Abstract
Since the discovery of the TP63 gene in 1998, many studies have demonstrated that ΔNp63, a p63 isoform of the p53 gene family, is involved in multiple functions during skin development and in adult stem/progenitor cell regulation. In contrast, TAp63 studies have been mostly restricted to its apoptotic function and more recently as the guardian of oocyte integrity. TAp63 endogenous expression is barely detectable in embryos and adult (except in oocytes), presumably because of its rapid degradation and the lack of antibodies able to detect weak expression. Nevertheless, two recent independent studies have demonstrated novel functions for TAp63 that could have potential implications to human pathologies. The first discovery is related to the protective role of TAp63 on premature aging. TAp63 controls skin homeostasis by maintaining dermal and epidermal progenitor/stem cell pool and protecting them from senescence, DNA damage and genomic instability. The second study is related to the role of TAp63, expressed by the primitive endoderm, on heart development. This unexpected role for TAp63 has been discovered by manipulation of embryonic stem cells in vitro and confirmed by the severe cardiomyopathy observed in brdm2 p63-null embryonic hearts. Interestingly, in both cases, TAp63 acts in a cell-nonautonomous manner on adjacent cells. Here, we discuss these findings and their potential connection during development.
Keywords: TP63, TAp63, heart development, SKPs, embryonic stem cells, aging
Facts
TAp63 (p63 containing the transactivation domain) maintains epidermal and dermal stem cells in a healthy and functional state with intact genomes.
In TAp63−/− mice, skin-derived precursors (SKPs) exhibit rapid signs of premature aging, senescence, DNA damage and genomic instability. Lack of TAp63 in the dermal stem cell compartment of the hair follicle is correlated with cellular senescence and hair loss.
TAp63 controls endodermal factors that exert, in a cell-nonautonomous manner, a cardiogenic effect on mesocardiac progenitors.
p63-null embryos (brdm2 strain) suffer from severe cardiomyopathy.
Open Questions
Do human SKPs age like in mice and depend on TAp63?
To prevent aging, should we increase the number of SKPs or just stimulate the residual cells?
Do SKPs cells and TAp63-endodermal cells originate from the same neural crest progenitor?
What is the molecular circuitry controlled by TAp63 during heart development?
Do patients affected by p63-related ectodermal dysplasia (ED) syndromes suffer from mild congenital cardiomyopathy?
p63 belongs to the p53 family of transcription factors, together with p53 and p73.1 By encoding two different N-termini (TA and ΔN) and multiple C-termini by alternative splicing, the TP63 gene generates multiple-protein isoforms.2, 3 All p63 isoforms contain identical DNA binding and oligomerization domains and are able to transactivate p53-responsive and specific target genes.4 Moreover, p53 gene members functions sometimes in an interdependent way.5, 6 Similar to p53, TAp63 (p63 containing the transactivation domain) is related to DNA damage response function, inducing cell-cycle arrest and apoptosis.7, 8 In adults, TAp63 is mainly expressed in oocytes, apparently to protect them from toxic insults9 and, similar to p53, induced in response to cellular stress 4, including wound healing.10 Low levels of TAp63 are detected in other tissues, including the suprabasal layers of the epidermis, suggesting a function in terminal differentiation.11 Conversely, ΔNp63 has been implicated in cell proliferation and cell adhesion12, 13, 14 and is a determinant of the homeostasis of the epidermal stem cell/progenitor niche.13, 15 Recently, whole-genome array analyses by Chip-seq have demonstrated that endogenous p63 binds to thousand of gene promoters, suggesting highly complex interactions with a large number of pathways.16, 17, 18 In addition, p63 is regulated by micro-RNAs,19 but can also regulate micro-RNAs20, 21 and Dicer.21 Numerous reports strongly suggest a significant role of p63 isoform in cancer progression and tumor suppression.4 ΔNp63 is often highly expressed in human tumors while nonsense and missense mutations on p63 have been recently identified in about 8% of 84 squamous cell carcinoma tumor samples.22 Moreover, in vivo studies demonstrated that TAp63 isoform regulates senescence and suppresses tumorigenesis23 and metastasis.21
Function and Regulation of ΔNp63 in Epithelial Development
During mouse development, the amino-deleted p63 isoform (ΔNp63) is expressed from E10.5 to birth in the surface epithelium, hair follicles and teeth, as revealed by in situ hybridization.24 Knockout mouse models engineered to study the function of p63 in vivo revealed a fundamental role for p63 in epithelial development.25, 26 Mice lacking all p63 isoforms die at birth and show severe developmental abnormalities, including lack of skin, limbs and epidermal appendages. The epithelial phenotype is due to a lack of commitment of the immature ectoderm to epidermal lineages,25, 27 a step driven specifically by ΔNp63.28, 29 In this regard, ectodermal organs such as hairs, whiskers, teeth and several glands, including mammary, salivary and lacrimal glands, are lacking in p63-deficient mice.25, 26 A key function for ΔNp63 in defining a squamous epithelial phenotype is to actively suppress mesodermal cell fates during the early development of Xenopus29 and mouse.30 Consistent with the role of p63 in epidermal commitment of ectodermal progenitors, heterozygous germline mutations in TP63 result in human ED syndromes involving defective development of the limbs, and/or ED characterized by defects in skin and its associated structures.31 Several recent reviews have focused upon the function of ΔNp63 in embryogenesis and in the adult,32, 33, 34 as well as in cancer.4 Here, we review two new functions recently attributed to TAp63 in premature aging of the skin35 and during cardiac development36 and discuss their potential implications to human pathologies.
