Abstract
A variable screening procedure via correlation learning was proposed in Fan and Lv (2008) to reduce dimensionality in sparse ultra-high dimensional models. Even when the true model is linear, the marginal regression can be highly nonlinear. To address this issue, we further extend the correlation learning to marginal nonparametric learning. Our nonparametric independence screening is called NIS, a specific member of the sure independence screening. Several closely related variable screening procedures are proposed. Under general nonparametric models, it is shown that under some mild technical conditions, the proposed independence screening methods enjoy a sure screening property. The extent to which the dimensionality can be reduced by independence screening is also explicitly quantified. As a methodological extension, a data-driven thresholding and an iterative nonparametric independence screening (INIS) are also proposed to enhance the finite sample performance for fitting sparse additive models. The simulation results and a real data analysis demonstrate that the proposed procedure works well with moderate sample size and large dimension and performs better than competing methods.
Keywords: Additive model, independent learning, nonparametric regression, sparsity, sure independence screening, nonparametric independence screening, variable selection
1 Introduction
With rapid advances of computing power and other modern technology, high-throughput data of unprecedented size and complexity are frequently seen in many contemporary statistical studies. Examples include data from genetic, microarrays, proteomics, fMRI, functional data and high frequency financial data. In all these examples, the number of variables p can grow much faster than the number of observations n. To be more specific, we assume log p = O(na) for some a ∈ (0, 1/2). Following Fan and Lv (2009), we call it non-polynomial (NP) dimensionality or ultra-high dimensionality. What makes the under-determined statistical inference possible is the sparsity assumption: only a small set of independent variables contribute to the response. Therefore, dimension reduction and feature selection play pivotal roles in these ultra-high dimensional problems.
The statistical literature contains numerous procedures on the variable selection for linear models and other parametric models, such as the Lasso (Tibshirani, 1996), the SCAD and other folded-concave penalty (Fan, 1997; Fan and Li, 2001), the Dantzig selector (Candes and Tao, 2007), the Elastic net (Enet) penalty (Zou and Hastie, 2005), the MCP (Zhang, 2010) and related methods (Zou, 2006; Zou and Li, 2008). Nevertheless, due to the “curse of dimensionality” in terms of simultaneous challenges on the computational expediency, statistical accuracy and algorithmic stability, these methods meet their limits in ultra-high dimensional problems.
Motivated by these concerns, Fan and Lv (2008) introduced a new frame-work for variable screening via correlation learning with NP-dimensionality in the context of least squares. Hall et al. (2009) used a different marginal utility, derived from an empirical likelihood point of view. Hall and Miller (2009) proposed a generalized correlation ranking, which allows nonlinear regression. Huang et al. (2008) also investigated the marginal bridge regression in the ordinary linear model. These methods focus on studying the marginal pseudo-likelihood and are fast but crude in terms of reducing the NP-dimensionality to a more moderate size. To enhance the performance, Fan and Lv (2008) and Fan et al. (2009) introduced some methodological extensions including iterative SIS (ISIS) and multi-stage procedures, such as SIS-SCAD and SIS-LASSO, to select variables and estimate parameters simultaneously. Nevertheless, these marginal screening methods have some methodological challenges. When the covariates are not jointly normal, even if the linear model holds in the joint regression, the marginal regression can be highly nonlinear. Therefore, sure screening based on nonparametric marginal regression becomes a natural candidate.
In practice, there is often little prior information that the effects of the covariates take a linear form or belong to any other finite-dimensional parametric family. Substantial improvements are sometimes possible by using a more flexible class of nonparametric models, such as the additive model , introduced by Stone (1985). It increases substantially the flexibility of the ordinary linear model and allows a data-analytic transform of the covariates to enter into the linear model. Yet, the literature on variable selection in nonparametric additive models are limited. See, for example, Koltchinskii and Yuan (2008), Ravikumar et al. (2009), Huang et al. (2010) and Meier et al. (2009). Koltchinskii and Yuan (2008) and Ravikumar et al. (2009) are closely related with COSSO proposed in Lin and Zhang (2006) with fixed minimal signals, which does not converge to zero. Huang et al. (2010) can be viewed as an extension of adaptive lasso to additive models with fixed minimal signals. Meier et al. (2009) proposed a penalty which is a combination of sparsity and smoothness with a fixed design. Under ultra-high dimensional settings, all these methods still suffer from the aforementioned three challenges as they can be viewed as extensions of penalized pseudo-likelihood approaches to additive modeling. The commonly used algorithm in additive modeling such as backfitting makes the situation even more challenging, as it is quite computationally expensive.
In this paper, we consider independence learning by ranking the magnitude of marginal estimators, nonparametric marginal correlations, and the marginal residual sum of squares. That is, we fit p marginal nonparametric regressions of the response Y against each covariate Xi separately and rank their importance to the joint model according to a measure of the goodness of fit of their marginal model. The magnitude of these marginal utilities can preserve the non-sparsity of the joint additive models under some reasonable conditions, even with converging minimum strength of signals. Our work can be regarded as an important and nontrivial extension of SIS procedures proposed in Fan and Lv (2008) and Fan and Song (2010). Compared with these papers, the minimum distinguishable signal is related with not only the stochastic error in estimating the nonparametric components, but also approximation errors in modeling nonparametric components, which depends on the number of basis functions used for the approximation. This brings significant challenges to the theoretical development and leads to an interesting result on the extent to which the dimensionality can be reduced by nonparametric independence screening. We also propose an iterative nonparametric independence screening procedure, INIS-penGAM, to reduce the false positive rate and stabilize the computation. This two-stage procedure can deal with the aforementioned three challenges better than other methods, as will be demonstrated in our empirical studies.
We approximate the nonparametric additive components by using a B-spline basis. Hence, the component selection in additive models can be viewed as a functional version of the grouped variable selection. An early literature on the group variable selection using group penalized least-squares is Antoniadis and Fan (2001) (see page 966), in which blocks of wavelet coefficients are either killed or selected. The group variable selection was more thoroughly studied in Yuan and Lin (2006), Kim et al. (2006), Wei and Huang (2007) and Meier et al. (2009). Our methods and results have important implications on the group variable selections, as in additive regression, each component can be expressed as a linear combination of a set of basis functions, whose coefficients have to be either killed or selected simultaneously.
The rest of the paper is organized as follows. In Section 2, we introduce the nonparametric independence screening (NIS) procedure in additive models. The theoretical properties for NIS are presented in Section 3. As a methodological extension, INIS-penGAM and its greedy version g-INIS-penGAM are outlined in Section 4. Monte Carlo simulations and a real data analysis in Section 5 demonstrate the effectiveness of the INIS method. We conclude with a discussion in Section 6 and relegate the proofs to Section 7.
2 Nonparametric independence screening
Suppose that we have a random sample from the population
| (1) |
in which X = (X1, … , Xp)T, ε is the random error with conditional mean zero. To expeditiously identify important variables in model (1), without the “curse-of-dimensionality”, we consider the following p marginal nonparametric regression problems:
| (2) |
where P denotes the joint distribution of (X, Y) and L2(P) is the class of square integrable functions under the measure P. The minimizer of (2) is fj = E(Y|Xj), the projection of Y onto Xj. We rank the utility of covariates in model (1) according to, for example, and select a small group of covariates via thresholding.
