Abstract
MicroRNAs (miRNA) are important regulators of gene expression. They are involved in many physiological processes ensuring the cellular homeostasis of human cells. Alterations of the miRNA expression have increasingly been associated with pathophysiologic changes of cancer cells making miRNAs currently to one of the most analyzed molecules in cancer research. Here, we provide an overview of miRNAs in lung cancer. Specifically, we address biological functions of miRNAs in lung cancer cells, miRNA signatures generated from tumor tissue and from patients’ body fluids, the potential of miRNAs as diagnostic and prognostic biomarker for lung cancer, and its role as therapeutic target.
Keywords: microRNA, lung cancer, body fluids, blood, biomarker, diagnosis, prognosis, therapy
Introduction
Besides housekeeping genes, the expression of all other genes is mostly regulated through a complex mechanism that enables a cell type specific and time specific expression. Regulations can occur during each step of gene expression, e.g., during chromatin remodeling, transcription and translation, RNA transport, or on the post-transcriptional level. The main gene expression regulators are proteins or enzymes, e.g., histones, transcription factors, and polymerases. Gene expression can also be regulated by antisense or sense nucleic acids (Helene and Toulme, 1990). MicroRNAs (miRNAs) are a highly conserved family of small RNAs (17–22 nt) that regulate the expression of their target genes usually on the post-transcriptional level by binding to complementary sequences on target messenger RNA transcripts (mRNAs) mostly resulting in gene silencing. Since the first description of miRNAs in 1993 by Victor Ambros, Rosalind Lee, and Rhonda Feinbaum in C. elegans (Lee et al., 1993) more than 1500 different human miRNAs (see miRBase V18, http://www.mirbase.org) have already been identified. As each miRNA can regulate hundreds of target genes, it is assumed that the majority of the 20,000–25,000 human genes may be regulated by specific miRNAs (van Kouwenhove et al., 2011). Silencing of the target genes is obviously the main regulation mechanism – either by translational repression or by mRNA degradation. Perfect matching of the miRNA to the 3′ UTR of its target mRNA results in direct mRNA degradation whereas imperfect matching – with nucleotides 2–7 of the miRNA (called “seed region”) still perfectly complementary – leads to translational repression. Bartel and colleagues analyzed the relative contribution of these two outcomes and found that degradation of the mRNA by miRNAs is with more than 80% of cases the predominant reason for a reduced protein output (Guo et al., 2010). Recently, miRNAs were also shown to up-regulate target gene expression either directly through binding to the target mRNA (Vasudevan et al., 2007) or indirectly through repressing nonsense-mediated RNA decay (Bruno et al., 2011). According to their function miRNAs play an essential role in cellular processes as development, proliferation, and apoptosis ensuring the cellular homeostasis of healthy human cells. An alteration of this cellular homeostasis through aberrant expression of miRNAs likely contributes to many human pathologies including cancer. Calin et al. (2002) revealed for the first time a possible correlation between miRNA deregulation and cancer. Subsequently, a multitude of studies about miRNA expression changes and cancer has been reported.
Lung cancer is worldwide the leading cause of cancer related deaths. The 5-year overall survival rate strongly correlates with the time of diagnosis and varies between 60 and 80% in clinical stage I to only 1% in clinical stage IV. Unfortunately, lung cancer is mostly diagnosed in late stages. Currently, no appropriate biomarker exists to detect lung cancer at early stages. Takamizawa et al. (2004) were the first to relate miRNA expression to lung cancer. Since then the number of publications dealing with the relation between miRNA expression and lung cancer has raised to above 400.
In this review we place emphasis on the current status of miRNA research in lung cancer. Specifically, we focus on current findings on the molecular role of miRNAs in lung cancer development and progression. In addition, we address the potential of miRNA research for tumor diagnosis and therapy.
Detection of miRNAs in Human Samples
Most of the miRNA expression data have been generated by the analysis of tissue samples with the main focus on cancer tissue. The data collected from tissue samples may provide the best insights into the involvement of miRNAs in a disease state. As miRNAs are markedly stable against degradation, stored formalin-fixed paraffin embedded (FFPE) tissue can be used for miRNA isolation (Liu and Xu, 2011). Lu et al. (2005) showed that tissue miRNA expression profiles are highly cell type specific and that they reflect the developmental lineage and the differentiation state. MiRNA expression data derived from 40 different tissue samples from healthy individuals revealed both a group of universally expressed miRNAs and groups of tissue specific expressed miRNAs (Liang et al., 2007). Besides tissues, sources for miRNAs can be body fluids such as whole blood, serum, plasma, urine, cerebrospinal fluid (CSF), and – especially in the case of lung cancer research – saliva, sputum, or bronchoalveolar lavage (BAL) (Weber et al., 2010; Tzimagiorgis et al., 2011). MiRNA profiles of body fluids are useful for the analysis of disease states especially when the disease does not originate from one distinct type of cell and when the tissue is not readily accessible, e.g., in neurological disorders (e.g., Schizophrenia, Lai et al., 2011; Alzheimer’s Disease, Schipper et al., 2007), in heart failure (Voellenkle et al., 2010; Meder et al., 2011), in autoimmune diseases (e.g., Lupus, Wang et al., 2011), and in respiratory tract diseases (e.g., COPD, Pottelberge et al., 2011). In general, miRNA profiles of body fluids, including urine (Hanke et al., 2010), serum (Mitchell et al., 2008; Otaegui et al., 2009), saliva (Park et al., 2009), sputum (Xing et al., 2010), CSF (Baraniskin et al., 2011) have been discussed as future non-invasive biomarkers. How miRNAs enter the body fluids is still a largely unsolved question. One possibility is that cancer cells without metastatic potential enter the blood stream and release their cell content including miRNAs after passing through a suicide program (Mehes et al., 2001). Alternatively, miRNAs packed in microvesicles or exosomes are actively released in the bloodstream (Hunter et al., 2008; Rabinowits et al., 2009). Notably, miRNAs measured in body fluids frequently reflect different cell types. For example, urine of a bladder cancer patient contains apoptotic or necrotic cells, non-malignant exfoliated urothelial cells, and leukocytes besides tumor cells (Hanke et al., 2010), all of which may contribute to the miRNA expression profile. Saliva contains blood cells, microorganisms, and apoptotic or detached living epithelial cells. Cell-free nucleic acids actively released by cancer and epithelial cells or inactively by apoptotic cells and micro-wounds have also been found in saliva (Park et al., 2006). Likewise, different cell types contribute to miRNA profiles in sputum (Thunnissen, 2003; Xie et al., 2010; Yu et al., 2010), BAL (Ahrendt et al., 1999; Schmidt et al., 2005), and CSF (Karlsson et al., 2001; Reiber and Peter, 2001).
In conclusion, the measurement of miRNAs in body fluid has high potential for future non-invasive diagnostic tests especially for cancer. There are, however, various hurdles to be overcome to turn a miRNA signature into a diagnostic tool. Among others, the amount of specific miRNAs may be limited in certain body fluids, the availability of body fluid may also be limited, standardized protocols have not yet been established for the isolation and analysis of RNA from body fluids, and detection methods have to be optimized. As for the latter, microarray experiments are both cost-intensive and time-consuming while qRT-PCR lacks reliable endogenous controls for body fluids.
The Molecular Biology of miRNAs in Lung Cancer
The first aberrantly expressed miRNA in lung cancer was identified in 2004 (Takamizawa et al., 2004). By analyzing 143 potentially curative resected lung cancer samples Takamizawa et al. (2004) showed that a reduced let-7 expression is correlated with a shorter post-operative survival. They confirmed their results by introducing let-7 into the adenocarcinoma cell line A549. The observed overexpression resulted in growth inhibition of the cells. These findings laid the basis for further studies on the molecular mechanisms of the tumor suppressor function of let-7. The 3′ UTR of HRAS, KRAS, and NRAS that are members of the RAS GTPase family, contain multiple putative let-7 binding sites. The expression of let-7 in lung cancer was inversely correlated to RAS expression. On the basis of these results Johnson et al. (2005) concluded that let-7 is a negative regulator of the oncogene RAS. Microarray analysis revealed additional genes whose expressions were altered in the presence of excess let-7 (Johnson et al., 2007). These genes include key cell cycle proto-oncogenes such as CDC25a, CDK16, and cyclin D that are involved in the G1/S transition. These findings gave further support to the assumption that let-7 functions as tumor suppressor miRNA. Recently, let-7 was shown to target BCL-2, thereby inhibiting the growth of A549 cells (Xiong et al., 2011). As BCL-2 is a proto-oncogene involved in regulation of apoptosis, a negative regulation through let-7 may result in growth suppression and apoptosis induction of A549 cells. Esquela-Kerscher et al. (2008) confirmed that let-7 reduces in vivo tumor growth of lung cancer cell xenografts in immunodeficient mice.
Hayashita et al. (2005) found an overexpressed intronic miRNA cluster (miR-17-92) encompassing seven different miRNAs namely hsa-miR-17-5p, hsa-miR-17-3p, hsa-miR-18a, hsa-miR-19a, hsa-miR-19b-1, hsa-miR-20a, and hsa-miR-92 in the amplified chromosomal region 13q31.3 in lung cancer, mostly in small cell lung cancer. This polycistronic miRNA cluster was first described by He et al. (2005) in B-cell lymphomas. Antisense oligonucleotides against mir-17-5p and miR-20a were shown to induce apoptosis in mir-17-92 overexpressing lung cancer cells (Matsubara et al., 2007). Recently, Kanzaki et al. (2011) were able to identify several direct targets of the miR-17-92 oncogene. A summary of the various roles of the miR-17-92 cluster was given by Joshua T. Mendell in Cell (Mendell, 2008).
Besides hsa-let-7 and the miRNAs of the miR-17-92 cluster, there are numerous reports on other miRNAs that are deregulated in lung cancer tissue, e.g., hsa-miR-21 whose overexpression was suggested to be an independent negative prognostic factor for the overall survival in NSCLC patients (Markou et al., 2008; Gao et al., 2010). Hsa-miR-21 targets tumor suppressor genes such as programmed cell death 4 (Pdcd4; Lu et al., 2008) and PTEN (Zhang et al., 2010). There is evidence that the expression of miRNA-21 is up regulated by epidermal growth factor receptor (EGFR)-signaling in lung cancer (Seike et al., 2009). In 15% of all lung cancer patients, mostly never-smokers, EGFR contained a mutation resulting in constitutive activation of tyrosine kinase (TK), which in turn leaded to tumor progression (da Cunha Santos et al., 2011). The inhibition of the EGFR signaling by a tyrosine kinase inhibitor (TKI) resulted in a reduced expression of miR-21 (Seike et al., 2009). But, since miR-21 is also deregulated in several other cancer types, it seems to be a general oncogenic miRNA without tissue specificity (Ciafre et al., 2005; Volinia et al., 2006; Iorio et al., 2007; Meng et al., 2007).
Impact of miRNA Research on Clinical Oncology
The 5-year survival of lung cancer patients is 15% for all stages combined. Early detection of lung cancer in high-risk patients is likely to improve the prognosis. Currently, less than 20% of lung cancer patients are diagnosed with a locally confined tumor (Jemal et al., 2009). This low detection rate calls for the identification of new reliable biomarkers to allow non-invasive early detection of locally confined lung cancers. The markers should also contribute to the distinction between benign and malignant lesions.