TAp63 in skin aging
p63+/− mice have a decreased life span, link with senescence.37 More recently, the function has been attributed to TAp63, as TAp63-null mice suffer from premature aging.35 TAp63 is expressed by a subset of dermal stem cells skin-derived precursors (SKPs) that originate from Sox2+ dermal precursors localized in the dermal papilla and the dermal sheath of the hair follicle (Figure 3). This population of self-renewing cells is capable of clonal and multilineage differentiation,38, 39 functions to induce hair growth and contributes to dermis homeostasis especially during wound healing.40 These hair follicle dermal precursor cells share properties with neural crest precursors, differentiating into mesenchymal derivatives such as adipocytes and skeletogenic cells and peripheral neural crest cell types such as Schwann cells. Lineage tracing analysis allowed the authors to conclude that SKPs, and the endogenous dermal precursors from which they derive, originate from the neural crest in the face and the somites in the dorsal trunk, but despite these different origins, they both generate precursors with a highly similar phenotype.41
In TAp63−/− mice, SKPs exhibit rapid signs of premature aging, including a marked increase in senescence, DNA damage and genomic instability, resulting in an exhaustion of these cells.35 Lack of TAp63 in the dermal stem cell compartment of the hair follicle (dermal papilla and dermal sheath) results in senescence-associated-β-gal positivity, a marker of the terminal growth-arrest response known as cellular senescence. This senescence response is associated with hair loss and is also observed in normal aging of wild-type mice.
At the cellular level, the data provide a number of TAp63-dependent mechanisms that explain depletion of adult stem-cell function. TAp63−/− SKPs showed significantlly increased proliferation and self-renewal in culture, a phenotype that correlated with decreased expression of p57Kip2, a cell-cycle inhibitor that is a direct transcriptional target of p63.42 As adult stem cell populations are not immortal, a large enhancement of proliferation of follicle-associated SKPs would be predicted to lead to stem cell depletion. Interestingly, re-expression of p57Kip2 in TAp63−/− SKPs rescued the hyperproliferative phenotype of TAp63−/− SKPs, indicating that regulation of p57Kip2 by TAp63 is involved in the maintenance of these dermal precursors.
Although the ubiquitous knockdown of TAp63 resulted in senescence of epidermal stem cells and significant genetic instability in the epidermis, the conditional ablation of TAp63 in the epidermal stem cells (using K14-Cre transgene) did not recapitulate this phenotype, suggesting a critical role for the dermal stem cells in regulating the epidermal compartment. Furthermore, cultured SKPs (isolated from dorsal and whisker pad skin) and epidermal cells bereft of TAp63 undergo senescence, as well as enhanced DNA damage and significant genetic instability. Thus, TAp63 is critical for skin homeostasis, maintaining epidermal and dermal stem cells in a healthy and functional state with intact genomes. The method used by Su et al.35, 43 did not allow separating epidermal stem cell from hair and skin, which leaves open the question whether TAp63 has a major role in the bulge epidermal stem cells (Figure 1). In the rodent HF, two different neural crest stem cell-like (NCSC) populations with a unique genetic signature that distinguishes them from other hair follicle stem cells can be derived from the outer root sheath: the epidermal neural crest stem cells (Epi-NCSC)44 and the Nestin+K15− cells.45 Interestingly, the genes composing the Epi-NCSC signature in rodent are similarly activated in hair follicle stem cells/neural crest cells and display comparable signature and embryonic origin with SKPs, suggesting that these two populations are equivalent (reviewed by Biernaskie46). Thus, the similarities between SKPs and certain outer root sheath/bulge stem cells suggest that TAp63 may also mediate age-related changes in the epithelial hair follicle compartment.