To obtain a sample version of the marginal nonparametric regression, we employ a B-Spline basis. Let Sn be the space of polynomial splines of degree l ≥ 1 and {Ψjk, k = 1, … , dn} denote a normalized B-Spline basis with ∥Ψjk∥∞ ≤ 1, where ∥ · ∥∞ is the sup norm. For any , we have
for some coefficients . Under some smoothness conditions, the non-parametric projections can well be approximated by functions in . The sample version of the marginal regression problem can be expressed as
| (3) |
where Ψj ≡ Ψj(Xj) = (Ψ1(Xj), ⋯ , Ψdn(Xj))T denotes the dn dimensional basis functions and is the expectation with respect to the empirical measure , i.e., the sample average of . This univariate nonparametric smoothing can be rapidly computed, even for NP-dimensional problems. We correspondingly define the population version of the minimizer of the componentwise least square regression,
where E denotes the expectation under the true model.
We now select a set of variables
| (4) |
where and νn is a predefined threshold value. Such an independence screening ranks the importance according to the marginal strength of the marginal nonparametric regression. This screening can also be viewed as ranking by the magnitude of the correlation of the marginal nonparametric estimate with the response , since . In this sense, the proposed NIS procedure is related to the correlation learning proposed in Fan and Lv (2008).
Another screening approach is to rank according to the descent order of the residual sum of squares of the componentwise nonparametric regressions, where we select a set of variables:
with is the residual sum of squares of the marginal fit and γn is a predefined threshold value. It is straightforward to show that . Hence, the two methods are equivalent.
The nonparametric independence screening reduces the dimensionality from p to a possibly much smaller space with model size or . It is applicable to all models. The question is whether we have mistakenly deleted some active variables in model (1). In other words, whether the procedure has a sure screening property as postulated by Fan and Lv (2008). In the next section, we will show that the sure screening property indeed holds for nonparametric additive models with a limited false selection rate.
3 Sure Screening Properties
In this section, we establish the sure screening properties for additive models with results presented in three steps.
3.1 Preliminaries
We now assume that the true regression function admits the additive structure:
| (5) |
For identifiability, we assume have mean zero. Consequently, the response Y has zero mean, too. Let be the true sparse model with non-sparsity size . We allow p to grow with n and denote it as pn whenever needed.
The theoretical basis of the sure screening is that the marginal signal of the active components does not vanish, where . The following conditions make this possible. For simplicity, let [a, b] be the support of Xj.
- The nonparametric marginal projections belong to a class of functions whose rth derivative f(r) exists and is Lipschitz of order α:
for some positive constant K, where r is a non-negative integer and α ∈ (0, 1] such that d = r + α ≥ 0.5. The marginal density function gj of Xj satisfies 0 < K1 ≤ gj(Xj) ≤ K2 < ∞ on [a, b] for 1 ≤ j ≤ p for some constants K1 and K2.
E{E(Y|Xj)2} ≥ c1dnn−2k, for some 0 < k < d/(2d + 1) and c1 > 0.
Under conditions A and B, the following three facts hold when l ≥ d and will be used in the paper. We state them here for readability.
- Fact 1. There exists a positive constant C1 such that (Stone, 1985)
(6) - Fact 3. There exist some positive constants D1 and D2 such that (Zhou et al., 1998)
(8)
The following lemma shows that the minimum signal of is at the same level of the marginal projection, provided that the approximation error is negligible.
- Lemma 1. Under conditions A–C, we have
provided that for some ξ ∈ (0, 1).
A model selection consistency result can be established with nonparametric independence screening under the partial orthogonality condition, i.e., is independent of . In this case, there is a separation between the strength of marginal signals ∥fnj∥2 for active variables and inactive variables , which are zero. When the separation is sufficiently large, these two sets of variables can be easily identified.
3.2 Sure Screening
In this section, we establish the sure screening properties of the nonparametric independence screening (NIS). We need the following additional conditions:
-
D
∥m∥∞ < B1 for some positive constant B1, where ∥ · ∥∞ is the sup norm.
-
E
The random error are i.i.d. with conditional mean zero and for any B2 > 0, there exists a positive constant B3 such that E[exp(B2|εi|)|Xi] < B3.
-
F
There exist a positive constant c1 and ξ ∈ (0, 1) such that .
The following theorem gives the sure screening properties. It reveals that it is only the size of non-sparse elements sn that matters for the purpose of sure screening, not the dimensionality pn. The first result is on the uniform convergence of to ∥fnj∥2.
Theorem 1. Suppose that Conditions A, B, D and E hold.
- For any c2 > 0, there exist some positive constants c3 and c4 such that
(9) - If, in addition, Conditions C and F hold, then by taking νn = c5dnn−2k with c5 ≤ c1ξ/2, we have
Note that the second part of the upper bound in Theorem 1 is related to the uniform convergence rates of the minimum eigenvalues of the design matrices. It gives an upper bound on the number of basis dn = o(n1/3) in order to have the sure screening property, whereas Condition F requires dn ≥ B4n2k/(2d+1), where B4 = (c1(1 − ξ)/C1)−1/(2d+1).
It follows from Theorem 1 that we can handle the NP-dimensionality:
| (10) |
Under this condition,
i.e., the sure screening property. It is worthwhile to point out that the number of spline basis dn affects the order of dimensionality, comparing with the results of Fan and Lv (2008) and Fan and Song (2010) in which univariate marginal regression is used. Equation (10) shows that the larger the minimum signal level or the smaller the number of basis functions, the higher dimensionality the nonparametric independence screening (NIS) can handle. This is in line with our intuition. On the other hand, the number of basis functions can not be too small, since the approximation error can not be too large. As required by Condition F, dn ≥ B4n2k/(2d+1); the smoother the underlying function, the smaller dn we can take and the higher the dimension that the NIS can handle. If the minimum signal does not converge to zero, as in Lin and Zhang (2006), Koltchinskii and Yuan (2008) and Huang et al. (2010), then k = 0. In this case, dn can be taken to be finite as long as it is sufficiently large so that minimum signal in Lemma 1 exceeds the noise level. By taking dn = n1/(2d+1), the optimal rate for nonparametric regression (Stone, 1985), we have log pn = o(n2(d−1)/(2d+1)). In other words, the dimensionality can be as high as exp{o(n2(d−1)/(2d+1))}.
3.3 Controlling false selection rates
The sure screening property, without controlling false selection rates, is not insightful. It basically states that the NIS has no false negatives. An ideal case for the vanishing false positive rate is that
so that there is a gap between active variables and inactive variables in model (1) when using the marginal nonparametric screener. In this case, by Theorem 1(i), if (9) tends to zero, with probability tending to one that
Hence, by the choice of νn as in Theorem 1(ii), we can achieve model selection consistency:
We now deal with the more general case. The idea is to bound the size of the selected set by using the fact that var(Y) is bounded. In this part, we show that the correlations among the basis functions, i.e., the design matrix of the basis functions, are related to the size of selected models.
Theorem 2. Suppose Conditions A–F hold and var(Y) = O(1). Then, for any νn = c5dnn−2k, there exist positive constants c3 and c4 such that
where Σ = EΨΨT and Ψ = (Ψ1 ⋯ , Ψpn)T.
