Abstract
In the developing spinal cord, motor neurons (MNs) and oligodendrocytes arise sequentially from a common pool of progenitors. However, the genetic network responsible for this neurogenesis to gliogenesis switch is largely unknown. A transcriptome analysis identified the Notch ligand Jagged2 (JAG2) as a Sonic hedgehog-regulated factor transiently expressed in MN progenitors (pMNs). In vivo loss- and gain-of-function experiments show that JAG2 schedules the differentiation of the pMN progenitors. At early developmental stages, Olig2 expressing pMN progenitors that enter the differentiation pathway exclusively generate MNs. At these times, the activation of the Notch pathway by JAG2 maintains selected pMN progenitors in an undifferentiated state by two mechanisms; first it inhibits MN generation by reducing Olig2 proteins levels, and second it directly inhibits the premature generation of oligodendrocyte progenitors (OLPs) by maintaining high levels of Hes5. Later, extinction of JAG2 from the pMN results in the loss of Hes5 expression, heralding the gliogenic phase of pMN progenitors. Strikingly, downregulation of JAG2 in pMN progenitors is sufficient to promote the precocious generation of OLPs. Together these data provide evidence that JAG2 is a key regulator of the timely and ordered generation of two of the defining cell types in the spinal cord, MNs and OLPs.
Keywords: sonic hedgehog, Notch, Olig2, Hes5, spinal cord, neurogenesis
In many developing tissues, most notably the central nervous system, the identity of a cell is linked to the time at which it is generated. However, the mechanisms that underpin the temporal specification of cell fate are poorly understood. A well-defined example of this problem is the sequential generation of motor neurons (MNs) and oligodendrocytes (OLPs) in the developing spinal cord. These two cell types arise from a common domain of progenitors, termed the MN progenitor (pMN) domain, which is located in the ventral neural tube. Initially, the pMN domain produces MNs that send axonal trajectories out of the CNS to innervate peripheral targets. Subsequently, progenitors in the same region produce oligodendrocytes, the myelinating cells of the CNS. The pMN domain is specified by the instructive activity of the secreted protein Sonic hedgehog (Shh), which, signalling through the canonical Gli pathway, provides positional identity to neural progenitors by directing a transcription network in responding cells.1 Amongst the members of this network is the basic helix-loop-helix (bHLH) transcription factor Olig2, which is expressed in the pMN domain and provides a molecular identifier for the pMN domain.2, 3
Early studies showed that Shh is both necessary and sufficient for the generation of spinal MNs.4, 5, 6 Subsequently, Shh was shown to be required for OLPs to emerge at the appropriate developmental stages.7, 8 This indicated a dual role for Shh in both MN and OLP generation. Consistent with this, Shh induces the expression of Olig2 in the pMN domain. MNs and OLPs are depleted or eliminated in the spinal cord of mice and zebrafish lacking Olig2.9, 10, 11, 12 In addition, Olig2 promotes the expression of the neurogenic bHLH factor, Ngn2, that contributes to the terminal differentiation of MNs.13, 14 These data led to a model in which MNs and OLPs derive from a common progenitor that expresses Olig2, which switches from MN to OLP generation at a defined developmental time point. However, the factors regulating the temporal switch from MN to OLP and maintaining the population of progenitors undifferentiated to ensure that the appropriate numbers of each cell type are generated, remain elusive.
The Notch pathway maintains the balance between progenitor self renewal and differentiation.15 Ligands of the Notch pathway (Dll1, Jagged 1 (JAG1)) are expressed in distinct patterns along the dorsal–ventral (DV) axis in the developing spinal cord.16, 17, 18, 19 Notch signalling is a cell–cell communication system, and activation of Notch signalling results from the interaction between the Notch receptor and its ligands, Delta and JAG, expressed by adjacent cells. The ligand receptor interaction induces a proteolytic cleavage of the Notch protein and nuclear translocation of its intracellular domain (Notch ICD), which interacts with the DNA-binding protein CSL (RBP-Jk) to activate Notch downstream genes, such as the Hes transcription factors.15 These proteins have been demonstrated to mediate their functions, at least in part, by repressing the neurogenic bHLH factor Ngn2 that contributes to the terminal differentiation of MNs,13, 14 and by inhibiting the maturation of OLPs, at least in the optic nerve.20 Thus, a plausible model might entail Notch activity preserving undifferentiated progenitors in the pMN during the period of MN generation so that sufficient numbers are available when oligogenesis supervenes. Supporting this view, in the zebrafish embryo, Notch signalling is required during embryogenesis to maintain radial glia and permit oligodendrocyte development.21 However, whether Notch signalling has a direct role in determining the timing, and the switch, from neurogenesis to gliogenesis is unknown.