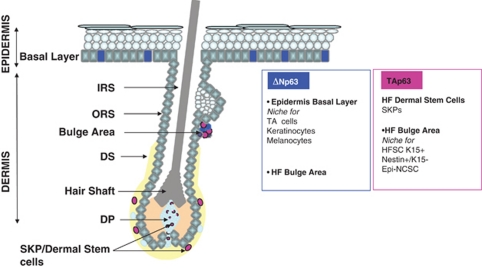
Figure 1.
Summary of the stem cells niche location in skin and hair follicle and their p63 isoforms expression. ΔNp63 is expressed in the basal layer of the epidermis, known as stem cell niches for keratinocytes and melanocytes, as well as in the hair follicle (HF)-bulge area, where hair follicle stem cells (HFSC)/K15+cells, Nestin+/K15− cells and epidermal-neural crest stem cells (Epi-NCSC) reside. TAp63 is expressed in the HF dermal stem cells and in the HF bulge area. Whether or not each of the stem cells localized in the HF-bulge area expresses p63 isoform still remains to be investigated. DP, dermal papilla; DS, dermal sheath; IRS, inner root sheath; ORS, outer root sheath; SKPs, skin-derived precursors; TA cells, transient amplifying cells
The findings described above strongly suggest that depletion of adult stem cells may be a major cause of tissue aging47 and raise the possibility that functional depletion of adult stem cells is responsible for the phenotypes of humans with dysregulated TAp63, including skin and hair conditions.48 In this regard, the TAp63−/− mice display a phenotype similar to defects in humans with Hay–Wells syndrome where patients with mutations in p63 develop dermatitis and alopecia.49 We suggest that these patients have defects in dermal stem cell maintenance, which we propose could be reversed by drug discovery efforts that enhance dermal stem cell or SKPs self-renewal.
Aging is accompanied by a decline in the homeostatic and regenerative capacity on all tissues and organs. In mammalian, the correlation between aging and stem cells, or the ‘stem-cell hypothesis for aging', has been described in a variety of tissues.47 In this hypothesis, the age-related stem cells changes are due to a depletion of stem cells, chronological aging, change in local systemic environment or intrinsic change in aging stem cells (DNA damage, telomere shortening).50, 51 A key regulator may be the p53 family, where levels of family members such as p53 and TAp63 have been shown to be crucially important in aging in rodents. Evolutionary theory argues that evolution selects for genes that are beneficial in early life even if they are detrimental later. p53 family members are a notable example and their study with regard to age-related changes in stem cell function is important to understand how abnormal stem cell fate, including senescence can influence tissue aging.50
There are numerous key questions that skin, aging and stem cells leave us for the future. TAp63 is expressed by SKPs, which either derive from somites or originate from the neural crest cells.41 Contrary to what had been observed with hair follicles SKPs, downregulation of TAp63 did not influence the proliferation capacity and differentiation potential of human bone marrow-derived mesenchymal stem cells while protecting them against apoptosis.52 This observation suggests two distinct roles for TAp63 in mesenchymal stem cells versus neural crest-derived stem cells, opening doors for further investigations. Another fundamental question is related to the TAp63-driven senescence of the dermal stem cells. Could it be a way to prevent cancer in a vulnerable aging tissue and if so, what could be the link with the tumor suppressor role of TAp63 described in other tissues.35
Moreover, if targeting stem cells is a potential strategy for the war against human aging, there are several points to be explored. As p63 is a story of mice and men,34 we must test whether human SKPs age as they do prematurely in TAp63 knockout mice. Recent studies reported a dermal stem cell depletion with aging53 and a premature senescence of the dermal papilla cell in balding scalps.54 Nevertheless, what could be the role of TAp63 in these processes? What would be the optimal level of manipulation of TAp63 in the SKPs as we age? We would certainly have to fine-tune the risk between too little expression leading to hyper proliferation and exhaustion of the dermal stem cells and too much which could potentially stop their proliferation. What could be the best strategy? Should we identify factors secreted by the niche that may be affected during aging? We might find a way to increase the number of SKPs (in case of depletion) or just stimulate the residual cells. Should we force the senescent cells to reenter into cell cycle?
TAp63 in heart development
When aggregated as embryoid bodies, embryonic stem (ES) cells spontaneously recapitulate in vitro the main steps of mesoderm and cardiac lineages leading to mature cardiomyocytes, as visualized by the appearance of beating areas in the cell culture. In vivo and in vitro cardiac lineages can be followed by the sequential appearance of specific markers, as illustrated in Figure 2. In the course of our study on the role of p63 during ectodermal commitment of pluripotent stem cells, we surprisingly observed that stable inactivation of TP63 prevented the production of beating areas.36 By the use of transient siRNAs raised against each of the p63 isoforms, we found that specific knockdown of TAp63 prevented the formation of beating cardiomyocytes. Although markers of the mesodermal fate (brakyury and mesp1) were not affected by the absence of TAp63, we observed a significant reduction of early cardiac progenitor markers (tbx5, nkx2.5, isl1) and mature cardiomyocyte markers (troponinT, α-actinin and mlc2v). These data suggest that TAp63 controls cardiogenesis, as soon as the mesocardiac cells become cardiac progenitors from both the first (tbx5) and second (isl1) heart lineages (see Figure 2).