The significance of the result is that when λmax(Σ) = O(nτ), the selected model size with the sure screening property is only of polynomial order, whereas the original model size is of NP-dimensionality. In other words, the false selection rate converges to zero exponentially fast. The size of the selected variables is of order O(n2k + τ. This is of the same order as in Fan and Lv (2008). Our result is an extension of Fan and Lv (2008), even in this very specific case without the condition 2κ + τ < 1. The results are also consistent with that in Fan and Song (2010): the number of selected variables is related to the correlation structure of the covariance matrix.
In the specific case where the covariates are independent, then the matrix Σ is block diagonal with j-th block Σj. Hence, it follows from (8) that .
4 INIS Method
4.1 Description of the Algorithm
After variable screening, the next step is naturally to select the variables using more refined techniques in the additive model. For example, the penalized method for additive model (penGAM) in Meier et al. (2009) can be employed to select a subset of active variables. This results in NIS-penGAM. To further enhance the performance of the method, in terms of false selection rates, following Fan and Lv (2008) and Fan et al. (2009), we can iteratively employ the large-scale screening and moderate-scale selection strategy, resulting in the INIS-penGAM.
Given the data {(Xi, Yi)}, i = 1, ⋯ , n, for each component fj(·), j = 1, ⋯ , p, we choose the same truncation term dn = O(n1/5). To determine a data-driven thresholding for independence screening, we extend the random permutation idea in Zhao and Li (2010), which allows only 1 − q proportion (for a given q ∈ [0, 1]) of inactive variables to enter the model when X and Y are not related (the null model). The random permutation is used to decouple Xi and Yi so that the resulting data (Xπ(i), Yi) follow a null model, where π(1), ⋯ , π(n) are a random permutation of the index 1, ⋯ , n. The algorithm works as follows:
- Step 1: For every j ∈ {1, ⋯ , p}, we compute
Randomly permute the rows of X, yielding . Letω(q) be the qth quantile of , where
Then, NIS selects the following variables:
In our numerical examples, we use q = 1 (i.e., take the maximum value of the empirical norm of the permuted estimates). Step 2: We apply further the penalized method for additive model (penGAM) in Meier et al. (2009) on the set to select a subset . Inside the penGAM algorithm, the penalty parameter is selected by cross validation.
- Step 3: For every , we minimize
with respect to for all and . This regression reflects the additional contribution of the j-th components conditioning on the existence of the variable set . After marginally screening as in the first step, we can pick a set of indices. Here the size determination is the same as in Step 1, except that only the variables not in are randomly permuted. Then we apply further the penGAM algorithm on the set to select a subset .(11) Step 4: We iterate the process until or .
Here are a few comments about the method. In Step 2, we use the penGAM method. In fact, any variable selection method for additive models will work such as the SpAM in Ravikumar et al. (2009) and also the adaptive group LASSO for additive models in Huang et al. (2010). A similar sample splitting idea as described in Fan et al. (2009) can be applied here to further reduce false selection rate.
4.2 Greedy INIS (g-INIS)
We now propose a greedy modification to the INIS algorithm to speed up the computation and to enhance the performance. Specifically, we restrict the size of the set in the iterative screening steps to be at most p0, a small positive integer, and the algorithm stops when none of the variables is recruited, i.e., exceeding the thresholding for the null model. In the numerical studies, p0 is taken to be one for simplicity. This greedy version of the INIS algorithm is called “g-INIS”.
When p0 = 1, the g-INIS method is connected with the forward selection (Efroymson, 1960; Draper and Smith, 1966). Recently, Wang (2009) showed that under certain technical conditions, forward selection can also achieve the sure screening property. Both g-INIS and forward selection recruit at most one new variable into the model at a time. The major difference is that unlike the forward selection which keeps a variable once selected, g-INIS has a deletion step via penalized least-squares that can remove multiple variables. This makes the g-INIS algorithm more attractive since it is more flexible in terms of recruiting and deleting variables.
The g-INIS is particularly effective when the covariates are highly correlated or conditionally correlated. In this case, the original INIS method tends to select many unimportant variables that have high correlation with important variables as they, too, have large marginal effects on the response. Although greedy, the g-INIS method is better at choosing true positives due to more stringent screening and improves the chance of the remaining important variables to be selected in subsequent stages due to less false positives at each stage. This leads to conditioning on a smaller set of more relevant variables and improve the overall performance. From our numerical experience, the g-INIS method outperforms the original INIS method in all examples in terms of higher true positive rate, smaller false positive rate and smaller prediction error.
5 Numerical Results
In this section, we will illustrate our method by studying the performance on the simulated data and a real data analysis. Part of the simulation settings are adapted from Fan and Lv (2008), Meier et al. (2009), Huang et al. (2010), and Fan and Song (2010).
5.1 Comparison of Minimum Model Size
We first illustrate the behavior of the NIS procedure under different correlation structures. Following Fan and Song (2010), the minimum model size(MMS) required for the NIS procedure and the penGAM procedure to have the sure screening property, i.e., to contain the true model , is used as a measure of the effectiveness of a screening method. We also include the correlation screening of Fan and Lv (2008) for comparison. The advantage of the MMS method is that we do not need to choose the thresholding parameter or penalized parameters. For NIS, we take dn = [n1/5] + 2 = 5. We set n = 400 and p = 1000 for all examples.
Example 1. Following Fan and Song (2010), let be i.i.d standard normal random variables and
where are standard normally distributed. We consider the following linear model as a specific case of the additive model: Y = β*T X + ε, in which ε ~ N(0, 3) and β* = (1, −1, ε)T has s non-vanishing components, taking values ±1 alternately.
Example 2. In this example, the data is generated from the simple linear regression Y = X1+X2+X3+ε, where ε ~ N(0, 3). However, the covariates are not normally distributed: {Xk}k≠2 are i.i.d standard normal random variables whereas , where . In this case, E(Y|X1) and E(Y|X2) are nonlinear.
The minimum model size(MMS) for each method and its associated robust estimate of the standard deviation(RSD = IQR/1.34) are shown in Table 1. The column “NIS”, “penGAM”, and “SIS” summarizes the results on the MMS based on 100 simulations, respectively for the nonparametric independence screening in the paper, penalized method for additive model of Meier et al. (2009), and the linear correlation ranking method of Fan and Lv (2008). For Example 1, when the nonsparsity size s > 5, the irrepresentable condition required for the model selection consistency of LASSO fails. For these cases, penGAM fails even to include the true model until the last step. In contrast, the proposed nonparametric independence screening performs reasonably well. It is also worth noting that SIS performs better than NIS in the first example, particularly for s = 24. This is due to the fact that the true model is linear and the covariates are jointly normally distributed, which implies that the marginal projection is also linear. In this case, NIS selects variables from pdn parameters whereas SIS selects only from p parameters. However, for the nonlinear problem like Example 2, both nonlinear method NIS and penGAM behave nicely, whereas SIS fails badly even though the underlying true model is indeed linear.
Table 1.