MicroRNAs play an essential role in lung development (Tomankova et al., 2010). Due to the different expression pattern in healthy lung tissue compared to lung cancer tissue it seemed legitimate to assume that aberrant miRNA expression may be involved in the onset of lung cancer (Mascaux et al., 2009; Megiorni et al., 2011). By microarray analyses of the miRNA expression in 104 pairs of primary lung cancers and corresponding non-cancerous lung tissues Yanaihara et al. (2006) identified a specific miRNA profile, encompassing 43 differentially expressed miRNAs. Volinia et al. (2006) performed a large-scale analysis of the miRNA profiles of 540 samples, encompassing 363 samples from patients with six different types of solid tumors including lung cancer and 177 normal tissue samples. They identified a cancer miRNA signature with mostly overexpressed miRNAs. Besides the identification of cancer type specific miRNA signatures, research is also aiming at the identification of specific miRNAs that are suited to differentiate between histological lung cancer subclasses. As treatment depends on the histological subtype, such miRNAs are likely to be useful for decision-making in clinical treatment. Lebanony et al. (2009) were able to provide a highly accurate subclassification of NSCLC patients. They identified miR-205 as suitable marker for squamous cell lung carcinoma by comparing the miRNA expression pattern between 122 adenocarcinoma and squamous NSCLC samples (Lebanony et al., 2009). MiRNA signatures also appeared suitable to distinguish SCLC cells from NCLC cells (Du et al., 2010). Vosa et al. (2011) provided evidence for miR-374 as a potential marker for early stage NSCLC. Recently, different groups were able to identify miRNAs that differentiated NSCLC patients with brain metastases from patients without brain metastases (Arora et al., 2011; Nasser et al., 2011). Biomarkers that allow identification of NSCLC patients with increased risk for brain metastases will be of great value for the decision-making in preventive radiation treatment.
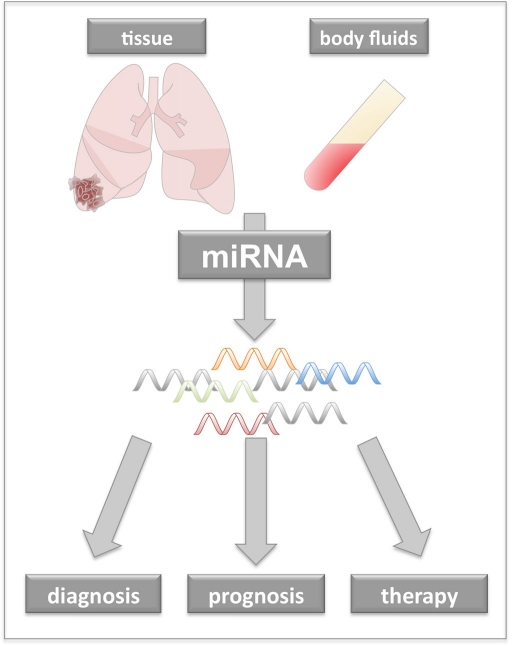
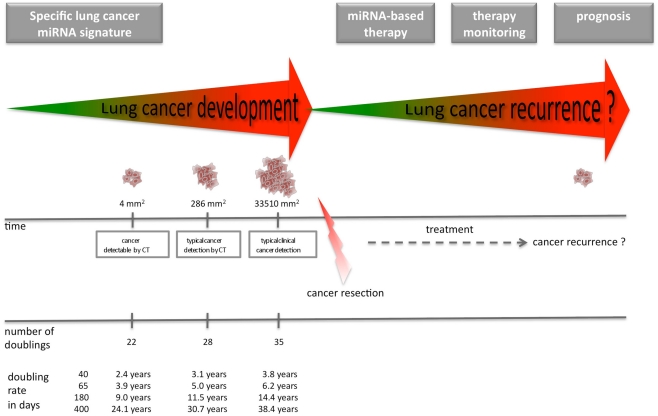
As miRNAs are very stable not only in tissue but also in body fluids, they offer themselves as potential biomarker for non-invasive early detection of lung cancer. Especially, for lung cancer the poor survival time and the high relapse rates after surgery call for new methods to detect the disease at early stage. MiRNA expression patterns have also the potential to be a useful prognostic tool. In addition, there is growing interest to use miRNAs as therapeutic agent. Especially the increasing knowledge about the role of miRNAs as tumor suppressors or activators of oncogenes, will help to develop novel miRNA-based therapeutic approaches. Figure 1 provides an overview about potential different clinical applications of miRNAs in oncology. Figure 2 gives an overview of the potential time points for the application of lung cancer specific miRNAs. Table A1 in Appendix provides information on miRNAs associated with lung cancer.
Figure 1.
The impact of miRNA research on clinical oncology. Alterations in miRNA expression are associated with pathophysiological changes in cancer cells. As miRNA signatures from cancer tissue or patients’ body fluids differ from those of healthy individuals miRNA signatures may likely to contribute to improved early diagnosis or patients’ prognosis. Through altered expression single miRNAs may act as oncogene or they may loose their tumor suppressor properties. Those miRNAs could be utilized for anti-cancer therapy.
Figure 2.
Time points for the application of lung cancer specific miRNAs. Specific tumor-associated miRNA signatures might detect the tumor prior to CT. After cancer resection and/or therapy (radiotherapy or chemotherapy) miRNAs might help in therapy monitoring and prognosis. MiRNAs may also be used for anti-cancer therapy instead of or in combination with radiotherapy or chemotherapy. The timeline at the bottom of the Figure shows the growths rate of a lung tumor with several doubling rates (adapted from Bach et al., 2007).
miRNAs as potential diagnostic biomarker for lung cancer
It is well known that the onset of cancer impacts the immune system leading to changes in the gene expression of blood cells (Pardoll, 2003; Kossenkov et al., 2011). Jeong et al. (2011) showed that the expression of let-7a is reduced not only in lung cancer tissue, but also in blood of lung cancer patients compared to healthy individuals. In our recent studies, we were able to separate blood samples of lung cancer patients from blood samples of healthy individuals by miRNA signatures with a specificity of 98.1% and a sensitivity of 92.5% (Keller et al., 2009). In addition we reported miRNA signatures that differentiated blood samples of lung cancer patients from blood samples of patients with non-malignant chronic obstructive pulmonary disease with 89.2% specificity, and 91.7% sensitivity (Leidinger et al., 2011). Recently, we showed in a multicenter study that different types of cancer or non-cancer diseases could be differentiated by blood-borne miRNA profiles (Keller et al., 2011a).
As above mentioned, miRNAs are also present in other body fluids. Yu et al. (2010) showed that miRNAs were stably present in sputum. They were able to differentiate lung adenocarcinoma patients from healthy individuals by using a panel of four sputum miRNAs namely miR-486, miR-21, miR-200b, and miR-375, with high sensitivity (80.6%) and specificity (91.7%; Yu et al., 2010). The same group identified three sputum miRNAs, namely miR-205, miR-210, and miR-708 that distinguished squamous cell lung carcinoma patients from healthy individuals with 73% sensitivity and 96% specificity (Xing et al., 2010).
Since the first study that demonstrated larger amounts of stable miRNAs in serum and plasma, several studies proved that the serum or plasma miRNAs show great promise as novel non-invasive biomarkers for the early diagnosis of various cancers and other diseases (Chen et al., 2008; Mitchell et al., 2008). Chen et al. (2011) identified in a genome-wide serum miRNA expression study a specific panel of 10 miRNAs that was able to distinguish NSCLC cases from controls with high sensitivity and specificity and that correlated with the stage of NSCLC. Furthermore, this 10-serum miRNA profile could accurately classify serum samples collected up to 3 years prior to the clinical NSCLC diagnosis. By expression analysis of two serum miRNAs (hsa-miR-1254 and hsa-miR-574-5p), Foss et al. (2011) were able to discriminate early stage NSCLC samples from controls with a sensitivity of 82% and a specificity of 77% in a training cohort and with a sensitivity of 73% and a specificity of 71% in a validation cohort. Shen et al. (2011) recently identified a panel of four miRNAs namely miR-21, miR-126, miR-210, and miR-486-5p, that distinguished NSCLC patients from the healthy controls with 86.22% sensitivity and 96.55% specificity. Furthermore, the panel of miRNAs identified stage I NSCLC patients with 73.33% sensitivity and 96.55% specificity. Interestingly, two of these miRNAs, namely miR-21 and miR-486, show an overlap with the study on sputum by Yu et al. (2010). Rabinowits et al. (2009) investigated the expression of 12 specific miRNAs including hsa-miR-17-3p, hsa-miR-21, hsa-miR-106a, hsa-miR-146, hsa-miR-155, hsa-miR-191, hsa-miR-192, hsa-miR-203, hsa-miR-205, hsa-miR-210, hsa-miR-212, and hsa-miR-214, in circulating exosomes. The authors suggest that circulating exosomal miRNA might be useful in a screening test for lung adenocarcinoma. In a recent study, we reported serum miRNA profiles as a non-invasive method to detect lung cancer at an early stage (Keller et al., 2011b). We analyzed miRNA signatures in serum from lung cancer patient samples, which were collected prior and after diagnosis. We found that most obvious changes in miRNA expression profiles occur at a time close to diagnosis possibly indicating increased tumor development. Likewise, Boeri et al. (2011) were able to predict lung cancer in plasma samples 1–2 years prior to diagnosis using CT. For the time being, however, the source of circulating miRNAs is elusive. As indicated above it has been suggested that they are released due to apoptosis or active exocytosis processes (Kosaka et al., 2010). This hypothesis is supported by the study by Rabinowits et al. (2009) that showed a similarity between the circulating exosomal miRNA and the lung tumor-derived miRNA patterns. In contrast, miRNAs deregulated in lung cancer tissue were rarely detected in plasma samples from lung cancer patients in the study of Boeri et al. (2011) that compared the expression of deregulated miRNAs in lung cancer tissue with the expression in plasma specimens. To draw further conclusions about the relationship between miRNA profiles in body fluids and the tumor, more insight is required into both the exact role of miRNAs as molecular regulators in tumor cells and the mechanisms underlying the release of miRNA into body fluids.
miRNAs as potential prognostic biomarker for lung cancer
Various up- and downregulated miRNAs have been associated with patients’ survival. Vosa et al. (2011) showed that low expression of miR-374 could be associated with patients’ low survival time. Yanaihara et al. (2006) demonstrated that high expression of hsa-miR-155 and low expression of hsa-let-7a-2 were associated with lung cancer patients’ poor survival time. Raponi et al. (2009) showed that miR-146b was associated with reduced survival time in squamous cell carcinoma tissues (SCC). Recently, a study by Landi et al. (2010) presented a miRNA signature including let-7e, miR-34a, miR-34c-5p, miR-25, and miR-191, which was associated with a prognosis of poor survival among male smokers suffering from stage I to IIIa SCC. A miR-21 overexpression in NSCLC that was detected by Markou et al. (2008) has been suggested as future negative prognostic factor. Hu et al. (2010) analyzed miRNA expression profiles in sera of 303 patients with stage I to IIIA NSCLC. They detected miRNA levels altered between patients with shorter survival and longer survival time. Moreover, their results revealed that four miRNAs, including miR-486, miR-30d, miR-1, and miR-499 correlated with overall survival. Furthermore, increased levels of miR-25 and miR-223 in serum may in the future serve as potential markers for NSCLC (Chen et al., 2008). By using microarray, Liu et al. (2011) analyzed the miRNA expression of six paired lung cancer and normal tissues and identified three differentially expressed miRNAs namely miR-21, miR-141, and miR-200c. High expressions of miR-21 and miR-200c in the tumor and of miR-21 in serum were associated with a poor survival in NSCLC patients (Liu et al., 2011).
Although the above studies clearly show that miRNA expression can be associated with patients’ survival, the specific miRNAs associated with patient survival differ substantially between studies. These discrepancies are due to (i) different sources of miRNA, i.e., tissue or body fluid, (ii) different methods applied for miRNA analysis, i.e., microarray, qRT-PCR, and deep sequencing, (iii) varying numbers of analyzed miRNAs, and (iv) the criteria used to select patient cohorts, i.e., ethnicity, clinicopathologic features, and therapy. Future studies of miRNA signatures should benefit from largely standardized protocols.
miRNAs as potential therapeutic agent
An increasing number of studies examined the therapeutic potential of miRNAs. Major emphasis is given to the analysis of hsa-let-7, which was the first miRNA associated with lung cancer development (Takamizawa et al., 2004; Johnson et al., 2005, 2007). Exogenous delivery of let-7 inhibited lung cancer growth both in mouse models and in human lung cancer cell lines (Esquela-Kerscher et al., 2008; Trang et al., 2010). As for the therapeutic potential of other miRNAs, a very recent study by Frezzetti et al. (2011) showed that upregulation of miR-21 is controlled by the oncogene RAS and that a locked nucleic acid (LNA) against miR-21 decelerates tumor growth in mice. MiRNA-145 that is known to function as tumor suppressor in several types of cancer (Akao et al., 2007; Porkka et al., 2007; Nam et al., 2008) was recently shown to inhibit cell proliferation in NSCLC cells. Specifically, exogenous miRNA-145 inhibited cell proliferation in NSCLC cells by targeting Myc (Chen et al., 2010). MiR-93, miR-98, and miR-197 all of which are overexpressed in lung cancer, interact with the 3′ UTR of the tumor suppressor gene FUS1. This interaction results in a downregulation of the protein expression making these miRNAs crucial for tumor progression (Du et al., 2009). MiR-128b was shown to directly regulate EGFR in NSCLC cell lines. In addition, loss of heterozygosity of miR-128b was associated with both the clinical response and patients’ survival (Weiss et al., 2008). Overexpression of miR-192, which is weakly expressed in lung cancer, resulted in decreased retinoblastoma 1 (rb1) mRNA and Rb1 protein expression. These data indicate that miR-192 induces cell apoptosis through the caspase pathway (Feng et al., 2011).