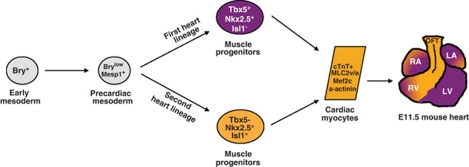
Figure 2.
Cellular hierarchy of cardiac progenitor cells and their lineage diversification. The earliest precursors for heart-forming cells are produced in the lateral posterior region of the epiblasts (E6.5) and then in the mesoderm expressing the T-box transcription factor brachyury T (Bry). A few hours later (E7.0), the precardiac mesoderm is formed with cells transiently expressing the mesoderm posterior (Mesp)1 marker. Mesp1+ cells then migrate to the anterior region by ingressing through the primitive streak. Cardiac progenitors are then determined expressing Nkx2.5, GATAs and Mef2c. The progenitor cells expand rapidly forming the cardiac crescent following the fusion of the bilateral heart-forming areas in the splanchnic mesoderm. Ultimately, cells segregate into two spatially and temporally distinct cardiogenic heart fields exhibiting unique time courses of differentiation and distinct regional contributions to the embryonic heart. Cells then express cardiac transcription factors such as tbx5, GATA4, Nkx2.5 and Islet-1 (Isl1). Fully differentiated cardiomyocytes are identified by Troponin-T, MLC2v and α-actinin. Progenitor cells from the first heart lineage (FHF; blue) contribute primarily to the left and right atria (LA and RA) and the left ventricle (LV) of the primitive mammalian heart while the second heart field (SHF; red) contributes to the outflow tract (OFT) and right ventricle (RV)
By immunofluorescence staining during cardiac differentiation of ES cells, we observed that TAp63 is not detected in cells of the mesocardiac lineage but coexpressed with sox17, a specific marker of endodermal cells. Accordingly, p63 transcripts were detected in the endoderm of E8.5 embryos by in situ hybridization.36 At an early stage of mammalian embryogenesis, the primitive and extra-embryonic endoderm that expresses sox17 is in close proximity to the primitive mesoderm and has an inductive role in adjacent mesocardiac progenitors. It has been shown that expression of the transcription factor sox-17 in the mesendoderm is essential for the specification of cardiac mesoderm in ES cells.55 By Chip-PCR experiments, promoter/reporter assays and chimeric co-culture rescue assays, we demonstrated that TAp63 is apparently not expressed by precardiac mesoderm cells but by endodermal derivatives to stimulate cardiac progenitor lineages in a cell-nonautonomous manner. Altogether, TAp63 controls the inductive role of the endoderm on cardiac progenitor fate.
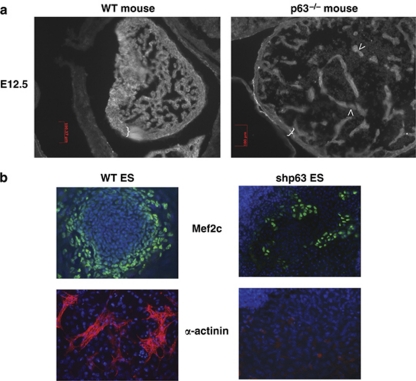
Our surprising data prompted us to examine the embryos deficient for p63 that have been produced more than a decade ago. We analyzed p63−/− embryos of the brdm2 strain available from the Jackson Laboratory (Bar Harbor, ME, USA).25 Histological sections revealed a severe dilated cardiomyopathy in p63-null heart (Figure 3). Both left and right ventricular chambers showed deficient trabeculation and thin ventricular walls.36 Cardiomyocytes displayed pronounced disorganization of the myofibrillar apparatus and mitochondria. In contrast to spontaneous Ca2+ spiking in the WT myocardium, weak Ca2+ spikes were observed in p63-deficient embryos, highlighting the defective cardiac activity of the homozygous mutant heart. Therefore, the cardiac phenotype of the p63-null embryos fits well the defective cardiac differentiation of ES cells in the absence of TAp63 (Figure 3).
Figure 3.