Minimum model size and robust estimate of standard deviations (in parentheses).
| Model | NIS | PenGAM | SIS |
|---|---|---|---|
| Ex 1 (s = 3; SNR ≈ 1:01) | 3(0) | 3(0) | 3(0) |
| Ex 1 (s = 6; SNR ≈ 1:99 ) | 56(0) | 1000(0) | 56(0) |
| Ex 1 (s = 12; SNR ≈ 4:07) | 66(7) | 1000(0) | 62(1) |
| Ex 1 (s = 24; SNR ≈ 8:20) | 269(134) | 1000(0) | 109(43) |
| Ex 2 (SNR ≈ 0:83) | 3(0) | 3(0) | 360(361) |
5.2 Comparison of Model Selection and Estimation
As in the previous section, we set n = 400 and p = 1000 for all the examples to demonstrate the power of our newly proposed methods INIS and g-INIS. Here in the NIS step, we fix dn = 5 as in the last subsection. The number of simulations is 100. Here, we use five-fold cross validation in Step 2 of the INIS algorithm. For simplicity of notations, we let
and
Example 3. Following Meier et al. (2009), we generate the data from the following additive model:
The covariates X = (X1, ⋯ , Xp)T are simulated according to the random effect model
where W1, ⋯ , Wp and U are i.i.d. Unif(0, 1) and ε ~ N(0, 1). When t = 0, the covariates are all independent, and when t = 1 the pairwise correlation of covariates is 0.5.
Example 4. Again, we adapt the simulation model from Meier et al. (2009). This example is a more difficult case than Example 3 since it has 12 important variables with different coefficients.
where ε ~ N(0, 1). The covariates are simulated as in Example 3.
Example 5. We follow the simulation model of Fan et al. (2009), in which Y = β1X1 + β2X2 + β3X3 + β4X4 + ε is simulated, where ε ~ N(0, 1). The covariates X1, ⋯ , Xp are jointly Gaussian, marginally N(0, 1), and with for all i ≠ 4 and corr(Xi, Xj) = 1/2 if i and j are distinct elements of {1, ⋯ , p}\{4}. The coefficients β1 = 2, β2 = 2, β3 = 2, β4 = , and βj = 0 for j > 4 are taken so that X4 is independent of Y, even though it is the most important variable in the joint model, in terms of the regression coefficient.
For each example, we compare the performances of INIS-penGAM, g-INIS-penGAM proposed in the paper, penGAM(Meier et al., 2009), and ISIS-SCAD (Fan et al., 2009) which aims for sparse linear model. Their results are shown respectively in the rows “INIS”, “g-INIS”, “penGAM” and “ISIS” of Table 2, in which the True Positives(TP), False Positives(FP), Prediction Error(PE) and Computation Time (Time) are reported for each method. Here the prediction error is calculated on an independent test data set of size n/2.
Table 2.
Average values of the numbers of true (TP) and false (FP) positives, prediction error (PE), and Time (in seconds). Robust standard deviations are given in parentheses.
| Model | Method | TP | FP | PE | Time |
|---|---|---|---|---|---|
| Ex 3(t = 0) (SNR ≈ 9.02) |
INIS | 4. 00(0. 00) | 2.58(2.24) | 3. 02(0. 34) | 18. 50(7. 22) |
| g-INIS | 4. 00(0. 00) | 0. 67(0. 75) | 2. 92(0. 30) | 25. 03(4. 87) | |
| penGAM | 4. 00(0. 00) | 31. 86(23. 51) | 3. 30(0. 40) | 180. 63(6. 92) | |
| ISIS | 3. 03(0. 00) | 29.97(0.00) | 15.95(1.74) | 12. 95(4. 18) | |
|
| |||||
| Ex 3(t = 1) (SNR ≈ 7.58) |
INIS | 3. 98(0. 00) | 15.76(6.72) | 2. 97(0. 39) | 78. 80(26. 91) |
| g-INIS | 4. 00(0. 00) | 0. 98(1. 49) | 2. 61(0. 26) | 33. 89(9. 99) | |
| penGAM | 4. 00(0. 00) | 39. 21(24. 63) | 2. 97(0. 28) | 254. 06(13. 06) | |
| ISIS | 3. 01(0. 00) | 29.99(0.00) | 12. 91(1. 39) | 18. 59(4. 37) | |
|
| |||||
| Ex 4(t = 0) (SNR ≈ 8.67) |
INIS | 11. 97(0. 00) | 3.22(1.49) | 0.97(0.11) | 73.60(25.77) |
| g-INIS | 12. 00(0. 00) | 0. 73(0. 75) | 0.91(0.10) | 160. 75(19. 94) | |
| penGAM | 11. 99(0. 00) | 80. 10(18. 28) | 1. 27(0. 14) | 233. 72(10. 25) | |
| ISIS | 7.96(0.75) | 25.04(0.75) | 4.70(0.40) | 12. 89(5. 00) | |
|
| |||||
| Ex 4(t = 1) (SNR ≈ 10.89) |
INIS | 10. 01(1. 49) | 15.56(0.93) | 1.03(0.13) | 125.11(39.99) |
| g-INIS | 10. 78(0. 75) | 1.08(1.49) | 0.87(0.11) | 156. 37(28. 58) | |
| penGAM | 10. 51(0. 75) | 62. 11(26. 31) | 1. 13(0. 12) | 278.61(16.93) | |
| ISIS | 6.53(0.75) | 26.47(0.75) | 4.30(0.44) | 17.02(4.01) | |
|
| |||||
| Ex 5 (SNR ≈ 6.11) | INIS | 3. 99(0. 00) | 21.96(0.00) | 1. 62(0. 18) | 94. 50(7. 12) |
| g-INIS | 4. 00(0. 00) | 1. 04(1. 49) | 1. 16(0. 12) | 39.78(12.45) | |
| penGAM | 3. 00(0. 00) | 195. 03(21. 08) | 1.93(0.28) | 1481. 12(181. 93) | |
| ISIS | 4. 00(0. 00) | 29.00(0.00) | 1.40(0.17) | 17. 78(3. 85) | |
First of all, for the greedy modification, g-INIS-penGAM, the number of false positive variables is approximately 1 for all examples and the number of false positive for both INIS-penGAM and ISIS-SCAD are much smaller than that for penGAM. In terms of false positives, we can see that in Examples 3 and 4, INIS-penGAM and penGAM have similar performance, whereas penGAM misses one variable most of the time in Example 5. The linear method ISIS-SCAD missed important variables in the nonlinear models in Examples 3 and 4.
One may notice that in Example 4 (t = 1), even INIS and g-INIS miss more than one variables on average. To explore the reason, we took a close look at the iterative process for this example and find out the variable X1 and X2 are missed quite often. The explanation is that although the overall SNR (Signal to Noise Ratio) for this example is around 10.89, the individual contributions to the total signal vary significantly. Now, let us introduce the notion of individual SNR. For example, var(m1(X1))/var(ε) in the additive model
is the individual SNR for the first component under the oracle model where m2, ⋯ , mp are known. In Example 4 (t = 1), the variance of all 12 components are as follows:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.08 | 0.09 | 0.21 | 0.26 | 0.19 | 0.20 | 0.47 | 0.58 | 0.33 | 0.36 | 0.84 | 1.03 |
We can see that the variance varies a lot among the 12 components, which leads to very different marginal SNRs. For example, the individual SNR for the first component is merely 0.08/0.518 = 0.154, which is very challenging to be detected. With the overall SNR fixed, the individual SNRs play an important role in measuring the difficulty for selecting individual variables
In the perspective of the prediction error, INIS-penGAM, g-INIS-penGAM and penGAM outperforms ISIS-SCAD in the nonlinear models whereas their performances are worse than ISIS-SCAD in the linear model, Example 5. Overall, it is quite clear that the greedy modification g-INIS is a competing variable selection method in ultra-high dimensional additive models where we have very low false selection rate, small prediction errors, and fast computation.
