed] [Google Scholar]
- 43.Jacob E, et al. Influence of hypoxia and of hypoxemia on the development of cardiac activity in zebrafish larvae. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;283:R911–R917. doi: 10.1152/ajpregu.00673.2001. [DOI] [PubMed] [Google Scholar]
- 44.Huisken J, et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 45.Kopp R, et al. Cardiac performance in the zebrafish breakdance mutant. J. Exp. Biol. 2005;208:2123–2134. doi: 10.1242/jeb.01620. [DOI] [PubMed] [Google Scholar]
- 46.Liebling M, et al. Four-dimensional cardiac imaging in live embryos via postacquisition synchronization of nongated slice sequences. J. Biomed. Opt. 2005;10:054001. doi: 10.1117/1.2061567. [DOI] [PubMed] [Google Scholar]
- 47.Forouhar AS, et al. The embryonic vertebrate heart tube is a dynamic suction pump. Science. 2006;312:751–753. doi: 10.1126/science.1123775. [DOI] [PubMed] [Google Scholar]
- 48.Liebling M, et al. Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development. Dev. Dyn. 2006;235:2940–2948. doi: 10.1002/dvdy.20926. [DOI] [PubMed] [Google Scholar]
- 49.Lu J, et al. Three-dimensional real-time imaging of cardiac cell motions in living embryos. J. Biomed. Opt. 2008;13:014006. doi: 10.1117/1.2830824. [DOI] [PubMed] [Google Scholar]
- 50.Burns CG, et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- 51.Lu G, et al. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes & Dev. 2007;21:784–796. doi: 10.1101/gad.1499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y-X, et al. Cardiac neural crest in zebrafish embryos contributes to myocardial cell lineage and early heart function. Dev. Dyn. 2003;226:540–550. doi: 10.1002/dvdy.10264. [DOI] [PubMed] [Google Scholar]
- 50.Zhao L, et al. Heart-specific isoform of tropomyosin4 is essential for heartbeat in zebrafish embryos. Cardiovasc. Res. 2008 doi: 10.1093/cvr/cvn177. Epub 2008 July 14. [DOI] [PubMed] [Google Scholar]
- 51.Bartman T, et al. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2:673–681. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rottbauer W, et al. VEGF-PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes & Dev. 2005;19:1624–1634. doi: 10.1101/gad.1319405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sultana N, et al. Zebrafish early cardiac connexin, Cx36.7/Ecx, regulates myofibril orientation and heart morphogenesis by establishing Nkx2.5 expression. Proc. Natl. Acad. Sci. 2008;105:4763–4768. doi: 10.1073/pnas.0708451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mably JD, et al. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]