Here we show that in the chick and the mouse embryo, the expression of the Notch ligand JAG2 is restricted to the pMN domain during the period of MN generation, and that this pattern of expression depends on Shh activity. In vivo experiments demonstrate that loss of JAG2 resulted in the accelerated differentiation of MNs and the premature generation of OLPs. Conversely, GOF experiments demonstrate that JAG2 activity prevents the differentiation of pMNs, at least in part by lowering the levels of Olig2 protein, thus preserving Olig2+ progenitors for later oligodendrogenesis. In search for the Notch-dependent effector regulating this cell fate switch, we found that Hes5, a Notch target and direct repressor of OLP specific genes, is regulated and mediates the activity of JAG2. Together the data provide new insight into the genetic network responsible for the timely generation of spinal MNs and oligedendrocyte progenitors.
Results
Shh signalling directs expression of JAG2 to the pMN domain
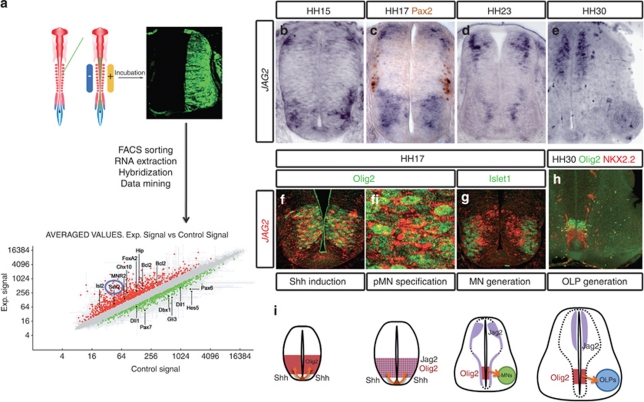
An in vivo transcriptome analysis of neural progenitors responding to Shh activity (see Materials and Methods),22 focussed our attention on the Notch pathway, and the possibility that Notch signalling has a role in patterning the neural tube. In response to activators of the Shh pathway, expression of the Notch ligand Serrate2/JAG2 appeared upregulated (Figure 1a).
Figure 1.
The Notch ligand Jagged2 is transiently expressed in the ventral spinal cord. (a) A transcriptional profiling strategy for Shh identified patterning determinants and components of the Notch pathway. Chick embryos were electroporated with activator or inhibitors of the Shh pathway (see Materials and Methods), transfected GFP-expressing cells were FACS sorted, and extracted RNA was used to hybridise full genome genechips. In response to activators of the Shh pathway (Gli3Act), genes expressed in the ventral NT were upregulated, whereas dorsal genes appeared downregulated. In addition, expression of the Notch ligand Serrate2/Jagged2 appeared upregulated. (b–e) Expression of Jagged2 revealed by in situ hybridisation in sections of chick embryos at the indicated stages. (c) Shows double labelling with the intermediate neuronal subtypes (Pax2). (f, g) Expression of Jagged2 revealed by fluorescent in situ (red), double labelled with the pMN marker Olig2 (f) or the MN marker islet1 (g). (h) Double immnunohistochemistry for Olig2/Nkx2.2 shows the generation of OLP. (i) Summary of the expression of Jagged2 in relation to the generation of MNs and OLPs from the pMN domain
To examine the distribution of JAG2 in the developing spinal cord, we mapped its expression in relation to proteins defining different DV. The onset of expression occurred after neural tube closure in scattered cells (data not shown). From HH15 to HH17 JAG2 was expressed within the motor neuron progenitor domain (pMN; Figures 1b and c). At these stages, JAG2 was co-expressed with the MN progenitor protein Olig2 in a salt-and-pepper pattern (Figure 1f), and was excluded from the differentiated MNs (Figure 1g). In addition, JAG2 was expressed in a subset of dorsal progenitors (Figures 1b–d). Expression of JAG2 in the pMN was transient, however, was maintained in the transition zone (TZ) by HH23/24 (Figure 1d) and was extinguished by HH30 (Figures 1e and i). Dorsal expression of JAG2 is maintained at HH30, together with lower expression levels in some sub-populations of differentiated MNs. Interestingly, the downregulation of JAG2 expression in the pMN domain correlates with the time OLP generation supervenes (Figures 1e, h and i).