5.3 dn and SNR
In this subsection, we conduct simulation study to investigate the performance of INIS-penGAM estimator under different SNR settings using different number (dn) of basis functions.
Example 6. We generate the data from the following additive model:
where the covariates X = (X1, ⋯ , Xp)T are simulated according to Example 3. Here C takes a series of different values (C2 = 2, 1, 0.5, 0.25) to make the corresponding SNR = 0.5, 1, 2, 4. We report the results of using number of basis functions dn = 2, 4, 6, 8, in Tables 4 and 5 in the Appendix.
From Table 4 in the Appendix where all the variables are independent, both methods have very good true positives under various SNR when dn is not too large. However, for the case of SNR = 0.5 and dn = 16, the INIS and penGAM perform poorly in terms of low true positive rate. This is due to the fact that when dn is large, the estimation variance will be large and this makes it difficult to differentiate the active variables from inactive ones when the signals are weak.
Now let us have a look at the more difficult case in Table 5 (in the Appendix) where pairwise correlation between variables is 0.5. We can see that INIS have a competitive performance under various SNR values except when dn = 16. When SNR = 0.5, we can not achieve sure screening under the current sample size and configuration for the aforementioned reasons.
5.4 An analysis on Affymetric GeneChip Rat Genome 230 2.0 Array
We use the data set reported in Scheetz et al. (2006) and analyzed by Huang et al. (2010) to illustrate the application of the proposed method. For this data set, 120 twelve-week-old male rats were selected for tissue harvesting from the eyes and for microarray analysis. The microarrays used to analyze the RNA from the eyes of these animals contain over 31,042 different probe sets (Affymetric GeneChip Rat Genome 230 2.0 Array). The intensity values were normalized using the robust multi-chip averaging method (Irizarry et al., 2003) method to obtain summary expression values for each probe set. Gene expression levels were analyzed on a logarithmic scale.
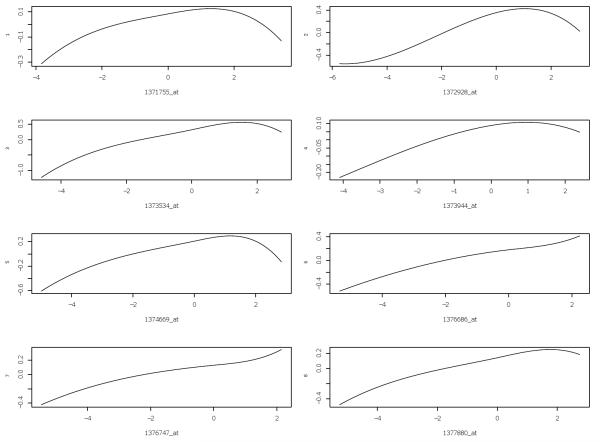
Following Huang et al. (2010), we are interested in finding the genes that are related to the gene TRIM32, which was recently found to cause Bardet-Biedl syndrome (Chiang et al., 2006), and is a genetically heterogeneous disease of multiple organ systems including the retina. Although over 30,000 probe sets are represented on the Rat Genome 230 2.0 Array, many of them are not expressed in the eye tissue. We only focus on the 18975 probes which are expressed in the eye tissue. We use our INIS-penGAM method directly on this dataset, where n = 120 and p = 18975, and the method is denoted as INIS-penGAM (p = 18975). Direct application of penGAM approach on the whole dataset is too slow. Following Huang et al. (2010), we use 2000 probe sets that are expressed in the eye and have highest marginal correlation with TRIM32 in the analysis. On the subset of the data (n = 120, p = 2000), we apply the INIS-penGAM and penGAM to model the relation between the expression of TRIM32 and those of the 2000 genes. For simplicity, we did not implement g-INIS-penGAM. Prior to the analysis, we standardize each probe to be of mean 0 and variance 1. Now, we have three different estimators, INIS-penGAM (p = 18975), INIS-penGAM (p = 2000) and penGAM (p = 2000). The INIS-penGAM (p = 18975) selects the following 8 probes: 1371755_at, 1372928_at, 1373534_at, 1373944_at, 1374669_at, 1376686_at, 1376747_at, 1377880_at. The INIS-penGAM (p = 2000) selects the following 8 probes: 1376686_at, 1376747_at, 1378590_at, 1373534_at, 1377880_at, 1372928_at, 1374669_at, 1373944_at. On the other hand, the penGAM (p = 2000) selects 32 probes. The residual sum of squares (RSS) for these fittings are 0.24, 0.26 and 0.1 for INIS-penGAM (p = 18975), INIS-penGAM (p = 2000) and penGAM (p = 2000), respectively.
In order to further evaluate the performances of the two methods, we use cross-validation and compare the prediction mean square error (PE). We randomly partition the data into a training set of 100 observations and a test set of 20 observations. We compute the number of probes selected using the 100 observations and the prediction errors on these 20 test sets. This process is repeated 100 times. Table 3 gives the average values and their associated robust standard deviations over 100 replications. It is clear in the table that by applying the INIS-penGAM approach, we select far fewer genes and give smaller prediction error. Therefore, in this example, the INIS-penGAM provides the biological investigator a more targeted list of probe sets, which could be very useful in further study.
Table 3.
Mean Model Size (MS) and Prediction Error (PE) over 100 repetitions and their robust standard deviations(in parentheses) for INIS (p = 18975), INIS (p = 2000) and penGAM (p = 2000).
| Method | MS | PE |
|---|---|---|
| INIS (p = 18975) | 7.73(0.00) | 0.47(0.13) |
| INIS (p = 2000) | 7.68(0.75) | 0.44(0.15) |
| penGAM (p = 2000) | 26.71(14.93) | 0.48(0.16) |
6 Remarks
In this paper, we studied the nonparametric independence screening (NIS) method for variable selection in additive models. B-spline basis functions are used for fitting the marginal nonparametric components. The proposed marginal projection criteria is an important extension of the marginal correlation. Iterative NIS procedures are also proposed such that variable selection and coefficient estimation can be achieved simultaneously. By applying the INIS-penGAM method, we can preserve the sure screening property and substantially reduce the false selection rate. A greedy modification of the method g-INIS-penGAM is proposed to further reduce the false selection rate. Moreover, we can deal with the case where some variable is marginally uncorrelated but jointly correlated with the response. The proposed method can be easily generalized to generalized additive model with appropriate conditions.
As the additive components are specifically approximated by truncated series expansions with B-spline bases in this paper, the theoretical results should hold in general and the proposed framework can be readily adaptive to other smoothing methods with additive models (Horowitz et al., 2006; Silverman, 1984), such as local polynomial regression (Fan and Jiang, 2005), wavelets approximations(Antoniadis and Fan, 2001; Sardy and Tseng, 2004) and smoothing spline (Speckman, 1985). This is an interesting topic for future research.