To fully benefit from the large number of miRNAs and their potential targets, computational algorithms like PicTar, miRanda, and Target Scan have been developed to detect adequate miRNA targets (Sethupathy et al., 2006; Backes et al., 2010). In addition, suitable delivery systems are being developed to transport miRNAs or antagomirs – specific oligomers used as miRNA antagonists – to their potential targets (Landen et al., 2005; Shahzad et al., 2011). Antagomirs have been shown to reduce levels of corresponding miRNAs in vivo (Krutzfeldt et al., 2005). Using a lipid-based delivery system and chemically synthesized miR-34a, Wiggins et al. (2010) demonstrated growth inhibition of subcutaneous NSCLC cells in mice. The same group used this delivery system in Kras-activated autochthonous mouse models of NSCLC. They found that systemic application of complexes consisting of synthetic miRNA-mimics for let-7 and miR-34a and of the neutral lipid emulsion are preferentially directed to tumor sites and significantly decreased tumor burden (Trang et al., 2011). Most recently, a cationic lipid-based miRNA system was used to condense miRNA miR-133b to form lipoplexes in order to enhance cellular uptake and pharmacological effectiveness in vitro and in vivo (Wu et al., 2011). As a tumor suppressor that directly targets the pro-survival gene MCL-1, hsa-miR-133b seems to be a potential therapeutic target to influence both cell survival and sensitivity of lung cancer cells to chemotherapeutic agents (Wu et al., 2011). Although these examples underline the therapeutic perspectives of miRNAs, several challenges have to be addressed on the road toward clinical application. First, delivery systems without toxic side effects are required to effectively and selectively transport miRNA-based therapeutics to the tumor site. As of now, lipid-based carrier (Wiggins et al., 2010), nanoparticles (Shi et al., 2011) or viral delivery systems (Zhang et al., 2006), have in vitro or in vivo been investigated for cancer therapy. Second, the ability of a single miRNA to regulate of up to several 100 targets genes complicates specific targeting and might readily result in unspecific effects. For further information on the therapeutic potential of miRNAs we would like to refer the reader to the reviews of Kasinski and Slack (2011) in Nature Reviews and McDermott et al. (2011) in Pharmaceutical Research, which provide a comprehensive summary of the current in vivo and/or in vitro studies.
Conclusion
Since their discovery in the 1990s miRNAs have increasingly been recognized as significant not only for the understanding of cancer growth and progression, but also as potential cancer biomarkers. Lung cancer as a disease with poor prognosis and a death toll of thousands of cases per year is one of the prime cancers that call for new markers to allowing early diagnosis. As summarized there is considerable progress in developing miRNA signatures into new biomarkers for lung cancer. MiRNA signatures are likely to contribute to improved early diagnosis, patients’ prognosis, or anti-cancer therapy. It is to be, however, recognized, that the overwhelming majority of studies is still in the field of basic science and numerous hurdles need to be overcome before any of the miRNA-based approaches can be introduced in clinical practice. As for cancer detection, the major challenge will be the implementation of standardized protocols for the isolation and the analysis of miRNAs. For cancer therapy, robust and specific delivery systems have to be developed for the transport of miRNAs to the tumor site. Finally, both cancer detection and cancer therapy will greatly benefit from a better understanding of the biological role of miRNAs in cancer cells.
Conflict of Interest Statement
Siemens Healthcare employs Andreas Keller.
Acknowledgments
Funding was obtained from Hedwig Stalter foundation, Homburger Forschungsförderungsprogramm (HOMFOR), and Deutsche Forschungsgemeinschaft (DFG).
Appendix
Table A1.
Summary of miRNAs associated with lung cancer.
| miRNA | miRNA function in lung neoplasia | Reference |
|---|---|---|
| hsa-mir-125b-1 | Deletion | Calin et al. (2004) |
| hsa-let-7a-1 | Reduced expression, decreased abundance; negatively regulate the RAS gene | Takamizawa et al. (2004) |
| hsa-let-7a-2 | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7a-3 | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7b | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7c | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7d | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7e | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7f-1 | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7f-2 | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7g | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-let-7i | Reduced expression, decreased abundance; negatively regulate the RAS gene | |
| hsa-mir-17 | Overexpressed | Hayashita et al. (2005) |
| hsa-mir-18a | Overexpressed | |
| hsa-mir-19a | Overexpressed | |
| hsa-mir-19b-1 | Overexpressed | |
| hsa-mir-20a | Overexpressed | |
| hsa-mir-92a-1 | Overexpressed | |
| hsa-mir-128b | Overexpressed | Volinia et al. (2006) |
| hsa-mir-155 | Overexpressed | |
| hsa-mir-17 | Overexpressed | |
| hsa-mir-191 | Overexpressed | |
| hsa-mir-199a-1 | Overexpressed | |
| hsa-mir-21 | Overexpressed | |
| hsa-mir-101-1 | Downregulation | Yanaihara et al. (2006) |
| hsa-mir-106a | Upregulation | |
| hsa-mir-124a-1 | Downregulation | |
| hsa-mir-124a-3 | Downregulation | |
| hsa-mir-125a-precursor | Downregulation | |
| hsa-mir-125a | Downregulation | |
| hsa-mir-126* | Downregulation | |
| hsa-mir-126 | Downregulation | |
| hsa-mir-140 | Downregulation | |
| hsa-mir-143 | Downregulation | |
| hsa-mir-145 | Downregulation | |
| hsa-mir-146 | Upregulation | |
| hsa-mir-150 | Upregulation | |
| hsa-mir-155 | Upregulation | |
| hsa-mir-17-3p | Upregulation | |
| hsa-mir-181c-precursor | Downregulation | |
| hsa-mir-191 | Upregulation | |
| hsa-mir-192-precursor | Downregulation | |
| hsa-mir-192 | Upregulation | |
| hsa-mir-197 | Upregulaion | |
| hsa-mir-198 | Downregulation | |
| hsa-mir-199b-precursor | Downregulation | |
| hsa-mir-203 | Upregulation | |
| hsa-mir-205 | Upregulation | |
| hsa-mir-21 | Upregulation | |
| hsa-mir-210 | Upregulation | |
| hsa-mir-212 | Upregulation | |
| hsa-mir-214 | Upregulation | |
| hsa-mir-216-precursor | Downregulation | |
| hsa-mir-218-2 | Downregulation | |
| hsa-mir-219-1 | Downregulation | |
| hsa-mir-220 | Downregulation | |
| hsa-mir-224 | Downregulation | |
| hsa-mir-24-2 | Upregulation | |
| hsa-mir-26a-1-precursor | Downregulation | |
| hsa-mir-27b | Downregulation | |
| hsa-mir-29b-2 | Downregulation | |
| hsa-mir-30a-5p | Downregulation | |
| hsa-mir-32 | Downregulation | |
| hsa-mir-33 | Downregulation | |
| hsa-mir-9 | Downregulation | |
| hsa-mir-95 | Downregulation | |
| hsa-let-7a-2-precursor | Downregulation | |
| hsa-mir-132 | miR-132, previously shown to be differentially upregulated in six solid cancer types (breast, colon, lung, pancreas, prostate, and stomach carcinomas) | Lee et al. (2007) |
| hsa-mir-29a | miR-29 family (29a,b,c) reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B | Fabbri et al. (2007) |
| hsa-mir-29b-1 | miR-29 family (29a,b,c) reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B | |
| hsa-mir-29c | miR-29 family (29a,b,c) reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B | |
| hsa-mir-128b | Increased | Weiss et al. (2008) |
| hsa-mir-126 | Inhibits invasion in non-small cell lung carcinoma cell lines | Crawford et al. (2008) |
| hsa-mir-1 | Downregulated | Nasser et al. (2008) |
| hsa-mir-183 | Potential metastasis-inhibitor | Wang et al. (2008) |
| hsa-let-7a | let-7a: A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk | Chin et al. (2008) |
| hsa-let-7b | let-7b: A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk | |
| hsa-let-7d | let-7d: A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk | |
| hsa-let-7g | let-7g: A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk | |
| hsa-mir-126 | Inhibits the growth of lung cancer cell line | Liu et al. (2009a) |
| hsa-mir-142-5p | Was repressed, overexpression can inhibit lung cancer growth | Liu et al. (2009b) |
| hsa-mir-145 | Was repressed, overexpression can inhibit lung cancer growth | |
| hsa-mir-34c | Was repressed, overexpression can inhibit lung cancer growth | |
| hsa-mir-205 | Highly specific marker for squamous cell lung carcinoma | Lebanony et al. (2009) |
| hsa-mir-196a-2 | Genetic variant is associated with increased susceptibility of lung cancer in Chinese | Tian et al. (2009) |
| hsa-miR-21 | Upregulated in lung cancer in never-smokers | Seike et al. (2009) |
| hsa-mir-141 | Upregulated in lung cancer in never-smokers | |
| hsa-mir-210 | Upregulated in lung cancer in never-smokers | |
| hsa-mir-200b | Upregulated in lung cancer in never-smokers | |
| hsa-mir-346 | Upregulated in lung cancer in never-smokers | |
| hsa-mir-126* | Downregulated in lung cancer in never-smokers | |
| hsa-mir-126 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-30a | Downregulated in lung cancer in never-smokers | |
| hsa-mir-30d | Downregulated in lung cancer in never-smokers | |
| hsa-mir-486 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-129 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-451 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-521 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-138 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-30b | Downregulated in lung cancer in never-smokers | |
| hsa-mir-30c | Downregulated in lung cancer in never-smokers | |
| hsa-mir-516a | Downregulated in lung cancer in never-smokers | |
| hsa-mir-520 | Downregulated in lung cancer in never-smokers | |
| hsa-mir-17-5p | miR-17–92 cluster may contribute to protect SCLC cells carrying RB inactivation from excessive DNA damage and paradoxical growth inhibitory effects, to a large extent by direct downregulation of E2F1 expression. Overexpression involved in fine-tuning ROS generation | Ebi et al. (2009) |
| hsa-mir-20a | miR-17–92 cluster may contribute to protect SCLC cells carrying RB inactivation from excessive DNA damage and paradoxical growth inhibitory effects, to a large extent by direct downregulation of E2F1 expression. Overexpression involved in fine-tuning ROS generation | |
| hsa-mir-145 | miR-34c, miR-145, or miR-142-5p expression markedly diminished proliferation of lung cancer cell lines, clinical implications discussed | Sempere et al. (2009) |
| hsa-mir-142-5p | miR-34c, miR-145, or miR-142-5p expression markedly diminished proliferation of lung cancer cell lines, clinical implications discussed | |
| hsa-mir-34c | miR-34c, miR-145, or miR-142-5p expression markedly diminished proliferation of lung cancer cell lines, clinical implications discussed | |
| hsa-mir-133b | Low expression, targets pro-survival molecules MCL-1 and BCL-2L2 | Crawford et al. (2009) |
| hsa-mir-98 | Regulate tumor suppressor gene FUS1 | Du et al. (2009) |
| hsa-mir-197 | Regulate tumor suppressor gene FUS1 | |
| hsa-mir-93 | Regulate tumor suppressor gene FUS1 | |
| hsa-mir-185 | Cell cycle arrest | Takahashi et al. (2009) |
| hsa-miR-107 | Cell cycle arrest | |
| hsa-mir-34a | Prognostic marker of relapse in surgically resected non-small cell lung cancer | Gallardo et al. (2009) |
| hsa-let-7g | let-7g was downregulated in radio-resistant H1299 cells; increased with response to ionizing radiation when knockdown LIN28B | Jeong et al. (2009) |
| hsa-mir-34a | miR-34a:MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells | Duan et al. (2010) |
| hsa-mir-133b | Downregulated | Navon et al. (2009) |
| hsa-mir-486-5p | Downregulated | |
| hsa-mir-629 | Upregulated | |
| hsa-let-7a | Inhibition of proliferation in non-small cell lung cancer | Zhong et al. (2010) |
| hsa-mir-126 | Inhibition of proliferation in non-small cell lung cancer | |
| hsa-mir-145 | Inhibition of proliferation in non-small cell lung cancer | |
| hsa-mir-181b | Modulates multidrug resistance by targeting BCL-2 in human cancer cell lines | Zhu et al. (2010a) |
| hsa-mir-486 | Levels of four miRNAs (i.e., miR-486, miR-30d, miR-1, and miR-499) were significantly associated with overall survival | Hu et al. (2010) |
| hsa-mir-30d | Levels of four miRNAs (i.e., miR-486, miR-30d, miR-1, and miR-499) were significantly associated with overall survival | |
| hsa-mir-499 | Levels of four miRNAs (i.e., miR-486, miR-30d, miR-1, and miR-499) were significantly associated with overall survival | |
| hsa-mir-1 | Levels of four miRNAs (i.e., miR-486, miR-30d, miR-1, and miR-499) were significantly associated with overall survival | |
| hsa-mir-21 | MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) | Zhang et al. (2010b) |
| hsa-mir-136 | We found that miR-136, miR-376a, and miR-31 were each prominently overexpressed in murine lung cancers | Liu et al. (2010) |
| hsa-mir-376a | We found that miR-136, miR-376a, and miR-31 were each prominently overexpressed in murine lung cancers | |