We tested whether the ventral expression of JAG2 depends on Shh activity. Analysis of the Shh−/− embryos showed the lack of JAG2 expression in the ventral hindbrain and spinal cord (Supplementary Figure 1). In addition, inhibition of Shh signalling by a dominant-negative version of Gli3 (Gli3Rep)23 resulted in the loss of JAG2 expression from pMN. Conversely, constitutive activation of the Shh pathway, using Gli3Act resulted in a dorsal expansion of JAG2 (Supplementary Figure 1). Together these results indicate that Shh signalling is required and sufficient for the ventral expression of JAG2.
Experimental evidence supports a model in which the specific distribution of Notch ligands endows distinct progenitor domains with the ability to influence the pace of cell differentiation in a domain- and cell-type-specific manner.19, 24, 25 This prompted us to ask whether JAG2 influenced the generation of either MNs and/or OLPs. We generated short-hairpin RNA (shRNA) to knockdown endogenous JAG2. Analysis by qPCR of target RNA in transfected neural cells indicated that the shJAG2 (loss-of-function: LOF-JAG2) reduces RNA levels to ∼50% of normal without reducing the closely related ligand JAG1 (Supplementary Figure 2). Knockdown of JAG2 did not affect the relative distribution of proliferating progenitors and differentiated neurons in the spinal cord, neither changed the identity of progenitors along the DV axis (Supplementary Figure 2 and data not shown) similar to the results recently reported for JAG1.19 This analysis indicated that shJAG2 did not affect the anatomical integrity of the neural tube, prompting us to analyse cell fate identities.
JAG2 activity regulates the timely generation of MNs
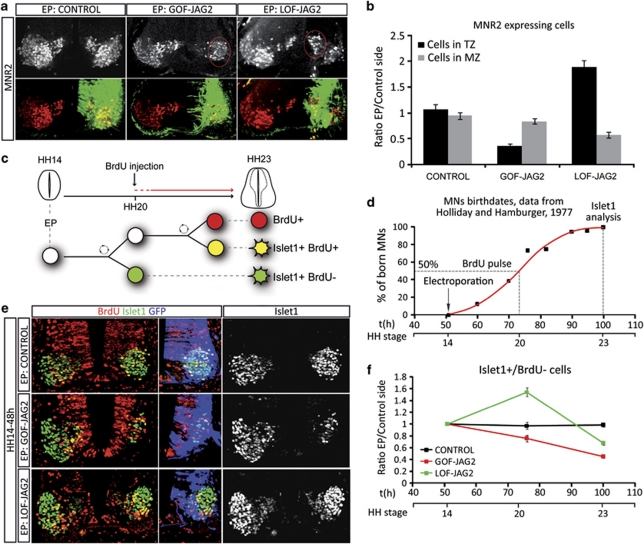
In the spinal cord, MNs are generated from the pMN domain in a narrow time–window (stage HH15 to stage HH23/24).26 To test the role of JAG2, brachial regions of embryos were electroporated at HH14, before the onset of MN generation, and assayed with the early MN marker MNR2/HB9.14 Quantification of MNs was performed relative to the ventricular lumen (Figure 2a). MNR2+ cells positioned adjacent to the ventricular zone (VZ) correspond to newly generated MNs (MNR2+ cells in the TZ), whereas those positioned towards the basal/pial surface correspond to terminally differentiated MNs (MNR2+ cells in the marginal zone). Knockdown of JAG2 (LOF-JAG2) markedly changed the distribution of MNR2+ cells: more cells were positioned adjacent to the VZ and less terminally differentiated MNs were present. By contrast, overexpression of JAG2 (GOF-JAG2) reduced the total number of MNR2+ cells at both locations (Figure 2b). This suggested JAG2 activity to regulate the tempo of MN generation and prompted us to analyse MN differentiation.
Figure 2.