7 Proofs
Proof of Lemma 1.
By the property of the least-squares, E(Y − fnj)fnj = 0 and E(Y − fnj)fnj = 0. Therefore,
It follows from this and the orthogonal decomposition fj = fnj + (fj − fnj) that
The desired result follows from Condition C together with Fact 1.
The following two types of Bernstein’s inequality in van der Vaart and Wellner (1996) will be needed. We reproduce them here for the sake of read-ability.
Lemma 2 (Bernstein’s inequality, Lemma 2.2.9, van der Vaart and Wellner (1996)). For independent random variables Y1, ⋯ , Yn with bounded ranges [−M, M] and zero means,
for v ≥ var(Y1 + ⋯ + Yn).
Lemma 3 (Bernstein’s inequality, Lemma 2.2.11, van der Vaart and Wellner (1996)). Let Y1, ⋯ , Yn be independent random variables with zero mean such that E|Yi|m ≤ m!Mm−2vi/2, for every m ≥ 2 (and all i) and some constants M and vi. Then
for v ≥ v1 + ⋯ vn.
The following two lemmas will be needed to prove Theorem 1.
Lemma 4. Under Conditions A, B and D, for any δ > 0, there exist some positive constants c6 and c7 such that
for k = 1, ⋯ , dn, j = 1, ⋯ , p.
Proof of Lemma 4.
Denote by Tjki = Ψjk(Xij)Yi − EΨjk(Xij)Yi. Since Yi = m(Xi) + εi, we can write Tjki = Tjki1 + Tjki2, where
and Tjki2 = Ψjk(Xij)εi.
By Conditions A, B, D and Fact 2, recalling ∥Ψjk∥∞ ≤ 1, we have
| (12) |
By Bernstein’s inequality (Lemma 2), for any δ1 > 0,
| (13) |
Next, we bound the tails of Tjki2. For every r ≥ 2,
where the last inequality utilizes Condition E and Fact 2. By Bernstein’s inequality (Lemma 3), for any δ2 > 0,
| (14) |
Combining (13) and (14), the desired result follows by taking and c7 = max(2/3B1, .
Throughout the rest of the proof, for any matrix A, let be the operator norm and ∥A∥∞ = maxi,j |Aij| be the infinity norm. The next lemma is about the tail probability of the eigenvalues of the design matrix.
Lemma 5. Under Conditions A and B, for any δ > 0,
In addition, for any given constant c4, there exists some positive constant c8 such that
| (15) |
Proof of Lemma 5.
For any symmetric matrices A and B and any ∥x∥ = 1, where ∥ · ∥ is the Euclidean norm,
Taking minimum among ∥x∥ = 1 on the left side, we have
which is equivalent to λmin(A + B) ≥ λmin(A) + λmin(B).
Then we have
which is the same as
By switching the roles of A and B, we also have
In other words,
| (16) |
Let . Then, it follows from (16) that
| (17) |
We now bound the right-hand side of (17). Let be the (i, l) entry of Dj. Then, it is easy to see that for any ∥x∥ = 1,
| (18) |
Thus,
On the other hand, by using (18) again, we have
We conclude that
The same bound on |λmin(−Dj)| can be obtained by using the same argument. Thus, by (17), we have
| (19) |
We now use Bernstein’s inequality to bound the right-hand side of (19). Since ∥Ψjk∥∞ ≤ 1, and by using Fact 2, we have that
By Bernstein’s inequality (Lemma 2), for any δ > 0,
| (20) |
It follows from (19), (20) and the union bound of probability that
This completes the proof of the first inequality.
To prove the second inequality, let us take in (20), where c9 ∈ (0, 1). By recalling Fact 3, it follows that
| (21) |
for some positive constant c4. The second part of the lemma thus follows from the fact that λmin(H)−1 = λmax(H−1), if we establish
| (22) |
by using (21), where c8 = 1/(1 − c9) − 1.
We now deduce (22) from (21). Let and . Then A > 0 and B > 0. We aim to show for a ∈ (0,1),
where c = 1/(1 − a) − 1.
Since
we have
Note that for a ∈ (0, 1), we have 1 − 1/(1 + a) < 1/(1 − a) − 1. Then it follows that
which is equivalent to |A – B| ≥ aB.
This concludes the proof of the lemma.
Proof of Theorem 1.
We first show part (i). Recall that
and
Let , , a = EΨjY. and . By some algebra,
we have
| (23) |
where
Note that
| (24) |
By Lemma 4 and the union bound of probability,
| (25) |
Recall the result in Lemma 5 that, for any given constant c4, there exists a positive constant c8 such that
Since by Fact 3,
it follows that
| (26) |
Combining (24)–(26) and the union bound of probability, we have
| (27) |
To bound S2, we note that
| (28) |
Since by Condition D,
| (29) |
it follows from (25), (26), (28), (29) and the union bound of probability that
| (30) |
Now we bound S3. Note that
| (31) |
By the fact that ∥AB∥ ≤ ∥A∥ · ∥B∥, we have
| (32) |
For any ∥x∥ = 1 and dn-dimensional square matrix D,
| (33) |
Therefore, ∥D∥ ≤ dn∥D∥∞. We conclude that
By (20), (26), (29), (32), (33) and the union bound of probability, it follows that
| (34) |
It follows from (23), (27), (30), (34) and the union bound of probability that for some positive constants c10, c11 and c12,
| (35) |
In (35), let for any given c2 > 0, i.e., taking , there exist some positive constants c3 and c4 such that
The first part thus follows the union bound of probability.
To prove the second part, note that on the event
by Lemma 1, we have
| (36) |
Hence, by the choice of 03BD;n we have . The result now follows from a simple union bound:
This completes the proof.
Proof of Theorem 2. The key idea of the proof is to show that
| (37) |
If so, by definition and ∥Ψjk∥∞ ≤ 1, we have
This implies that the number of {j : ∥fnj∥2 > εdnn−2k} can not exceed O(n2k λmax(Σ)) for any ε > 0. Thus, on the set
the number of can not exceed the number of {j: ∥fnj∥2 > εdnn−2k}, which is bounded by O{n2k λmax(Σ)}. By taking ε = c5/2, we have
The conclusion follows from Theorem 1(i).
It remains to prove (37). Note that (37) is more related to the joint regression rather than the marginal regression. Let
which is the joint regression coefficients in the population. By the score equation of αn, we get
Hence
Now, it follows from the orthogonal decomposition that
Since var(Y) = O(1), we conclude that var(ΨT αn = O(1), i.e.
This completes the proof. □.
Figure 1.
Fitted regression functions for the 8 probes that are selected by INIS-penGAM (p = 18975).
A APPENDIX: Tables for Simulation Results of Section 5.3
Table 4.