Defective heart development and cardiogenesis in the absence of p63 (a) Dilated cardiomyopathy in E12.5 p63−/− embryonic heart (right panel), revealed by a thin ventricular wall (indicated by blue brackets) and an abnormal trabeculation (arrows), as compared with normal WT heart (left panel). (b) Defective production and migration of cardiomyocytes in the absence of p63 (right) as compared with WT ES cells (left) at day 9. Immunostaining with anti-Mef2c and α-actinin. Dapi in blue
The multiple defects in the two ventricular chambers and atrium strongly suggest that p63 controls the specification of cardiac progenitors of the first and second heart lineages, as suspected from our in vitro data. Islet-1 (isl1) is transiently expressed in a population of multipotential mesodermal precursors emerging from a second heart lineage that contribute to two-thirds of the heart56 Tbx5 transcripts are found in the endocardium and myocardium of the inflow region, the atria, the AVC and the left ventricle, but not in the right ventricle and OFT of the linear, looping and chambered heart, strongly suggesting that its expression is mainly associated with the first heart field and its derivatives.57 Analysis of isl1 expression at early stages showed that p63-deficient mice have a decreased pool of isl1-positive progenitor cells in the primary left ventricle, wall of the atrium, the sinus venosus, the epicardium and the outflow tract. In addition, p63-null embryos at E 8.5 displayed a lower expression level of isl1, nkx2.5 and tbx5 cardiac-related genes. Altogether, our finding strongly argue that TAp63 is a key endodermal cardiogenic factor that modulates, in a cell-nonautonomous manner, the specification and/or the proliferation/survival of cardiac progenitors that migrate from the cardiac crescent and this could explain the defective ventricular wall and trabeculation (Figure 4). As we observed less sox-17 endodermal cells during ES cell commitment, it could suggest that TAp63 could function as a self-renewal gene for endodermal cardiogenic progenitors. Interestingly, two TA-null mice have been produced recently. For the first one, no embryonic development defect was reported.9 However, the second KO mouse strain displayed a strong proportion of embryonic lethality.35 To circumvent this phenotype that could be due to heart failure, the authors have backcrossed their mice to an alternative genetic background. These data suggest that the function of TAp63 in heart development could be compensated by alternative pathways and/or that, depending on the genetic background of the mouse strains, some modifier genes may amplify or neutralize its effect.
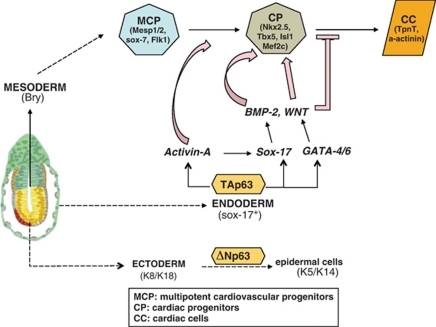
Figure 4.
Dual functions of TAp63 and ΔNp63 in development. At early developmental event, pluripotent epiblast (extreme left) produces the three germ layers, ectoderm, mesoderm and endoderm. We and others have shown that ΔNp63 is mandatory for in vivo27 and in vitro28, 29 epidermal commitment of ectodermal cells. More recently, we found that TAp63 is expressed in vitro and in vivo by primitive endodermal cells and controls the expression of endodermal factors (sox-17, GATA) that allow the production and secretion of cardiogenic factors (Activin-A, BMP-2, Wnt) for proper cardiogenesis in a cell-nonautonomous manner.36 MCP, multipotent cardiovascular progenitors; CP, cardiac progenitors; CC, cardiac cells
As p63 is recognized as a major epithelial transcription factor, we were puzzled by a potential link between p63 and cardiogenesis. As a matter of fact, sparse pieces of evidence can be found in the literature of the last years that point to a connection between p63 and heart development and/or physiology. Among them, the original papers on p63 reported the presence of p63 transcripts in heart tissue and in the branchial arches of E8.5 embryos.3, 26 Moreover, Chip-seq and Chip-on-chip analysis experiments performed on somatic epithelial cells identified p63-target genes related to cardiac development.17, 58 Finally, a specific activity of a strong p63 enhancer was reported in the embryonic heart area of transgenic mice.59
As stated above, p63 expression was detected by in situ hybridization in the branchial (pharyngeal) arches, where cardiac progenitors are located and reside close to ectodermal and endodermal progenitor cells before their migration to the cardiac tube.60 The embryonic pharyngeal arch contain tbx1-positive cells that originate from the neural crest and have a vital role in heart development.61 Interestingly, we found out that p63 was in the list of genes that were specifically repressed at E9.5 in pharyngeal arch of tbx1 KO embryos.61
Although the KO p63 mice have been described a decade ago and used broadly by several independent research groups, we have discovered that the p63 null embryos suffer from a severe congenital heart failure that could explain their lethality before or at birth. Actually, the p63-null embryos at mid-gestation have abnormal epithelia tissues but the defective heart appears macroscopically normal. The ES cell model prompted us to have a new look at the heart histology of p63 null embryos and found this novel and unexpected role for TAp63 in the cardiac differentiation and heart development. These studies provide a strong rationale for using ES cells as a cellular model to study development in vitro.