Jagged2 activity controls the timely differentiation of MNs. (a) Embryos electroporated (EP) at HH14, with the indicated DNAs, were analysed at 72 h post EP with the early MN marker MNR2 (red). EP side is shown to the right, GFP (green) shows transfected cells. Quantification of motor neurons and their spatial distribution was performed analysing confocal images. (b) Histograms represent the number of MNR2-positive cells depending on the distance to the lumen; TZ, transition zone; MZ, mantle zone. (c) Schematic representation of the MN birth dating experiments following Jagged2 manipulations. Embryos EP at HH14 were labelled with a single BrdU pulse 24 h PE, and analysed at 48 h PE for BrdU/MN markers. (d) Schematic representation of motor neurons birth dates in the brachial spinal cord of the chick embryo, data adapted from Hollyday and Hamburguer.26 (e) Example sections showing double staining for the MN marker Islet1 (green) and BrdU (red). EP side is shown to the right, GFP (blue) shows transfected cells. (f) Quantitative data showing the proportion of born MNs, plots correspond to the ratio of EP versus non-EP side in each experimental condition (control, black dots; GOF-Jag2, red dots; LOF-Jag2, green dots). Time is represented as hours of incubation and HH stages. First dot corresponds to the EP time, second to the BrdU−/Islet1+ cells (MNs born at the time of the BrdU pulse), third dot represents the total Islet1+ cells (MNs terminally differentiated 48 h PE). Bars correspond to the standard error (S.E.M.)
To unequivocally identify the birth date of MNs, we administered a single dose of bromodeoxyuridine (BrdU) at HH20 (24 h PE, the time at which ∼50% should be post mitotic26) and harvested the embryos at HH23/24 (48 h PE). Double immunohistochemistry for BrdU and the MN marker Islet1 allowed us to identify MNs (Islet1+/BrdU−) that had exited the cell cycle and initiated differentiation before HH20 (Figures 2c and d). Embryos electroporated at HH14 (before the onset of MN generation) with the control vector contained 43±0.015% of Islet1+/BrdU− cells, confirming that ∼50% of MNs were generated before HH20 (Figures 2e and f). By contrast, in embryos in which JAG2 activity was depleted from HH14, the percentage of Islet1+/BrdU− MNs increased significantly to 61±0.017% (∼1.5-fold increase) Furthermore, the total number of Islet1+ cells appeared reduced, 48 h PE of LOF-JAG2 (Figures 2e and f). This suggests that loss of JAG2 activity promotes the premature differentiation of MNs.
Conversely, gain-of-function (GOF) experiments with JAG2 (GOF-JAG2) resulted in the decrease in MN differentiation and the percentage of Islet1+/BrdU− decreased to 33±0.02% (∼1.5-fold decrease; Figures 2e and f). Importantly, these experiments indicated that the ability of JAG2 to activate Notch is limited to pMN progenitors, as differentiation of other neuronal identities was largely unaffected (Supplementary Figure 3). This is in concordance to the recently reported roles for Dll1 and JAG1.19, 25
JAG2 activity inhibits the differentiation of Olig2+ progenitors
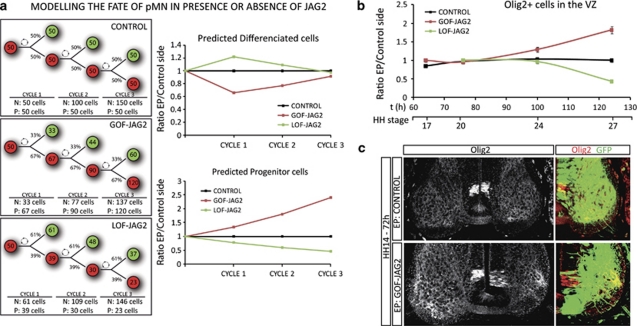
Intriguingly both LOF and GOF in vivo experiments resulted in a reduction of the final numbers of differentiated MNs. To understand this result, we took advantage of the data generated by the determination of the percentages of born MNs (Figure 2c), to model the behaviour of the pMN domain for three cell divisions (∼48 h). We took the simplifying assumption that normally 50% of divisions were asymmetric. We examined the effect of the changes in the rate of neurogenesis measured in the LOF and the GOF experiments. Plotting the ratios of predicted differentiated MNs in each experimental condition showed the initial increase in MN in the absence of JAG2, whereas at the end of the 48 h period, the final MN number decreased in both conditions. These profiles were similar to those obtained empirically (Figure 3a). Moreover, the model predicted specific changes in progenitor numbers, prompting us to experimentally test this prediction.