Average values of the numbers of true (TP), false (FP) positives, prediction error (PE), computation time (Time) for Example 6 (t = 0). Robust standard deviations are given in parentheses.
| SNR | dn | Method | TP | FP | PE | Time |
|---|---|---|---|---|---|---|
| 0.5 | 2 | INIS | 3.96(0.00) | 2.28(1.49) | 7.74(0.79) | 16.09(5.32) |
| penGAM | 4.00(0.00) | 27.85(16.98) | 8.07(0.92) | 354.46(31.48) | ||
| 4 | INIS | 3.93(0.00) | 2.29(1.68) | 7.90(0.81) | 21.68(8.95) | |
| penGAM | 3.99(0.00) | 25.61(13.62) | 8.21(0.84) | 421.17(35.71) | ||
| 8 | INIS | 3.81(0.00) | 2.59(2.24) | 8.16(1.08) | 33.10(15.79) | |
| penGAM | 3.95(0.00) | 34.59(20.34) | 8.49(0.82) | 484.17(179.70) | ||
| 16 | INIS | 3.38(0.75) | 2.02(1.49) | 8.60(1.13) | 42.69(20.13) | |
| penGAM | 3.74(0.00) | 33.48(23.88) | 9.04(0.93) | 685.97(267.43) | ||
| 1.0 | 2 | INIS | 4.00(0.00) | 2.16(2.24) | 3.98(0.34) | 16.03(5.74) |
| penGAM | 4.00(0.00) | 26.51(14.18) | 4.20(0.46) | 284.85(20.30) | ||
| 4 | INIS | 4.00(0.00) | 2.08(1.49) | 3.97(0.45) | 20.80(8.57) | |
| penGAM | 4.00(0.00) | 28.33(15.49) | 4.24(0.47) | 362.02(81.43) | ||
| 8 | INIS | 4.00(0.00) | 2.72(2.24) | 4.04(0.43) | 35.79(18.38) | |
| penGAM | 4.00(0.00) | 36.50(21.83) | 4.37(0.47) | 427.60(152.53) | ||
| 16 | INIS | 4.00(0.00) | 1.80(1.49) | 4.26(0.45) | 46.81(21.47) | |
| penGAM | 4.00(0.00) | 38.60(19.78) | 4.80(0.57) | 595.87(197.06) | ||
| 2.0 | 2 | INIS | 4.00(0.00) | 2.03(2.24) | 2.12(0.17) | 15.92(5.42) |
| penGAM | 4.00(0.00) | 25.89(13.06) | 2.25(0.24) | 235.69(13.32) | ||
| 4 | INIS | 4.00(0.00) | 2.38(2.24) | 2.06(0.22) | 23.54(9.08) | |
| penGAM | 4.00(0.00) | 30.37(17.16) | 2.21(0.26) | 341.13(19.44) | ||
| 8 | INIS | 4.00(0.00) | 2.79(2.24) | 2.03(0.21) | 38.56(19.58) | |
| penGAM | 4.00(0.00) | 38.51(16.42) | 2.24(0.26) | 396.84(20.51) | ||
| 16 | INIS | 4.00(0.00) | 1.77(1.49) | 2.17(0.25) | 48.40(24.65) | |
| penGAM | 4.00(0.00) | 42.58(16.60) | 2.54(0.30) | 540.89(165.39) | ||
| 4.0 | 2 | INIS | 4.00(0.00) | 2.06(2.24) | 1.19(0.13) | 17.74(6.42) |
| penGAM | 4.00(0.00) | 28.57(14.37) | 1.27(0.15) | 213.43(12.09) | ||
| 4 | INIS | 4.00(0.00) | 2.33(1.49) | 1.09(0.10) | 23.28(9.37) | |
| penGAM | 4.00(0.00) | 30.75(17.35) | 1.18(0.14) | 300.69(12.21) | ||
| 8 | INIS | 4.00(0.00) | 2.88(2.24) | 1.02(0.12) | 39.21(19.17) | |
| penGAM | 4.00(0.00) | 40.51(17.54) | 1.14(0.14) | 340.06(11.49) | ||
| 16 | INIS | 4.00(0.00) | 1.72(1.49) | 1.10(0.12) | 49.79(25.78) | |
| penGAM | 4.00(0.00) | 45.77(19.03) | 1.33(0.16) | 481.19(141.51) | ||
Table 5.
Average values of the numbers of true (TP), false (FP) positives, prediction error (PE), computation time (Time) for Example 6 (t = 1). Robust standard deviations are given in parentheses.
| SNR | dn | Method | TP | FP | PE | Time |
|---|---|---|---|---|---|---|
| 0.5 | 2 | INIS | 3.35(0.75) | 33.67(8.96) | 9.49(1.28) | 196.87(91.48) |
| penGAM | 3.10(0.00) | 17.74(15.11) | 7.92(0.89) | 1107.78(385.95) | ||
| 4 | INIS | 3.02(0.00) | 20.22(2.43) | 8.70(1.14) | 109.51(56.11) | |
| penGAM | 2.78(0.00) | 15.91(10.07) | 7.99(0.91) | 734.08(227.55) | ||
| 8 | INIS | 2.51(0.75) | 10.48(0.75) | 8.37(0.89) | 65.12(16.64) | |
| penGAM | 2.59(0.75) | 16.47(9.70) | 8.13(0.90) | 624.31(56.23) | ||
| 16 | INIS | 2.10(0.00) | 4.47(0.75) | 8.44(1.00) | 46.84(15.61) | |
| penGAM | 2.41(0.75) | 15.56(10.63) | 8.42(0.97) | 786.45(244.02) | ||
| 1.0 | 2 | INIS | 3.83(0.00) | 32.46(9.70) | 4.86(0.60) | 164.97(64.14) |
| penGAM | 3.64(0.75) | 24.61(21.08) | 4.19(0.49) | 849.23(294.03) | ||
| 4 | INIS | 3.56(0.75) | 20.53(1.68) | 4.42(0.52) | 118.14(43.97) | |
| penGAM | 3.46(0.75) | 22.07(16.04) | 4.18(0.49) | 614.93(97.36) | ||
| 8 | INIS | 3.09(0.00) | 10.67(0.75) | 4.28(0.49) | 71.16(32.10) | |
| penGAM | 3.12(0.00) | 19.92(10.63) | 4.30(0.50) | 548.60(33.88) | ||
| 16 | INIS | 2.68(0.75) | 4.18(0.75) | 4.45(0.52) | 46.08(15.35) | |
| penGAM | 2.95(0.00) | 16.39(11.19) | 4.57(0.55) | 710.56(199.86) | ||
| 2.0 | 2 | INIS | 3.99(0.00) | 29.45(11.57) | 2.55(0.38) | 139.67(70.45) |
| penGAM | 3.97(0.00) | 36.57(22.57) | 2.25(0.28) | 626.84(210.44) | ||
| 4 | INIS | 3.93(0.00) | 19.12(3.73) | 2.26(0.24) | 111.01(21.82) | |
| penGAM | 3.91(0.00) | 31.31(20.52) | 2.19(0.23) | 481.87(52.11) | ||
| 8 | INIS | 3.50(0.75) | 10.29(0.75) | 2.21(0.23) | 78.06(32.23) | |
| penGAM | 3.71(0.75) | 27.06(19.03) | 2.28(0.29) | 448.38(26.63) | ||
| 16 | INIS | 2.93(0.00) | 4.07(0.00) | 2.42(0.32) | 51.69(1.10) | |
| penGAM | 3.22(0.00) | 19.51(12.13) | 2.53(0.30) | 661.93(46.27) | ||
| 4.0 | 2 | INIS | 4.00(0.00) | 29.47(11.38) | 1.45(0.21) | 144.22(72.54) |
| penGAM | 4.00(0.00) | 37.27(20.71) | 1.27(0.17) | 533.98(69.29) | ||
| 4 | INIS | 3.99(0.00) | 17.36(5.22) | 1.17(0.12) | 102.97(32.71) | |
| penGAM | 4.00(0.00) | 38.71(20.34) | 1.16(0.11) | 403.32(28.29) | ||
| 8 | INIS | 3.78(0.00) | 10.00(0.00) | 1.13(0.16) | 88.79(12.02) | |
| penGAM | 3.99(0.00) | 41.42(15.86) | 1.19(0.13) | 402.92(16.94) | ||
| 16 | INIS | 3.02(0.00) | 3.98(0.00) | 1.36(0.15) | 49.13(1.85) | |
| penGAM | 3.72(0.75) | 29.58(19.40) | 1.43(0.18) | 556.31(35.48) | ||
Footnotes
The financial support from NSF grants DMS-0714554, DMS-0704337, DMS-1007698 and NIH grant R01-GM072611 are greatly acknowledged. The authors are in deep debt to Dr. Lukas Meier for sharing the codes of penGAM. The authors thank the editor, the associate editor, and referees for their constructive comments.