Mutations of the p63 gene in humans are responsible for ED syndromes.31 Our findings suggest that these patients may suffer from mild congenital cardiomyopathy that has not yet been detected. The few recent reports of patients affected by both ED and arrhythmogenic right ventricular cardiomyopathy62 or atrial septal defect63 suggest that patients affected by p63-linked ED syndromes could have an increased risk of undetected heart failure. Moreover, duplication and deletion of the region 3q27-ter (where the p63 gene resides) are associated with congenital malformations, including dilated cardiomyopathy.64, 65, 66 However, we cannot exclude that the two events are unrelated. No mutation in TAp63-specific sequences has been described so far. The study described here suggests that such TAp63-specific mutations would be either lethal in utero or responsible for congenital cardiomyopathies for which no gene has yet been assigned. As a matter of fact, the frequency of p63−/− embryos was generally lower than expected from Mendelian segregation.36 Therefore, these data warrant a systematic analysis of potential genotype/phenotype correlations between p63 mutations and defects in cardiogenesis.
Our discovery opens the field to many questions to be raised: when exactly and in which cell types are TAp63 isoforms expressed during early mammalian development? What are the genes and signaling pathways controlled by TAp63 for proper cardiogenesis and heart development? Are there congenital cardiopathies in which p63 gene is mutated? What is the nature of the modifier(s) responsible for the discrepancy of cardiac phenotype between mouse strains? Are there additional tissues, whose development relays on endodermal inductive factors, affected by the absence of p63?
Do TAp63-Expressing Cells Share Common Origin and Function?
Except for the proapoptotic role of TAp63, most of the functions attributed to TP63 concern ΔNp63. These newly described functions of TAp63 in cardiogenesis and stem cell maintenance were unexpected. Interestingly, in both cases, TAp63 exerts its effects in cells with instructive functions on nearby cells of different origins, either dermal stem cells that instruct epidermal stem cells in the bulge region to grow hair or primitive endodermal cells that excert cardiogenic effect on cardiac progenitors. In addition, the expression of TAp63 in neural crest-derived stem cells such as some SKP populations provide clues to how this gene regulates cardiogenesis. Thus, TAp63 could function as a self-renewal gene for both endodermal cardiogenic cells36 and skin progenitors.35 This could be related to a similar function attributed to TAp73 on neural progenitors.67 To understand how TAp63 functions in these contexts, it will be necessary to identify its respective target genes by using comparative whole genome-wide map data from each cellular system.
Cardiac neural crest contributes to the heart stem cell pool68 and the embryonic branchial arches (where TAp63 is detected) contain mesendodermal cells of neural crest origin that participate to the heart development.69 As SKPs cells are also derived from the neural crest and can produce functional smooth muscle cells,70 it would be interesting to determine whether the TAp63-expressing endodermal cells with cardiogenic inductive role36 have a common embryonic origin with the dermal SKPs and whether the latter could have cardiogenic activity on mesocardiac progenitors. Again, a careful analysis of the microarray and Chip-seq analysis of SKPs cells may be informative.
The functions of TAp63 have been shown to be dependent on the genetic background of the mouse strain used to ablate TAp63.35 This highlights the variability between genetic background that could be explained by either the existence of p63-linked modifiers yet to be identified or that p63 is itself a modifier for other genes directly involved in these functions. As there is interdependence between the p53 gene members and their dozen isoforms, such discrepancies could also be explained by potential mutation or polymorphism on the related genes. Therefore, and as suggested in a recent review,6 the relative expression level of each gene must be carefully evaluated in different tissues during development and aging.
Finally, the new functions attributed to TAp63 raise the question of whether patients carrying mutations in p63-specific exons/introns could suffer from congenital cardiomyopathy and/or premature aging. A systematic analysis of potential genotype/phenotype correlations between p63 mutations/SNPs and defects in cardiogenesis and aging should be initiated.
Acknowledgments
We thank David Kaplan, Ruby Shalom-Feuerstein and Wim Declercq for their comments, suggestions and criticisms. Part of this work was supported for DA by the Sixth EEC Framework Program within the EPISTEM project (LSHB-CT-2005-019067), the Agence Nationale pour la Recherche (ANR BLANC 06 and GENOPAT 08) and the Israel Science Foundation.