Figure 3.
Overexpression of Jagged2 retains Olig2+ cells in a progenitor stage. (a) Model representing the fate of pMN progenitors along three cell cycles, in control situation in which 50% divisions are asymmetric, giving rise to one MN (green) and one progenitor cell (red). Percentages of asymmetric divisions are adapted from data provided by BrdU incorporation experiments in the presence or the absence of Jagged2. Drawing shows the resulting proportion of neurons (N) and progenitors (P). Plots represent the ratio of theoretical MNs generated in each condition, dots correspond to each cell cycle. Plots represent the ratio of theoretical progenitors that remain in each condition, dots correspond to each cell cycle. (b) Quantitative data showing the ratio of Olig2+ cells, plots correspond to the ratio of EP versus non-EP side in each experimental condition (control, black dots; GOF-Jag2, red dots; LOF-Jag2, green dots). Time is represented as hours of incubation and HH stages. Dots correspond to 12 h 24 h, 48 h and 72 h PE. Bars correspond to the standard error (S.E.M.). (c) Sample sections, EP with indicated DNAs, and immunostained with Olig2 (red). EP sides are shown to the right, GFP (green) shows transfected cells
Olig2 is a key component of the genetic network operating in the pMN for the generation of MNs.2, 3, 14 Consistent with the prediction of the model, the forced expression of JAG2 (GOF-JAG2) resulted in an increase in Olig2+ progenitor cells within the VZ (Figure 3b). Conversely, reduced JAG2 activity (LOF-JAG2) caused a gradual loss of Olig2+ cells from the VZ (Figure 3b). These experimental data match the changes in progenitor numbers predicted by our model. All together these data suggest that JAG2 controls MN differentiation through the regulation of Olig2+ cells.
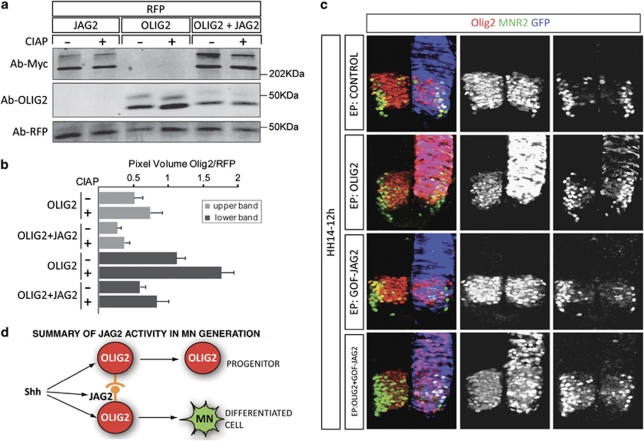
We observed that forced expression of JAG2 laterally inhibit Olig2+ cells that are retained as progenitors within the VZ (Figure 3c). It also came to our attention the fact that these Olig2+ cells appeared to express lower levels of Olig2 protein, when compared with the control contralateral Olig2+ progenitors (Figure 3c). To assess this observation, we quantified protein levels by western blot in neural tube extracts that were electroporated with either Olig2 alone or together with JAG2. Co-electroporation of JAG2 caused a ∼50% reduction on both Olig2 protein bands (Figures 4a,b). This observation suggested a mechanism by which JAG2 reduced Olig2 protein to a level below the threshold require to generate MNs, thus resulting in the lateral inhibition of MN differentiation. Indeed, overexpression of Olig2 alone was sufficient to promote MN generation, as shown by the ectopic MNR2+ cells (Figure 4c).14 However, exposure to JAG2 inhibits the MN differentiation activity of Olig2+ (Figure 4c). Altogether, our data suggest a model in which JAG2 functions in the pMN domain to inhibit laterally the Olig2-expressing progenitor from becoming MNs (Figure 4d). Olig2-expressing progenitors that do not contact with JAG2 were primed to enter the differentiation pathway to generate MNs. Lateral contact to JAG2 prevents MN differentiation, at least in part, by reducing the Olig2 protein levels.
Figure 4.