| hsa-mir-31 | We found that miR-136, miR-376a, and miR-31 were each prominently overexpressed in murine lung cancers | |
| hsa-mir-182 | Suppresses lung tumorigenesis through downregulation of RGS17 expression in vitro | Sun et al. (2010) |
| hsa-mir-148a | The silencing of mir-148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis | Hanoun et al. (2010) |
| hsa-mir-301a | Blocking of miR-301 in A549 cells leads to a decrease in the expression of the host gene, ska2 | Cao et al. (2010) |
| hsa-mir-34a | Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34 | Wiggins et al. (2010) |
| hsa-mir-103 | Significant overexpression of miR-103, miR-107, miR-301, and miR-338 in lung cancer cells as compared to HBECs | Du et al. (2010) |
| hsa-mir-107 | Significant overexpression of miR-103, miR-107, miR-301, and miR-338 in lung cancer cells as compared to HBECs | |
| hsa-mir-301 | Significant overexpression of miR-103, miR-107, miR-301, and miR-338 in lung cancer cells as compared to HBECs | |
| hsa-mir-338 | Significant overexpression of miR-103, miR-107, miR-301, and miR-338 in lung cancer cells as compared to HBECs | |
| hsa-mir-186* | May serve as a potential gene therapy target for refractory lung cancer that is sensitive to curcumin | Zhang et al. (2010a) |
| hsa-mir-206 | Associated with invasion and metastasis of lung cancer | Wang et al. (2010) |
| hsa-mir-497 | Modulates multidrug resistance of human cancer cell lines by targeting BCL-2 | Zhu et al. (2010b) |
| hsa-mir-145 | Inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1 | Cho et al. (2011) |
| hsa-mir-638 | Upregulation of mir-638 and mir-923 in bostrycin-treated lung adenocarcinoma cells | Chen et al. (2011b) |
| hsa-mir-923 | Upregulation of mir-638 and mir-923 in bostrycin-treated lung adenocarcinoma cells | |
| hsa-mir-34b | Suppresses the expression of α4 through specific binding to the 3′-untranslated region of α4 is downregulated in transformed or human lung tumors | Chen et al. (2011a) |
| hsa-let-7a-2 | 9-cis-RA, all-trans-RA, lithium chloride and CEBPα might play important regulatory roles in let-7a2 gene expression in A549 cells | Guan et al. (2011) |
| hsa-mir-200 | The notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice | Yang et al. (2011) |
| hsa-mir-9 | Enhances the sensitivity to ionizing radiation by suppression of NFKB1 | Arora et al. (2011a) |
| hsa-let-7g | Enhances the sensitivity to ionizing radiation by suppression of NFKB1 | |
| hsa-let-7g | Precursor let-7g microRNA can supress A549 lung cancer cell migration | Park et al. (2011) |
| hsa-mir-145 | Suppresses lung adenocarcinoma-initiating cell proliferation by targeting OCT4 | Yin et al. (2011) |
| hsa-mir-196a-2 | hsa-miR-196a2 rs11614913 polymorphism may contribute to the susceptibility of cancers. CC genotype might modulate lung cancer risk (OR = 1.25, 95% CI = 1.06–1.46, pheterogeneity = 0.958) | Chu et al. (2011) |
| hsa-mir-145 | Inhibits lung adenocarcinoma stem cells proliferation by targeting OCT4 gene | Zhang et al. (2011b) |
| hsa-mir-182 | Inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein | Zhang et al. (2011a) |
| hsa-mir-21 | High expression of serum miR-21 associated with poor prognosis in patients with lung cancer | Liu et al. (2011) |
| hsa-mir-200c | High expression of tumor miR-200c associated with poor prognosis in patients with lung cancer | |
| hsa-mir-222 | High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. | Zhang et al. (2011c) |
| hsa-mir-155 | Could significantly inhibit the growth of human lung cancer 95D cells in vitro, which might be closely related to miR-155 induced G0/G1 phase arrest | Qin et al. (2011) |
| hsa-mir-125a-5p | Induces apoptosis by activating p53 in lung cancer cells | Jiang et al. (2011) |
| hsa-mir-146b | Overexpression of the Lung Cancer-prognostic miR-146b has a minimal and negative effect on the malignant phenotype of A549 Lung cancer cells | Patnaik et al. (2011) |
| hsa-mir-101 | miR-101 DNA copy loss is a prominent subtype specific event in lung cancer | Thu et al. (2011) |
| hsa-mir-375 | miR-375 is activated by ASH1 and inhibits YAP1 in a lineage dependent manner in lung cancer | Nishikawa et al. (2011) |
| hsa-mir-26b | Expression of miR-26b was downregulated, and its target activating transcription factor 2 (ATF2) mRNA was up regulated in γ-irradiated H1299 cells | Arora et al. (2011b) |
| hsa-mir-155 | The levels of miR-155, miR-197, and miR-182 in the plasma of lung cancer including stage I patients were significantly elevated compared with controls (P < 0.001). The combination of these three miRNAs yielded 81.33% sensitivity and 86.76% specificity in discriminating lung cancer patients from controls. The levels of miR-155 and miR-197 were higher in the plasma from lung cancer patients with metastasis than in those without metastasis (P < 0.05) and were significantly decreased in responsive patients during chemotherapy (P < 0.001) | Zheng et al. (2011) |
| hsa-mir-197 | The levels of miR-155, miR-197, and miR-182 in the plasma of lung cancer including stage I patients were significantly elevated compared with controls (P < 0.001). The combination of these three miRNAs yielded 81.33% sensitivity and 86.76% specificity in discriminating lung cancer patients from controls. The levels of miR-155 and miR-197 were higher in the plasma from lung cancer patients with metastasis than in those without metastasis (P < 0.05) and were significantly decreased in responsive patients during chemotherapy (P < 0.001) | |
| hsa-mir-182 | The levels of miR-155, miR-197, and miR-182 in the plasma of lung cancer including stage I patients were significantly elevated compared with controls (P < 0.001). The combination of these three miRNAs yielded 81.33% sensitivity and 86.76% specificity in discriminating lung cancer patients from controls. The levels of miR-155 and miR-197 were higher in the plasma from lung cancer patients with metastasis than in those without metastasis (P < 0.05) and were significantly decreased in responsive patients during chemotherapy (P < 0.001) | |
| hsa-mir-183 | Expression levels of members of the miR-183 family in lung cancer tumor and sera were higher than that of their normal counterparts. The miR-96 expression in tumors was positively associated with its expression in sera. High expression of tumor and serum miRNAs of the miR-183 family were associated with overall poor survival in patients with lung cancer | Zhu et al. (2011) |
| hsa-mir-96 | Expression levels of members of the miR-183 family in lung cancer tumor and sera were higher than that of their normal counterparts. The miR-96 expression in tumors was positively associated with its expression in sera. High expression of tumor and serum miRNAs of the miR-183 family were associated with overall poor survival in patients with lung cancer | |
| hsa-mir-150 | Anti-miR-150 vector can regress A549 lung cancer tumors | Li et al. (2011) |
| hsa-mir-200b | Stably expressing microRNA-200b in As-p53lowHBECs (human bronchial epithelial cell) abolished Akt and Erk1/2 activation, and completely suppressed cell migration and invasion | Wang et al. (2011b) |
| hsa-mir-9 | Upregulated in NSCLC compared to non-tumorous tissue | Vosa et al. (2011) |
| hsa-mir-182 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-200a + | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-151 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-205 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-183 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-130b* | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-149 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-193b | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-339-5p | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-196b | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-224 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-31 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-196a | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-423-3p | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-708 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-106b* | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-210 | Upregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-1273 | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-206 | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-140-3p | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-338-3p | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-101 | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-144 | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-1285 | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-130a | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-486-5p | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-24-2* | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-144* | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-30a | Downregulated in NSCLC compared to non-tumorous tissue | |
| hsa-mir-20a | Upregulated in NSCLC serum compared to control serum | Chen et al. (2011c) |
| hsa-mir-24 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-25 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-145 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-152 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-199a-5p | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-221 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-222 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-223 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-320 | Upregulated in NSCLC serum compared to control serum | |
| hsa-mir-126 | Upregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | Wang et al. (2011a) |
| hsa-mir-let-7a | Upregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-495 | Upregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-451 | Upregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-128b | Upregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-130a | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-106b | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-19b | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-22 | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-15b | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-17-5p | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-mir-21 | Downregulated in radiotherapy sensitive patients compared to resistant patients of non-small cell lung cancer | |
| hsa-miR-126 | Downregulated in lung SCC compared to normal lung tissues | Yang et al. (2010) |
| hsa-miR-193a-3p | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-30d | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-30a | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-101 | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-let-7i | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-15a | Downregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-185 * | Upregulated in lung SCC compared to normal lung tissues | |
| hsa-miR-125a-5p | Upregulated in lung SCC compared to normal lung tissues | |
| hsa-let-7f | Decreased in plasma vesicles from 28 NSCLC patients and 20 controls. Plasma levels of mir-30e-3p and let-7f were associated with short disease-free survival and overall survival | Silva et al. (2011) |
| hsa-miR-20b | Decreased in plasma vesicles from 28 NSCLC patients and 20 controls | |
| hsa-miR-30e-3p | Decreased in plasma vesicles from 28 NSCLC patients and 20 controls. Plasma levels of mir-30e-3p and let-7f were associated with short disease-free survival and overall survival | |
| hsa-miR-21 | Overexpressed in cell lines for breast cancer, prostate cancer, glioblastoma, and lung cancer | Roa et al. (2010) |
| hsa-miR-182 | Overexpressed in cell lines for breast cancer, prostate cancer, glioblastoma, and lung cancer | |
| hsa-let-7-5a | Underexpressed in cell lines for breast cancer, prostate cancer, glioblastoma, and lung cancer | |
| hsa-miR-145 | Underexpressed in cell lines for breast cancer, prostate cancer, glioblastoma, and lung cancer | |
| hsa-miR-155 | ||
| hsa-miR-21 | Upregulated in lung carcinoma tissues and their corresponding normal lung tissues, high mir-21 expression and low mir-181a expression were associated with poor survival, independent of clinical covariates, including TNM staging, lymph note status | Gao et al. (2010) |
| hsa-mir-143 | Downregulated in lung carcinoma tissues and their corresponding normal lung tissues, low level expression of mir-143 was significantly correlated with smoking status | |
| hsa-mir-181a | Downregulated in lung carcinoma tissues and their corresponding normal lung tissues, high mir-21 expression and low mir-181a expression were associated with poor survival, independent of clinical covariates, including TNM staging, lymph note status | |
| hsa-let-7g | Upregulated in adenocarcinoma compared to squamous cell carcinoma | Landi et al. (2010) |