Glossary
- SKPs
skin-derived precursors
- TAp63
p63 containing the transactivation domain
- ΔNp63
amino-deleted p63 isoform
- ES cells
embryonic stem cells
- Ebs
embryoid bodies
- T Bry
T-box transcription factor brachyury
- Mesp1
mesoderm posterior1
- Isel1
Islet-1
- Chip
chromatin immunoprecipitation
- TF
transcription factor
- ED
ectodermal dysplasia syndromes
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Aylon Y, Oren M. New plays in the p53 theater. Curr Opin Genet Dev. 2011;21:86–92. doi: 10.1016/j.gde.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37:6092–6104. doi: 10.1093/nar/gkp674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, et al. p63 is an essential proapoptotic protein during neural development. Neuron. 2005;48:743–756. doi: 10.1016/j.neuron.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. p53 Isoforms: An Intracellular Microprocessor. Genes Cancer. 2011;2:453–465. doi: 10.1177/1947601911408893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 2000;60:4016–4020. [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, et al. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- Bamberger C, Hafner A, Schmale H, Werner S. Expression of different p63 variants in healing skin wounds suggests a role of p63 in reepithelialization and muscle repair. Wound Repair Regen. 2005;13:41–50. doi: 10.1111/j.1067-1927.2005.130106.x. [DOI] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Carroll DK, Carroll JS, Leong CO, Cheng F, Brown M, Mills AA, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton CE, Johnson KN, Mays DM, Boehnke K, Shyr Y, Boukamp P, et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Differ. 2010;1:e74. doi: 10.1038/cddis.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kettenbach A, Kapranov P, McKeon F, Gingeras TR, et al. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar DNA-binding profiles in vivo. PLoS One. 2010;5:e11572. doi: 10.1371/journal.pone.0011572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness' by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, et al. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, et al. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton CE, Tahinci E, Barbieri CE, Johnson KN, Hanson AJ, Jernigan KK, et al. DeltaNp63 antagonizes p53 to regulate mesoderm induction in Xenopus laevis. Dev Biol. 2009;329:130–139. doi: 10.1016/j.ydbio.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, McKeon F. Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med. 2002;8:133–139. doi: 10.1016/s1471-4914(01)02260-2. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Koster MI. p63 in skin development and ectodermal dysplasias. J Invest Dermatol. 2010;130:2352–2358. doi: 10.1038/jid.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. J Invest Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Medawar A, Hamon L, Shivtiel S, Wolchinsky Z, Zhou H, et al. TAp63 is important for cardiac differentiation of embryonic stem cells and heart development. Stem Cells. 2011;29:1672–1683. doi: 10.1002/stem.723. [DOI] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KJ, McKenzie IA, Mill P, Smith KM, Akhavan M, Barnabe-Heider F, et al. A dermal niche for multipotent adult skin-derived precursor cells. Nat Cell Biol. 2004;6:1082–1093. doi: 10.1038/ncb1181. [DOI] [PubMed] [Google Scholar]
- Toma JG, Akhavan M, Fernandes KJ, Barnabe-Heider F, Sadikot A, Kaplan DR, et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3:778–784. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- Biernaskie J, Paris M, Morozova O, Fagan BM, Marra M, Pevny L, et al. SKPs derive from hair follicle precursors and exhibit properties of adult dermal stem cells. Cell Stem Cell. 2009;5:610–623. doi: 10.1016/j.stem.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno H, Morozova O, Jones KL, Biernaskie JA, Paris M, Hosokawa R, et al. Convergent genesis of an adult neural crest-like dermal stem cell from distinct developmental origins. Stem Cells. 2010;28:2027–2040. doi: 10.1002/stem.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta C, Chiarelli A, Testoni B, Mantovani R, Guerrini L. Regulation of the cyclin-dependent kinase inhibitor p57Kip2 expression by p63. Cell Cycle. 2005;4:1625–1631. doi: 10.4161/cc.4.11.2135. [DOI] [PubMed] [Google Scholar]
- Beaudry VG, Attardi LD. SKP-ing TAp63: stem cell depletion, senescence, and premature aging. Cell Stem Cell. 2009;5:1–2. doi: 10.1016/j.stem.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Hu YF, Zhang ZJ, Sieber-Blum M. An epidermal neural crest stem cell (EPI-NCSC) molecular signature. Stem Cells. 2006;24:2692–2702. doi: 10.1634/stemcells.2006-0233. [DOI] [PubMed] [Google Scholar]