References
- Antoniadis A, Fan J. Regularization of wavelet approximations. Journal of the American Statistical Association. 2001;96:939–967. [Google Scholar]
- Candes E, Tao T. The dantzig selector: statistical estimation when p is much larger than n (with discussion) The Annals of Statistics. 2007;35:2313–2404. [Google Scholar]
- Chiang AP, Beck JS, Yen H-J, Tayeh MK, Scheetz TE, Swiderski R, Nishimura D, Braun TA, Kim K-Y, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, Sheffield VC. Homozygosity mapping with snp arrays identifies trim32, an e3 ubiquitin ligase, as a bardetcbiedl syndrome gene (bbs11) PNAS. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper NR, Smith H. Applied regression analysis. John Wiley & Sons Inc.; New York: 1966. [Google Scholar]
- Efroymson MA. Mathematical methods for digital computers. Wiley; New York: 1960. Multiple regression analysis; pp. 191–203. [Google Scholar]
- Fan J. Comments on “wavelets in statistics: A review” by a. antoniadis. j. Journal of the American Statistical Association. 1997;6:131–138. [Google Scholar]
- Fan J, Jiang J. Nonparametric inferences for additive models. Journal of the American Statistical Association. 2005;100:890–907. [Google Scholar]
- Fan J, Li R. Variable selection via nonconcave penalized like-lihood and its oracle properties. Journal of the American Statistical Association. 2001;96:1348–1360. [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Lv J. Non-concave penalized likelihood with np-dimensionality. Manuscript. 2009 doi: 10.1109/TIT.2011.2158486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Samworth R, Wu Y. Ultra-dimensional variable selection via independent learning: beyond the linear model. Journal of Machine Learning Research. 2009;10:1829–1853. [PMC free article] [PubMed] [Google Scholar]
- Fan J, Song R. Sure independence screening in generalized linear models with np-dimensionality. The Annals of Statistics. 2010 To appear. [Google Scholar]
- Hall P, Miller H. Using generalised correlation to effect variable selection in very high dimensional problems. The Journal of Computational and Graphical Statistics. 2009 To appear. [Google Scholar]
- Hall P, Titterington D, Xue J. Tilting methods for assessing the influence of components in a classifier. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2009;71:783–803. [Google Scholar]
- Horowitz J, Klemelä J, Mammen E. Optimal estimation in additive regression models. Bernoulli. 2006;12:271–298. [Google Scholar]
- Huang J, Horowitz J, Ma S. Asymptotic properties of bridge estimators in sparse high-dimensional regression models. The Annals of Statistics. 2008;36:587–613. [Google Scholar]
- Huang J, Horowitz J, Wei F. Variable selection in non-parametric additive models. The Annals of Statistics. 2010;38:2282–2313. doi: 10.1214/09-AOS781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim J, Kim Y. Blockwise sparse regression. Statistica Sinica. 2006;16:375–390. [Google Scholar]
- Koltchinskii V, Yuan M. Sparse recovery in large ensembles of kernel machines. In: Servedio RA, Zhang T, editors. In CLOT. Omni-press; 2008. pp. 229–238. [Google Scholar]
- Lin Y, Zhang HH. Component selection and smoothing in multivariate nonparametric regression. The Annals of Statistics. 2006;34:2272–2297. [Google Scholar]
- Meier L, Geer V, Bühlmann P. High-dimensional additive modeling. The Annals of Statistics. 2009;37:3779–3821. [Google Scholar]
- Ravikumar P, Liu H, Lafferty J, Wasserman L. Spam: Sparse additive models. Journal of the Royal Statistical Society: Series B. 2009;71:1009–1030. [Google Scholar]
- Sardy S, Tseng P. Amlet, ramlet, and gamlet: Automatic nonlinear fitting of additive models, robust and generalized, with wavelets. Journal of Computational and Graphical Statistics. 2004;13:283–309. [Google Scholar]
- Scheetz TE, Kim K-YA, Swiderski RE, Philp1 AR, Braun TA, Knudtson KL, Dorrance AM, DiBona GF, Huang J, Casavant TL, Sheffield VC, Stone EM. Regulation of gene expression in the mammalian eye and its relevance to eye disease. Proceedings of the National Academy of Sciences. 2006;103:14429–14434. doi: 10.1073/pnas.0602562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman B. Spline smoothing: The equivalent variable kernel method. The Annals of Statistics. 1984;12:898–916. [Google Scholar]
- Speckman P. Spline smoothing and optimal rates of convergence in nonparametric regression models. The Annals of Statistics. 1985;13:970–983. [Google Scholar]
- Stone C. Additive regression and other nonparametric models. The Annals of Statistics. 1985;13:689–705. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B. 1996;58:267–288. [Google Scholar]
- van der Vaart AW, Wellner JA. Weak Convergence and Empirical Processes. Springer; New York: 1996. [Google Scholar]
- Wang H. Forward regression for ultra-high dimensional variable screening. Journal of the American Statistical Association. 2009;104:1512–1524. [Google Scholar]
- Wei F, Huang J. Technical Report No.387. University of Iowa; 2007. Consistent group selection in high-dimensional linear regression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society: Series B. 2006;68:49–67. [Google Scholar]
- Zhang C-H. Nearly unbiased variable selection under minimax concave penalty. The Annals of Statisitics. 2010;38:894–942. [Google Scholar]
- Zhao DS, Li Y. Principled sure independence screening for cox models with ultra-high-dimensional covariates. Manuscript. 2010 doi: 10.1016/j.jmva.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Shen X, Wolfe DA. Local asymptotics for regression splines and confidence regions. The Annals of Statistics. 1998;26:1760–1782. [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2005;67:768–768. [Google Scholar]
- Zou H, Li R. One-step sparse estimates in nonconcave penalized likelihood models. The Annals of Statistics. 2008;36:1509–1533. doi: 10.1214/009053607000000802. [DOI] [PMC free article] [PubMed] [Google Scholar]