| hsa-let-7b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7c | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-29a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7f | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| has-miR-453 | Downregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7d | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-98 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7i | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-26a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-509-3p | Downregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-30b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-146b-5p | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-106b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-mir-663 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-30d | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-17 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-498 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-26b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-let-7e | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-mir-654-5p | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-181a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-103 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-195 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-191 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-20a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-106a | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-29c | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-29b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-491-5p | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-19b | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-107 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-miR-16 | Upregulated in adenocarcinoma compared to squamous cell carcinoma | |
| hsa-mir-129-5p | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | Patnaik et al. (2010) |
| hsa-mir-194 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-631 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-200b | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-585 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-623 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-617 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-622 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-638 | Underexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-24 | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-141 | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-27b | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-16 | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-21 | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-30c | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-106a | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-15b | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-23b | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-23b | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-130a | Overexpressed in recurrence vs. No recurrence case groups of stage I NSCLC | |
| hsa-mir-636 | Changed more than twofold by radiation doses of 20Gy | Shin et al. (2009) |
| hsa-mir-593 | Changed more than twofold by radiation doses of 20Gy | |
| hsa-mir-760 | Changed more than twofold by radiation doses of 20Gy | |
| hsa-mir-139-3p | Changed more than twofold by radiation doses of 20Gy | |
| hsa-mir-345 | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-885-3p | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-206 | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-516a-5p | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-16-2* | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-106a | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-548c-3p | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-127-3p | Changed more than twofold by radiation doses of 20Gy and 40Gy | |
| hsa-mir-1228* | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-30b* | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-376a | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-34b* | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-215 | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-183 | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-22* | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-34a | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-192 | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-30c-1* | Changed more than twofold by radiation doses of 40Gy | |
| hsa-mir-21 | Mir-21 expression in the sputum specimens was significantly higher in cancer patients than cancer-free individuals. overexpression of mir-21 showed highly discriminative receiver-operator characteristic (ROC) curve profile, clearly distinguishing cancer patients from cancer-free subjects Detection of mir-21 expression produced 69.66% sensitivity and 100.00% specificity in diagnosis of lung cancer, as compared with 47.82% sensitivity and 100.00% specificity by sputum cytology | Xie et al. (2010) |
| hsa-mir-140-3p | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | Bryant et al. (2011) |
| hsa-mir-628-5p | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-518f | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-636 | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-301a | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-34c | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-224 | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-197 | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-205 | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-135b | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-200b | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-200c | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-141 | Expression of 13 miRNA genes predicts response to EGFR inhibition in cancer cell lines and tumors, and discriminates primary from metastatic tumors | |
| hsa-mir-182 | Overexpressed in primary lung tumors vs. metastases to lung | Barshack et al. (2010) |
| hsa-mir-126 | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-200c | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-141 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-375 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-7 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-429 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-200a | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-370 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-451 | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-195 | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-200b | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-486-5p | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-214 | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-382 | Overexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mir-199a-5p | Underexpressed in primary lung tumors vs. metastases to lung | |
| hsa-mis-210 | Upregulated in lung SCC compared to normal tissue | Raponi et al. (2009) |
| hsa-mir-200c | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-17-5p | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-20a | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-203 | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-125a | Downregulated in lung SCC compared to normal tissue | |
| hsa-let-7e | Downregulated in lung SCC compared to normal tissue | |
| hsa-mir-200a | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-106b | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-93 | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-182 | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-183 | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-106a | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-20b | Upregulated in lung SCC compared to normal tissue | |
| hsa-mir-224 | Upregulated in lung SCC compared to normal tissue | |
| hsa-miR-126 | Downregulated in NSCLC blood compared to normal blood | Keller et al. (2009) |
| hsa-miR-423-5p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-let-7d | Downregulated in NSCLC blood compared to normal blood | |
| hsa-let-7i | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-15a | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-22 | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-98 | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-19a | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-20b | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-324-3p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-574-5p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-195 | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-25 | Upregulated in NSCLC blood compared to normal blood | |
| hsa-let-7e | Downregulated in NSCLC blood compared to normal blood | |
| hsa-let-7c | Downregulated in NSCLC blood compared to normal blood | |
| hsa-let-7f | Downregulated in NSCLC blood compared to normal blood | |
| hsa-let-7a | Downregulated in NSCLC blood compared to normal blood | |
| hsa-let-7g | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-140-3p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-339-5p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-361-5p | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-1283 | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-18a* | Upregulated in NSCLC blood compared to normal blood | |
| hsa-miR-26b | Downregulated in NSCLC blood compared to normal blood | |
| hsa-miR-641 | Downregulated in lung cancer vs. COPD | Leidinger et al. (2011) |
| hsa-miR-662 | Upregulated in lung cancer vs. COPD | |
| hsa-miR-369-5p | Downregulated in lung cancer vs. COPD | |
| hsa-miR-383 | Downregulated in lung cancer vs. COPD | |
| hsa-miR-636 | Upregulated in lung cancer vs. COPD | |
| hsa-miR-940 | Upregulated in lung cancer vs. COPD | |
| hsa-miR-26a | Downregulated in lung cancer vs. COPD | |
| hsa-miR-92a | Upregulated in lung cancer vs. COPD | |
| hsa-miR-328 | Upregulated in lung cancer vs. COPD | |
| hsa-let-7d* | Upregulated in lung cancer vs. COPD | |
| hsa-miR-1224-3p | Upregulated in lung cancer vs. COPD | |
| hsa-miR-513b | Downregulated in lung cancer vs. COPD | |
| hsa-miR-93* | Upregulated in lung cancer vs. COPD | |
| hsa-miR-675 | Upregulated in lung cancer vs. COPD |
This table only includes a selection of available publications.
References
- Ahrendt S. A., Chow J. T., Xu L. H., Yang S. C., Eisenberger C. F., Esteller M., Herman J. G., Wu L., Decker P. A., Jen J., Sidransky D. (1999). Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J. Natl. Cancer Inst. 91, 332–339 10.1093/jnci/91.4.332 [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y., Naoe T. (2007). MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26, 311–320 10.1089/dna.2006.0550 [DOI] [PubMed] [Google Scholar]
- Arora S., Ranade A. R., Tran N. L., Nasser S., Sridhar S., Korn R. L., Ross J. T., Dhruv H., Foss K. M., Sibenaller Z., Ryken T., Gotway M. B., Kim S., Weiss G. J. (2011). MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int. J. Cancer 129, 2621–2631 10.1002/ijc.25939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach P. B., Silvestri G. A., Hanger M., Jett J. R. (2007). Screening for lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 132(Suppl. 3), 69S–77S 10.1378/chest.07-1349 [DOI] [PubMed] [Google Scholar]
- Backes C., Meese E., Lenhof H. P., Keller A. (2010). A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 38, 4476–4486 10.1093/nar/gkq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniskin A., Kuhnhenn J., Schlegel U., Chan A., Deckert M., Gold R., Maghnouj A., Zollner H., Reinacher-Schick A., Schmiegel W., Hahn S. A., Schroers R. (2011). Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117, 3140–3146 10.1182/blood-2010-09-308684 [DOI] [PubMed] [Google Scholar]
- Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabro E., Croce C. M., Pastorino U., Sozzi G. (2011). MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3713–3718 10.1073/pnas.1100048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno I. G., Karam R., Huang L., Bhardwaj A., Lou C. H., Shum E. Y., Song H. W., Corbett M. A., Gifford W. D., Gecz J., Pfaff S. L., Wilkinson M. F. (2011). Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510 10.1016/j.molcel.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C. M. (2002). Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Zen K., Zhang C. Y. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006 10.1038/cr.2008.238 [DOI] [PubMed] [Google Scholar]
- Chen X., Hu Z., Wang W., Ba Y., Ma L., Zhang C., Wang C., Ren Z., Zhao Y., Wu S., Zhuang R., Zhang Y., Hu H., Liu C., Xu L., Wang J., Shen H., Zhang J., Zen K., Zhang C. Y. (2011). Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int. J. Cancer. [Epub ahead of print]. 10.1002/ijc.26177 [DOI] [PubMed] [Google Scholar]