- Amoh Y, Li L, Campillo R, Kawahara K, Katsuoka K, Penman S, et al. Implanted hair follicle stem cells form Schwann cells that support repair of severed peripheral nerves. Proc Natl Acad Sci USA. 2005;102:17734–17738. doi: 10.1073/pnas.0508440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernaskie J. Human hair follicles: ″bulging″ with neural crest-like stem cells. J Invest Dermatol. 2010;130:1202–1204. doi: 10.1038/jid.2009.449. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Gu X, Lundqvist EN, Coates PJ, Thurfjell N, Wettersand E, Nylander K. Dysregulation of TAp63 mRNA and protein levels in psoriasis. J Invest Dermatol. 2006;126:137–141. doi: 10.1038/sj.jid.5700010. [DOI] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nat Cell Biol. 2011;13:506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Lu S, Liang W, Li J, Dou X, Bian C, et al. TAp63alpha Mediates Chemotherapeutic Agent-Induced Apoptosis in Human Bone Marrow Mesenchymal Stem Cells. Stem Cells Dev. 2011;20:1319–1326. doi: 10.1089/scd.2010.0329. [DOI] [PubMed] [Google Scholar]
- Gago N, Perez-Lopez V, Sanz-Jaka JP, Cormenzana P, Eizaguirre I, Bernad A, et al. Age-dependent depletion of human skin-derived progenitor cells. Stem Cells. 2009;27:1164–1172. doi: 10.1002/stem.27. [DOI] [PubMed] [Google Scholar]
- Bahta AW, Farjo N, Farjo B, Philpott MP. Premature senescence of balding dermal papilla cells in vitro is associated with p16(INK4a) expression. J Invest Dermatol. 2008;128:1088–1094. doi: 10.1038/sj.jid.5701147. [DOI] [PubMed] [Google Scholar]
- Liu Y, Asakura M, Inoue H, Nakamura T, Sano M, Niu Z, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- Pozzi S, Zambelli F, Merico D, Pavesi G, Robert A, Maltere P, et al. Transcriptional network of p63 in human keratinocytes. PLoS One. 2009;4:e5008. doi: 10.1371/journal.pone.0005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonini D, Rossi B, Han R, Minichiello A, Di Palma T, Corrado M, et al. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol Cell Biol. 2006;26:3308–3318. doi: 10.1128/MCB.26.8.3308-3318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Dandonneau M, Mesbah K, Jarry T, Mattei MG, Kelly RG. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS One. 2009;4:e6267. doi: 10.1371/journal.pone.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Aggarwal VS, Nowotschin S, Bondarev A, Lipner S, Morrow BE. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev Biol. 2008;316:524–537. doi: 10.1016/j.ydbio.2008.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzise M, Arrigo T, De Luca F, Privitera A, Frigiola A, Carando A, et al. R298Q mutation of p63 gene in autosomal dominant ectodermal dysplasia associated with arrhythmogenic right ventricular cardiomyopathy. Eur J Med Genet. 2008;51:497–500. doi: 10.1016/j.ejmg.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Rinne T, Clements SE, Lamme E, Duijf PH, Bolat E, Meijer R, et al. A novel translation re-initiation mechanism for the p63 gene revealed by amino-terminal truncating mutations in Rapp-Hodgkin/Hay-Wells-like syndromes. Hum Mol Genet. 2008;17:1968–1977. doi: 10.1093/hmg/ddn094. [DOI] [PubMed] [Google Scholar]
- Chiodini BD, Lewis CM. Meta-analysis of 4 coronary heart disease genome-wide linkage studies confirms a susceptibility locus on chromosome 3q. Arterioscler Thromb Vasc Biol. 2003;23:1863–1868. doi: 10.1161/01.ATV.0000093281.10213.DB. [DOI] [PubMed] [Google Scholar]
- Chitayat D, Babul R, Silver MM, Jay V, Teshima IE, Babyn P, et al. Terminal deletion of the long arm of chromosome 3 [46,XX,del(3)(q27-->qter)] Am J Med Genet. 1996;61:45–48. doi: 10.1002/(SICI)1096-8628(19960102)61:1<45::AID-AJMG9>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Senzaki H, Inui M, Ban S, Masutani S, Morsy M, Kobayashi T, et al. Dilated cardiomyopathy in a 3-year-old girl with a terminal deletion, 46,XX,del(3)(q27-qter), of the long arm of chromosome 3. Eur J Pediatr. 2003;162:403–405. doi: 10.1007/s00431-003-1160-8. [DOI] [PubMed] [Google Scholar]
- Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, et al. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, et al. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. 2005;170:1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104:933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- Steinbach SK, El-Mounayri O, Dacosta RS, Frontini MJ, Nong Z, Maeda A, et al. Directed differentiation of skin-derived precursors into functional vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2011;31:2938–2948. doi: 10.1161/ATVBAHA.111.232975. [DOI] [PubMed] [Google Scholar]