- Chen Z., Zeng H., Guo Y., Liu P., Pan H., Deng A., Hu J. (2010). miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. 29, 151. 10.1186/1756-9966-29-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre S. A., Galardi S., Mangiola A., Ferracin M., Liu C. G., Sabatino G., Negrini M., Maira G., Croce C. M., Farace M. G. (2005). Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 334, 1351–1358 10.1016/j.bbrc.2005.07.030 [DOI] [PubMed] [Google Scholar]
- da Cunha Santos G., Shepherd F. A., Tsao M. S. (2011). EGFR mutations and lung cancer. Annu. Rev. Pathol. 6, 49–69 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- Du L., Schageman J. J., Subauste M. C., Saber B., Hammond S. M., Prudkin L., Wistuba I. I., Ji L., Roth J. A., Minna J. D., Pertsemlidis A. (2009). miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol. Cancer Res. 7, 1234–1243 10.1158/1541-7786.MCR-08-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Schageman J. J., Irnov Girard L., Hammond S. M., Minna J. D., Gazdar A. F., Pertsemlidis A. (2010). MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J. Exp. Clin. Cancer Res. 29, 75. 10.1186/1756-9966-29-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Trang P., Wiggins J. F., Patrawala L., Cheng A., Ford L., Weidhaas J. B., Brown D., Bader A. G., Slack F. J. (2008). The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7, 759–764 10.4161/cc.7.6.5834 [DOI] [PubMed] [Google Scholar]
- Feng S., Cong S., Zhang X., Bao X., Wang W., Li H., Wang Z., Wang G., Xu J., Du B., Qu D., Xiong W., Yin M., Ren X., Wang F., He J., Zhang B. (2011). MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 39, 6669–6678 10.1093/nar/gkr232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss K. M., Sima C., Ugolini D., Neri M., Allen K. E., Weiss G. J. (2011). miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J. Thorac. Oncol. 6, 482–488 10.1097/JTO.0b013e318208c785 [DOI] [PubMed] [Google Scholar]
- Frezzetti D., De Menna M., Zoppoli P., Guerra C., Ferraro A., Bello A. M., De Luca P., Calabrese C., Fusco A., Ceccarelli M., Zollo M., Barbacid M., Di Lauro R., De Vita G. (2011). Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene 30, 275–286 10.1038/onc.2010.416 [DOI] [PubMed] [Google Scholar]
- Gao W., Yu Y., Cao H., Shen H., Li X., Pan S., Shu Y. (2010). Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed. Pharmacother. 64, 399–408 10.1016/j.biopha.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 10.1038/nature09026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M., Hoefig K., Merz H., Feller A. C., Kausch I., Jocham D., Warnecke J. M., Sczakiel G. (2010). A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 28, 655–661 10.1016/j.urolonc.2009.01.027 [DOI] [PubMed] [Google Scholar]
- Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. (2005). A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65, 9628–9632 10.1158/0008-5472.CAN-05-2352 [DOI] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 10.1038/nature03338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helene C., Toulme J. J. (1990). Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim. Biophys. Acta 1049, 99–125 [DOI] [PubMed] [Google Scholar]
- Hu Z., Chen X., Zhao Y., Tian T., Jin G., Shu Y., Chen Y., Xu L., Zen K., Zhang C., Shen H. (2010). Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726 10.1200/JCO.2010.29.9487 [DOI] [PubMed] [Google Scholar]
- Hunter M. P., Ismail N., Zhang X., Aguda B. D., Lee E. J., Yu L., Xiao T., Schafer J., Lee M. L., Schmittgen T. D., Nana-Sinkam S. P., Jarjoura D., Marsh C. B. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3, e3694. 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P., Taccioli C., Volinia S., Liu C. G., Alder H., Calin G. A., Menard S., Croce C. M. (2007). MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009). Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 10.3322/caac.20006 [DOI] [PubMed] [Google Scholar]
- Jeong H. C., Kim E. K., Lee J. H., Lee J. M., Yoo H. N., Kim J. K. (2011). Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol. Med. Report 4, 383–387 [DOI] [PubMed] [Google Scholar]
- Johnson C. D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J., Chin L., Brown D., Slack F. J. (2007). The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 67, 7713–7722 10.1158/0008-5472.CAN-07-1133 [DOI] [PubMed] [Google Scholar]
- Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 10.1016/j.cell.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Kanzaki H., Ito S., Hanafusa H., Jitsumori Y., Tamaru S., Shimizu K., Ouchida M. (2011). Identification of direct targets for the miR-17-92 cluster by proteomic analysis. Proteomics 11, 3531–3539 10.1002/pmic.201000501 [DOI] [PubMed] [Google Scholar]
- Karlsson H., Bachmann S., Schroder J., McArthur J., Torrey E. F., Yolken R. H. (2001). Retroviral RNA identified in the cerebrospinal fluids and brains of individuals with schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 98, 4634–4639 10.1073/pnas.061021998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski A. L., Slack F. J. (2011). MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 11, 849–864 10.1038/nrc3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Leidinger P., Bauer A., ElSharawy A., Haas J., Backes C., Wendschlag A., Giese N., Tjaden C., Ott K., Werner J., Hackert T., Ruprecht K., Huwer H., Huebers J., Jacobs G., Rosenstiel P., Dommisch H., Schaefer A., Müller-Quernheim J., Wullich B., Keck B., Graf N., Reichrath J., Vogel B., Nebel A. J., SU Staehler P., Amarantos I., Boisguerin V., Staehler C., Beier M., Scheffler M., Büchler M., Wischhusen J., Haeusler S., Dietl J., Hofmann S., Lenhof H.-P., Schreiber S., Katus H., Rottbauer W., Meder B., Hoheisel J., Franke A., Meese E. (2011a). Toward the blood-borne miRNome of human diseases. Nat. Methods 8, 841–843 10.1038/nmeth.1682 [DOI] [PubMed] [Google Scholar]
- Keller A., Leidinger P., Gislefoss R., Haugen A., Langseth H., Staehler P., Lenhof H. P., Meese E. (2011b). Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol 8, 506–516 10.4161/rna.8.3.14994 [DOI] [PubMed] [Google Scholar]
- Keller A., Leidinger P., Borries A., Wendschlag A., Wucherpfennig F., Scheffler M., Huwer H., Lenhof H. P., Meese E. (2009). miRNAs in lung cancer – studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer 9, 353. 10.1186/1471-2407-9-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Ochiya T. (2010). Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 101, 2087–2092 10.1111/j.1349-7006.2010.01650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossenkov A. V., Vachani A., Chang C., Nichols C., Billouin S., Horng W., Rom W. N., Albelda S. M., Showe M. K., Showe L. C. (2011). Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin. Cancer Res. 17, 5867–5877 10.1158/1078-0432.CCR-11-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K. G., Tuschl T., Manoharan M., Stoffel M. (2005). Silencing of microRNAs in vivo with “antagomirs.” Nature 438, 685–689 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Yu S. L., Hsieh M. H., Chen C. H., Chen H. Y., Wen C. C., Huang Y. H., Hsiao P. C., Hsiao C. K., Liu C. M., Yang P. C., Hwu H. G., Chen W. J. (2011). MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 6, e21635. 10.1371/journal.pone.0021635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landen C. N., Jr., Chavez-Reyes A., Bucana C., Schmandt R., Deavers M. T., Lopez-Berestein G., Sood A. K. (2005). Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 65, 6910–6918 10.1158/0008-5472.CAN-05-0530 [DOI] [PubMed] [Google Scholar]
- Landi M. T., Zhao Y., Rotunno M., Koshiol J., Liu H., Bergen A. W., Rubagotti M., Goldstein A. M., Linnoila I., Marincola F. M., Tucker M. A., Bertazzi P. A., Pesatori A. C., Caporaso N. E., McShane L. M., Wang E. (2010). MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 16, 430–441 10.1158/1078-0432.CCR-09-1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebanony D., Benjamin H., Gilad S., Ezagouri M., Dov A., Ashkenazi K., Gefen N., Izraeli S., Rechavi G., Pass H., Nonaka D., Li J., Spector Y., Rosenfeld N., Chajut A., Cohen D., Aharonov R., Mansukhani M. (2009). Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 27, 2030–2037 10.1200/JCO.2008.19.4134 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Leidinger P., Keller A., Borries A., Huwer H., Rohling M., Huebers J., Lenhof H. P., Meese E. (2011). Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer 74, 41–47 10.1016/j.lungcan.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L., Chen C. (2007). Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166. 10.1186/1471-2164-8-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Xu X. (2011). MicroRNA isolation from formalin-fixed, paraffin-embedded tissues. Methods Mol. Biol. 724, 259–267 10.1007/978-1-61779-273-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. G., Zhu W. Y., Huang Y. Y., Ma L. N., Zhou S. Q., Wang Y. K., Zeng F., Zhou J. H., Zhang Y. K. (2011). High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834–838 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Lu Z., Liu M., Stribinskis V., Klinge C. M., Ramos K. S., Colburn N. H., Li Y. (2008). MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27, 4373–4379 10.1038/onc.2008.72 [DOI] [PubMed] [Google Scholar]
- Markou A., Tsaroucha E. G., Kaklamanis L., Fotinou M., Georgoulias V., Lianidou E. S. (2008). Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin. Chem. 54, 1696–1704 10.1373/clinchem.2007.101741 [DOI] [PubMed] [Google Scholar]
- Mascaux C., Laes J. F., Anthoine G., Haller A., Ninane V., Burny A., Sculier J. P. (2009). Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur. Respir. J. 33, 352–359 10.1183/09031936.00084108 [DOI] [PubMed] [Google Scholar]
- Matsubara H., Takeuchi T., Nishikawa E., Yanagisawa K., Hayashita Y., Ebi H., Yamada H., Suzuki M., Nagino M., Nimura Y., Osada H., Takahashi T. (2007). Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene 26, 6099–6105 10.1038/sj.onc.1210425 [DOI] [PubMed] [Google Scholar]
- McDermott A. M., Heneghan H. M., Miller N., Kerin M. J. (2011). The therapeutic potential of MicroRNAs: disease modulators and drug targets. Pharm. Res. 28, 3016–3029 10.1007/s11095-011-0550-2 [DOI] [PubMed] [Google Scholar]
- Meder B., Keller A., Vogel B., Haas J., Sedaghat-Hamedani F., Kayvanpour E., Just S., Borries A., Rudloff J., Leidinger P., Meese E., Katus H. A., Rottbauer W. (2011). MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 106, 13–23 10.1007/s00395-010-0123-2 [DOI] [PubMed] [Google Scholar]
- Megiorni F., Pizzuti A., Frati L. (2011). Clinical Significance of MicroRNA Expression profiles and polymorphisms in lung cancer development and management. Patholog. Res. Int. 2011, 780652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehes G., Witt A., Kubista E., Ambros P. F. (2001). Circulating breast cancer cells are frequently apoptotic. Am. J. Pathol. 159, 17–20 10.1016/S0002-9440(10)61667-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222 10.1016/j.cell.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 10.1053/j.gastro.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O’Briant K. C., Allen A., Lin D. W., Urban N., Drescher C. W., Knudsen B. S., Stirewalt D. L., Gentleman R., Vessella R. L., Nelson P. S., Martin D. B., Tewari M. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 10.1073/pnas.0803384105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E. J., Yoon H., Kim S. W., Kim H., Kim Y. T., Kim J. H., Kim J. W., Kim S. (2008). MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 14, 2690–2695 10.1158/1078-0432.CCR-07-1731 [DOI] [PubMed] [Google Scholar]
- Nasser S., Ranade A. R., Sridhar S., Haney L., Korn R. L., Gotway M. B., Weiss G. J. (2011). Biomarkers associated with metastasis of lung cancer to brain predict patient survival. Int. J. Data Min. Bioinform. 5, 287–307 10.1504/IJDMB.2011.040385 [DOI] [PubMed] [Google Scholar]
- Otaegui D., Baranzini S. E., Armananzas R., Calvo B., Munoz-Culla M., Khankhanian P., Inza I., Lozano J. A., Castillo-Trivino T., Asensio A., Olaskoaga J., Lopez de Munain A. (2009). Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE 4, e6309. 10.1371/journal.pone.0006309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D. (2003). Does the immune system see tumors as foreign or self? Annu. Rev. Immunol. 21, 807–839 10.1146/annurev.immunol.21.120601.141135 [DOI] [PubMed] [Google Scholar]
- Park N. J., Li Y., Yu T., Brinkman B. M., Wong D. T. (2006). Characterization of RNA in saliva. Clin. Chem. 52, 988–994 10.1373/clinchem.2006.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N. J., Zhou H., Elashoff D., Henson B. S., Kastratovic D. A., Abemayor E., Wong D. T. (2009). Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 15, 5473–5477 10.1158/1078-0432.CCR-08-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka K. P., Pfeiffer M. J., Waltering K. K., Vessella R. L., Tammela T. L., Visakorpi T. (2007). MicroRNA expression profiling in prostate cancer. Cancer Res. 67, 6130–6135 10.1158/0008-5472.CAN-07-0533 [DOI] [PubMed] [Google Scholar]
- Pottelberge G. R., Mestdagh P., Bracke K. R., Thas O., Durme Y. M., Joos G. F., Vandesompele J., Brusselle G. G. (2011). MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 183, 898–906 10.1164/rccm.201002-0304OC [DOI] [PubMed] [Google Scholar]
- Rabinowits G., Gercel-Taylor C., Day J. M., Taylor D. D., Kloecker G. H. (2009). Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 10, 42–46 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- Raponi M., Dossey L., Jatkoe T., Wu X., Chen G., Fan H., Beer D. G. (2009). MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 69, 5776–5783 10.1158/0008-5472.CAN-09-0587 [DOI] [PubMed] [Google Scholar]
- Reiber H., Peter J. B. (2001). Cerebrospinal fluid analysis: disease-related data patterns and evaluation programs. J. Neurol. Sci. 184, 101–122 10.1016/S0022-510X(00)00501-3 [DOI] [PubMed] [Google Scholar]
- Schipper H. M., Maes O. C., Chertkow H. M., Wang E. (2007). MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Bio. 1, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B., Engel E., Carstensen T., Weickmann S., John M., Witt C., Fleischhacker M. (2005). Quantification of free RNA in serum and bronchial lavage: a new diagnostic tool in lung cancer detection? Lung Cancer 48, 145–147 10.1016/j.lungcan.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Seike M., Goto A., Okano T., Bowman E. D., Schetter A. J., Horikawa I., Mathe E. A., Jen J., Yang P., Sugimura H., Gemma A., Kudoh S., Croce C. M., Harris C. C. (2009). MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U.S.A. 106, 12085–12090 10.1073/pnas.0905234106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P., Megraw M., Hatzigeorgiou A. G. (2006). A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods 3, 881–886 10.1038/nmeth954 [DOI] [PubMed] [Google Scholar]
- Shahzad M. M., Mangala L. S., Han H. D., Lu C., Bottsford-Miller J., Nishimura M., Mora E. M., Lee J. W., Stone R. L., Pecot C. V., Thanapprapasr D., Roh J. W., Gaur P., Nair M. P., Park Y. Y., Sabnis N., Deavers M. T., Lee J. S., Ellis L. M., Lopez-Berestein G., McConathy W. J., Prokai L., Lacko A. G., Sood A. K. (2011). Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia 13, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Todd N. W., Zhang H., Yu L., Lingxiao X., Mei Y., Guarnera M., Liao J., Chou A., Lu C. L., Jiang Z., Fang H., Katz R. L., Jiang F. (2011). Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab. Invest. 91, 579–587 10.1038/labinvest.2010.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. J., Zhong Z. R., Liu J., Zhang Z. R., Sun X., Gong T. (2011). Solid lipid nanoparticles loaded with anti-microRNA oligonucleotides (AMOs) for suppression of microRNA-21 functions in human lung cancer cells. Pharm. Res. 29, 97–109 10.1007/s11095-011-0514-6 [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Thunnissen F. B. (2003). Sputum examination for early detection of lung cancer. J. Clin. Pathol. 56, 805–810 10.1136/jcp.56.11.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomankova T., Petrek M., Kriegova E. (2010). Involvement of microRNAs in physiological and pathological processes in the lung. Respir. Res. 11, 159. 10.1186/1465-9921-11-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P., Medina P. P., Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A. G., Weidhaas J. B., Slack F. J. (2010). Regression of murine lung tumors by the let-7 microRNA. Oncogene 29, 1580–1587 10.1038/onc.2009.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P., Wiggins J. F., Daige C. L., Cho C., Omotola M., Brown D., Weidhaas J. B., Bader A. G., Slack F. J. (2011). Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19, 1116–1122 10.1038/mt.2011.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzimagiorgis G., Michailidou E. Z., Kritis A., Markopoulos A. K., Kouidou S. (2011). Recovering circulating extracellular or cell-free RNA from bodily fluids. Cancer Epidemiol. 35, 580–589 10.1016/j.canep.2011.02.016 [DOI] [PubMed] [Google Scholar]
- van Kouwenhove M., Kedde M., Agami R. (2011). MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 11, 644–656 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Voellenkle C., van Rooij J., Cappuzzello C., Greco S., Arcelli D., Di Vito L., Melillo G., Rigolini R., Costa E., Crea F., Capogrossi M. C., Napolitano M., Martelli F. (2010). MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genomics 42, 420–426 10.1152/physiolgenomics.00211.2009 [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosa U., Vooder T., Kolde R., Fischer K., Valk K., Tonisson N., Roosipuu R., Vilo J., Metspalu A., Annilo T. (2011). Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer 50, 812–822 10.1002/gcc.20902 [DOI] [PubMed] [Google Scholar]
- Wang G., Tam L. S., Li E. K., Kwan B. C., Chow K. M., Luk C. C., Li P. K., Szeto C. C. (2011). Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus 20, 493–500 10.1177/0961203310387179 [DOI] [PubMed] [Google Scholar]
- Weber J. A., Baxter D. H., Zhang S., Huang D. Y., Huang K. H., Lee M. J., Galas D. J., Wang K. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G. J., Bemis L. T., Nakajima E., Sugita M., Birks D. K., Robinson W. A., Varella-Garcia M., Bunn P. A., Jr., Haney J., Helfrich B. A., Kato H., Hirsch F. R., Franklin W. A. (2008). EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 19, 1053–1059 10.1093/annonc/mdn006 [DOI] [PubMed] [Google Scholar]
- Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A. G. (2010). Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930 10.1158/0008-5472.CAN-10-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Crawford M., Yu B., Mao Y., Nana-Sinkam S. P., Lee L. J. (2011). MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol. Pharm. 8, 1381–1389 10.1021/mp200300w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Todd N. W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S. A., Jiang F. (2010). Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 67, 170–176 10.1016/j.lungcan.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Todd N. W., Yu L., Fang H., Jiang F. (2010). Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod. Pathol. 23, 1157–1164 10.1038/modpathol.2010.111 [DOI] [PubMed] [Google Scholar]
- Xiong S., Zheng Y., Jiang P., Liu R., Liu X., Chu Y. (2011). MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int. J. Biol. Sci. 7, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., Calin G. A., Liu C. G., Croce C. M., Harris C. C. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- Yu L., Todd N. W., Xing L., Xie Y., Zhang H., Liu Z., Fang H., Zhang J., Katz R. L., Jiang F. (2010). Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int. J. Cancer 127, 2870–2878 10.1002/ijc.25169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. G., Wang J. J., Zhao F., Liu Q., Jiang K., Yang G. H. (2010). MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 411, 846–852 10.1016/j.cca.2010.06.030 [DOI] [PubMed] [Google Scholar]
- Zhang Y. A., Nemunaitis J., Samuel S. K., Chen P., Shen Y., Tong A. W. (2006). Antitumor activity of an oncolytic adenovirus-delivered oncogene small interfering RNA. Cancer Res. 66, 9736–9743 10.1158/0008-5472.CAN-06-0130 [DOI] [PubMed] [Google Scholar]
References
- Arora H., Qureshi R., Jin S., Park A. K., Park W. Y. (2011a). miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp. Mol. Med. 43, 298–304 10.3858/emm.2011.43.5.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora H., Qureshi R., Park A. K., Park W. Y. (2011b). Coordinated regulation of ATF2 by miR-26b in gamma-irradiated lung cancer cells. PLoS ONE 6, e23802. 10.1371/journal.pone.0023802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshack I., Lithwick-Yanai G., Afek A., Rosenblatt K., Tabibian-Keissar H., Zepeniuk M., Cohen L., Dan H., Zion O., Strenov Y., Polak-Charcon S., Perelman M. (2010). MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol. Res. Pract. 206, 578–584 10.1016/j.prp.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Bryant J. L., Britson J., Balko J. M., Willian M., Timmons R., Frolov A., Black E. P. (2011). A microRNA gene expression signature predicts response to erlotinib in epithelial cancer cell lines and targets EMT. Br. J. Cancer. [Epub ahead of print]. 10.1038/bjc.2011.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Huang B., Liu Z., Zhang J., Xu H., Xia W., Li J., Li S., Chen L., Ding H., Zhao Q., Fan M., Shen B., Shao N. (2010). Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem. Biophys. Res. Commun. 396, 978–982 10.1016/j.bbrc.2010.05.037 [DOI] [PubMed] [Google Scholar]
- Chen L. P., Lai Y. D., Li D. C., Zhu X. N., Yang P., Li W. X., Zhu W., Zhao J., Li X. D., Xiao Y. M., Zhang Y., Xing X. M., Wang Q., Zhang B., Lin Y. C., Zeng J. L., Zhang S. X., Liu C. X., Li Z. F., Zeng X. W., Lin Z. N., Zhuang Z. X., Chen W. (2011a). Alpha4 is highly expressed in carcinogen-transformed human cells and primary human cancers. Oncogene 30, 2943–2953 10.1038/onc.2011.20 [DOI] [PubMed] [Google Scholar]
- Chen W. S., Hou J. N., Guo Y. B., Yang H. L., Xie C. M., Lin Y. C., She Z. G. (2011b). Bostrycin inhibits proliferation of human lung carcinoma A549 cells via downregulation of the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 30, 17. 10.1186/1756-9966-30-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu Z., Wang W., Ba Y., Ma L., Zhang C., Wang C., Ren Z., Zhao Y., Wu S., Zhuang R., Zhang Y., Hu H., Liu C., Xu L., Wang J., Shen H., Zhang J., Zen K., Zhang C. Y. (2011c). Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int. J. Cancer. [Epub ahead of print]. 10.1002/ijc.26177 [DOI] [PubMed] [Google Scholar]
- Chin L. J., Ratner E., Leng S., Zhai R., Nallur S., Babar I., Muller R. U., Straka E., Su L., Burki E. A., Crowell R. E., Patel R., Kulkarni T., Homer R., Zelterman D., Kidd K. K., Zhu Y., Christiani D. C., Belinsky S. A., Slack F. J., Weidhaas J. B. (2008). A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 68, 8535–8540 10.1158/0008-5472.CAN-08-2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W. C., Chow A. S., Au J. S. (2011). MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol 8, 125–131 10.4161/rna.8.1.14259 [DOI] [PubMed] [Google Scholar]
- Chu H., Wang M., Shi D., Ma L., Zhang Z., Tong N., Huo X., Wang W., Luo D., Gao Y. (2011). Hsa-miR-196a2 Rs11614913 polymorphism contributes to cancer susceptibility: evidence from 15 case-control studies. PLoS ONE 6, e18108. 10.1371/journal.pone.0018108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M., Batte K., Yu L., Wu X., Nuovo G. J., Marsh C. B., Otterson G. A., Nana-Sinkam S. P. (2009). MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 388, 483–489 10.1016/j.bbrc.2009.07.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M., Brawner E., Batte K., Yu L., Hunter M. G., Otterson G. A., Nuovo G., Marsh C. B., Nana-Sinkam S. P. (2008). MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun. 373, 607–612 10.1016/j.bbrc.2008.06.090 [DOI] [PubMed] [Google Scholar]
- Du L., Schageman J. J., Irnov, Girard L., Hammond S. M., Minna J. D., Gazdar A. F., Pertsemlidis A. (2010). MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J. Exp. Clin. Cancer Res. 29, 75. 10.1186/1756-9966-29-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Schageman J. J., Subauste M. C., Saber B., Hammond S. M., Prudkin L., Wistuba II, Ji L., Roth J. A., Minna J. D., Pertsemlidis A. (2009). miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol. Cancer Res. 7, 1234–1243 10.1158/1541-7786.MCR-08-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., Gao L., Wu X., Wang L., Nana-Sinkam S. P., Otterson G. A., Villalona-Calero M. A. (2010). MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells. Int. J. Cancer 127, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H., Sato T., Sugito N., Hosono Y., Yatabe Y., Matsuyama Y., Yamaguchi T., Osada H., Suzuki M., Takahashi T. (2009). Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 28, 3371–3379 10.1038/onc.2009.201 [DOI] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., Volinia S., Guler G., Morrison C. D., Chan K. K., Marcucci G., Calin G. A., Huebner K., Croce C. M. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U.S.A. 104, 15805–15810 10.1073/pnas.0707628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo E., Navarro A., Vinolas N., Marrades R. M., Diaz T., Gel B., Quera A., Bandres E., Garcia-Foncillas J., Ramirez J., Monzo M. (2009). miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 30, 1903–1909 10.1093/carcin/bgp219 [DOI] [PubMed] [Google Scholar]
- Gao W., Yu Y., Cao H., Shen H., Li X., Pan S., Shu Y. (2010). Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed. Pharmacother. 64, 399–408 10.1016/j.biopha.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Guan H., Zhang P., Liu C., Zhang J., Huang Z., Chen W., Chen Z., Ni N., Liu Q., Jiang A. (2011). Characterization and functional analysis of the human microRNA let-7a2 promoter in lung cancer A549 cell lines. Mol. Biol. Rep. 38, 5327–5334 10.1007/s11033-010-0307-8 [DOI] [PubMed] [Google Scholar]
- Hanoun N., Delpu Y., Suriawinata A. A., Bournet B., Bureau C., Selves J., Tsongalis G. J., Dufresne M., Buscail L., Cordelier P., Torrisani J. (2010). The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin. Chem. 56, 1107–1118 10.1373/clinchem.2010.144709 [DOI] [PubMed] [Google Scholar]
- Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. (2005). A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65, 9628–9632 10.1158/0008-5472.CAN-05-2352 [DOI] [PubMed] [Google Scholar]
- Hu Z., Chen X., Zhao Y., Tian T., Jin G., Shu Y., Chen Y., Xu L., Zen K., Zhang C., Shen H. (2010). Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726 10.1200/JCO.2010.29.9487 [DOI] [PubMed] [Google Scholar]
- Jeong S. H., Wu H. G., Park W. Y. (2009). LIN28B confers radio-resistance through the posttranscriptional control of KRAS. Exp. Mol. Med. 41, 912–918 10.3858/emm.2009.41.6.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Huang Q., Chang J., Wang E., Qiu X. (2011). MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp. Lung Res. 37, 387–398 10.3109/01902148.2010.492068 [DOI] [PubMed] [Google Scholar]
- Keller A., Leidinger P., Borries A., Wendschlag A., Wucherpfennig F., Scheffler M., Huwer H., Lenhof H. P., Meese E. (2009). miRNAs in lung cancer – studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer 9, 353. 10.1186/1471-2407-9-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi M. T., Zhao Y., Rotunno M., Koshiol J., Liu H., Bergen A. W., Rubagotti M., Goldstein A. M., Linnoila I., Marincola F. M., Tucker M. A., Bertazzi P. A., Pesatori A. C., Caporaso N. E., McShane L. M., Wang E. (2010). MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 16, 430–441 10.1158/1078-0432.CCR-09-1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebanony D., Benjamin H., Gilad S., Ezagouri M., Dov A., Ashkenazi K., Gefen N., Izraeli S., Rechavi G., Pass H., Nonaka D., Li J., Spector Y., Rosenfeld N., Chajut A., Cohen D., Aharonov R., Mansukhani M. (2009). Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 27, 2030–2037 10.1200/JCO.2008.19.4134 [DOI] [PubMed] [Google Scholar]
- Lee J., Li Z., Brower-Sinning R., John B. (2007). Regulatory circuit of human microRNA biogenesis. PLoS Comput. Biol. 3, e67. 10.1371/journal.pcbi.0030067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidinger P., Keller A., Borries A., Huwer H., Rohling M., Huebers J., Lenhof H. P., Meese E. (2011). Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer 74, 41–47 10.1016/j.lungcan.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Li Y. J., Zhang Y. X., Wang P. Y., Chi Y. L., Zhang C., Ma Y., Lv C. J., Xie S. Y. (2011). Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol. Rep. 27, 129–134 [DOI] [PubMed] [Google Scholar]
- Liu B., Peng X. C., Zheng X. L., Wang J., Qin Y. W. (2009a). MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66, 169–175 10.1016/j.lungcan.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Liu X., Sempere L. F., Galimberti F., Freemantle S. J., Black C., Dragnev K. H., Ma Y., Fiering S., Memoli V., Li H., DiRenzo J., Korc M., Cole C. N., Bak M., Kauppinen S., Dmitrovsky E. (2009b). Uncovering growth-suppressive MicroRNAs in lung cancer. Clin. Cancer Res. 15, 1177–1183 10.1158/1078-0432.CCR-09-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sempere L. F., Ouyang H., Memoli V. A., Andrew A. S., Luo Y., Demidenko E., Korc M., Shi W., Preis M., Dragnev K. H., Li H., Direnzo J., Bak M., Freemantle S. J., Kauppinen S., Dmitrovsky E. (2010). MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J. Clin. Invest. 120, 1298–1309 10.1172/JCI39566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. G., Zhu W. Y., Huang Y. Y., Ma L. N., Zhou S. Q., Wang Y. K., Zeng F., Zhou J. H., Zhang Y. K. (2011). High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Nasser M. W., Datta J., Nuovo G., Kutay H., Motiwala T., Majumder S., Wang B., Suster S., Jacob S. T., Ghoshal K. (2008). Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 283, 33394–33405 10.1074/jbc.M804788200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Navon R., Wang H., Steinfeld I., Tsalenko A., Ben-Dor A., Yakhini Z. (2009). Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS ONE 4, e8003. 10.1371/journal.pone.0008003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa E., Osada H., Okazaki Y., Arima C., Tomida S., Tatematsu Y., Taguchi A., Shimada Y., Yanagisawa K., Yatabe Y., Toyokuni S., Sekido Y., Takahashi T. (2011). miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 71, 6165–6173 10.1158/0008-5472.CAN-11-1020 [DOI] [PubMed] [Google Scholar]
- Park S., Minai-Tehrani A., Xu C. X., Chang S. H., Woo M. A., Noh M. S., Lee E. S., Lim H. T., An G. H., Lee K. H., Sung H. J., Beck G. R., Cho M. H. (2011). Suppression of A549 lung cancer cell migration by precursor let-7g microRNA. Mol. Med. Rep. 3, 1007–1013 [DOI] [PubMed] [Google Scholar]
- Patnaik S. K., Kannisto E., Knudsen S., Yendamuri S. (2010). Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 70, 36–45 10.1158/1538-7445.AM10-36 [DOI] [PubMed] [Google Scholar]
- Patnaik S. K., Kannisto E., Mallick R., Yendamuri S. (2011). Overexpression of the lung cancer-prognostic miR-146b microRNAs has a minimal and negative effect on the malignant phenotype of A549 lung cancer cells. PLoS ONE 6, e22379. 10.1371/journal.pone.0022379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Zhou Y., Sheng M., Fei G., Ren T., Xu L. (2011). Effects of microRNA-155 on the growth of human lung cancer cell line 95D in vitro. Zhongguo Fei Ai Za Zhi 14, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M., Dossey L., Jatkoe T., Wu X., Chen G., Fan H., Beer D. G. (2009). MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 69, 5776–5783 10.1158/0008-5472.CAN-09-0587 [DOI] [PubMed] [Google Scholar]
- Roa W., Brunet B., Guo L., Amanie J., Fairchild A., Gabos Z., Nijjar T., Scrimger R., Yee D., Xing J. (2010). Identification of a new microRNA expression profile as a potential cancer screening tool. Clin. Invest. Med. 33, E124. [DOI] [PubMed] [Google Scholar]
- Seike M., Goto A., Okano T., Bowman E. D., Schetter A. J., Horikawa I., Mathe E. A., Jen J., Yang P., Sugimura H., Gemma A., Kudoh S., Croce C. M., Harris C. C. (2009). MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U.S.A. 106, 12085–12090 10.1073/pnas.0905234106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Liu X., Dmitrovsky E. (2009). Tumor-suppressive microRNAs in Lung cancer: diagnostic and therapeutic opportunities. Sci. World J. 9, 626–628 10.1100/tsw.2009.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Cha H. J., Lee E. M., Lee S. J., Seo S. K., Jin H. O., Park I. C., Jin Y. W., An S. (2009). Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int. J. Oncol. 35, 81–86 [PubMed] [Google Scholar]
- Silva J., Garcia V., Zaballos A., Provencio M., Lombardia L., Almonacid L., Garcia J. M., Dominguez G., Pena C., Diaz R., Herrera M., Varela A., Bonilla F. (2011). Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 37, 617–623 10.1183/09031936.00029610 [DOI] [PubMed] [Google Scholar]
- Sun Y., Fang R., Li C., Li L., Li F., Ye X., Chen H. (2010). Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem. Biophys. Res. Commun. 396, 501–507 10.1016/j.bbrc.2010.04.170 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Forrest A. R., Maeno E., Hashimoto T., Daub C. O., Yasuda J. (2009). MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS ONE 4, e6677. 10.1371/journal.pone.0006677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. (2004). Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 10.1158/0008-5472.CAN-04-0637 [DOI] [PubMed] [Google Scholar]
- Thu K. L., Chari R., Lockwood W. W., Lam S., Lam W. L. (2011). miR-101 DNA copy loss is a prominent subtype specific event in lung cancer. J. Thorac. Oncol. 6, 1594–1598 10.1097/JTO.0b013e3182217d81 [DOI] [PubMed] [Google Scholar]
- Tian T., Shu Y., Chen J., Hu Z., Xu L., Jin G., Liang J., Liu P., Zhou X., Miao R., Ma H., Chen Y., Shen H. (2009). A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol. Biomarkers Prev. 18, 1183–1187 10.1158/1055-9965.EPI-08-0814 [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 10.1073/pnas.0510565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosa U., Vooder T., Kolde R., Fischer K., Valk K., Tonisson N., Roosipuu R., Vilo J., Metspalu A., Annilo T. (2011). Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer 50, 812–822 10.1002/gcc.20902 [DOI] [PubMed] [Google Scholar]
- Wang G., Mao W., Zheng S. (2008). MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 582, 3663–3668 10.1016/j.febslet.2008.01.055 [DOI] [PubMed] [Google Scholar]
- Wang X., Ling C., Bai Y., Zhao J. (2010). MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat. Rec. (Hoboken) 294, 88–92 10.1002/ar.21287 [DOI] [PubMed] [Google Scholar]
- Wang X. C., Du L. Q., Tian L. L., Wu H. L., Jiang X. Y., Zhang H., Li D. G., Wang Y. Y., Wu H. Y., She Y., Liu Q. F., Fan F. Y., Meng A. M. (2011a). Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer 72, 92–99 10.1016/j.lungcan.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Wang Z., Yang J., Fisher T., Xiao H., Jiang Y., Yang C. (2011b). Akt activation is responsible for enhanced migratory and invasive behavior of arsenic-transformed human bronchial epithelial cells. Environ. Health Perspect. [Epub ahead of print]. 10.1289/ehp.1104061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G. J., Bemis L. T., Nakajima E., Sugita M., Birks D. K., Robinson W. A., Varella-Garcia M., Bunn P. A., Jr., Haney J., Helfrich B. A., Kato H., Hirsch F. R., Franklin W. A. (2008). EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 19, 1053–1059 10.1093/annonc/mdn006 [DOI] [PubMed] [Google Scholar]
- Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A. G. (2010). Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930 10.1158/0008-5472.CAN-10-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Todd N. W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S. A., Jiang F. (2010). Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer 67, 170–176 10.1016/j.lungcan.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., Calin G. A., Liu C. G., Croce C. M., Harris C. C. (2006). Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ahn Y. H., Gibbons D. L., Zang Y., Lin W., Thilaganathan N., Alvarez C. A., Moreira D. C., Creighton C. J., Gregory P. A., Goodall G. J., Kurie J. M. (2011). The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J. Clin. Invest. 121, 1373–1385 10.1172/JCI43475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li X., Yang Q., Wang X., Zhou Y., Jiang T., Ma Q., Wang Y. J. (2010). The role of microRNA in human lung squamous cell carcinoma. Cancer Genet. Cytogenet. 200, 127–133 10.1016/j.cancergencyto.2010.03.014 [DOI] [PubMed] [Google Scholar]
- Yin R., Zhang S., Wu Y., Fan X., Jiang F., Zhang Z., Feng D., Guo X., Xu L. (2011). microRNA-145 suppresses lung adenocarcinoma-initiating cell proliferation by targeting OCT4. Oncol. Rep. 25, 1747–1754 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang T., Ti X., Shi J., Wu C., Ren X., Yin H. (2010a). Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem. Biophys. Res. Commun. 399, 1–6 10.1016/j.bbrc.2010.07.135 [DOI] [PubMed] [Google Scholar]
- Zhang J. G., Wang J. J., Zhao F., Liu Q., Jiang K., Yang G. H. (2010b). MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin. Chim. Acta 411, 846–852 10.1016/j.cca.2010.06.030 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu T., Huang Y., Liu J. (2011a). microRNA-182 inhibits the proliferation and invasion of human lung adenocarcinoma cells through its effect on human cortical actin-associated protein. Int. J. Mol. Med. 28, 381–388 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wu Y., Feng D., Zhang Z., Jiang F., Yin R., Xu L. (2011b). miR-145 inhibits lung adenocarcinoma stem cells proliferation by targeting OCT4 gene. Zhongguo Fei Ai Za Zhi 14, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ma T., Yang S., Xia M., Xu J., An H., Yang Y., Li S. (2011c). High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. Mol. Cell. Biochem. 357, 363–371 10.1007/s11010-011-0907-1 [DOI] [PubMed] [Google Scholar]
- Zheng D., Haddadin S., Wang Y., Gu L. Q., Perry M. C., Freter C. E., Wang M. X. (2011). Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 4, 575–586 [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Ma X., Sun C., Chen L. (2010). MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem. Biol. Interact. 184, 431–438 10.1016/j.cbi.2010.01.025 [DOI] [PubMed] [Google Scholar]
- Zhu W., Liu X., He J., Chen D., Hunag Y., Zhang Y. K. (2011). Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer 11, 393. 10.1186/1471-2407-11-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Shan X., Wang T., Shu Y., Liu P. (2010a). miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int. J. Cancer 127, 2520–2529 10.1002/ijc.25260 [DOI] [PubMed] [Google Scholar]
- Zhu W., Zhu D., Lu S., Wang T., Wang J., Jiang B., Shu Y., Liu P. (2010b). miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med. Oncol. [Epub ahead of print]. 10.1007/s12032-010-9797-4 [DOI] [PubMed] [Google Scholar]