Jagged2 reduces Olig2 protein levels and inhibits the Olig2 capacity to differentiate MNs. (a) Western blots from NT total lysates were immunobloted with anti-Olig2, anti-Myc and anti-RFP antibodies. Embryos co-EP at HH14 with Jagged2-Myc, Olig2 and pCAGGS-ires-RFP were dissected at 12 h PE. Note that Olig2 protein is distributed within two separate bands that were both shifted but maintained in the presence of calf intestine alkaline phosphatase (CIAP), showing that both forms are phosphoproteins. These two bands might reflect alternate translation initiation sites.2 Anti-Myc reveals the presence or absence of Jagged2, anti-RFP as EP control. (b) Quantitative analysis of upper and lower Olig2 bands, in the presence or the absence of Jagged2, normalised to RFP levels. Olig2 protein levels are significantly reduced in NT co-transfected with Jagged2. (c) The Olig2 capacity to differentiate MNs is prevented by Jagged2 activity. Embryos EP at HH14 with the indicated DNAs were analysed 12 h PE for the MN progenitor marker Olig2 (red) and the MN definitive marker MNR2 (green). GFP is shown in blue. EP of Olig2 is sufficient for the generation of ectopic MNR2+ MNs. Quantification of ratio of MNR2+ cells in EP versus control non-EP side revealed a ratio of 0.85±0.07 in control EP embryos, whereas in Olig2-EP increased to 2.02±0.03. EP of Jag2 prevents the generation of MNR2+ cells to 0.58±0.02, whereas co-EP of Olig2 and Jag2 also reduced the appearance of MNR2+ cells to 0.51±0.05 in the normal ventral domain, but also prevented the generation of ectopic dorsal MNs. (d) Summary model of Jagged2 activity in the pMN domain, Jagged2 activity splits Olig2+ progenitors into two different identities
JAG2 activity is necessary to prevent premature OLP generation
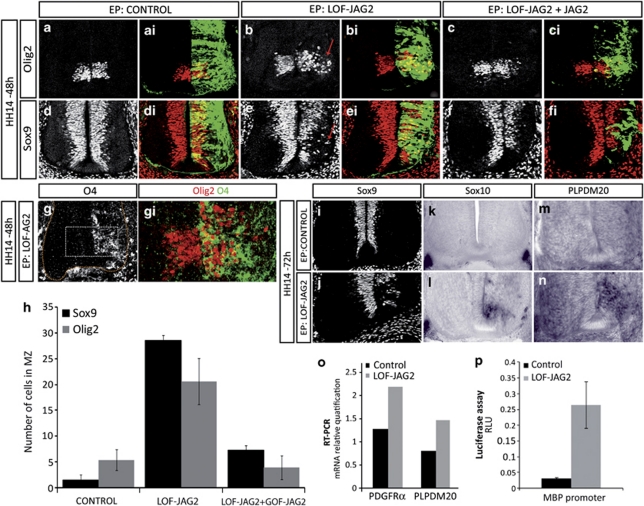
Once neurogenesis of MNs is terminated, progenitors in the pMN domain switch to the generation of OLPs. We analysed the effect of JAG2 depletion on the oligodendrogenesis. Strikingly, after knockdown of JAG2 function at HH14, Olig2-expressing cells were observed migrating away from the pMN (mean 19.14±1.99 cells/section, n=20; Figures 5a and b). This contrasted with control embryos in which Olig2 was robustly expressed in the pMN domain and no migratory Olig2+ cells were detectable. Furthermore, Olig2+ migratory cells were significantly reduced when the human JAG2 (GOF-JAG2) was co-electroporated, supporting the specificity of the phenotype (rescue experiments: mean 9±2.27 cells/section, n=15; Figures 5c and h). Olig2-expressing cells that migrate away from the VZ have been characterised as OLPs,2, 3, 9, 10 but these are not normally generated until after the period of MN generation. To characterise the molecular nature of these migratory Olig2+ cells, we co-immunostained with the OLP-specific marker O4.7 This revealed that most of the migratory Olig2+ cells co-expressed O4 (Figure 5g), consistent with these cells being OLPs.
Figure 5.
Depletion of Jagged2 activity at the time of MN generation results in premature OLP generation. (a–h) Embryos electroporated at HH14 with the indicated DNAs were analysed at 48 h PE with OLP markers. EP side is shown to the right, GFP (green) shows transfected cells. (a and b) Olig2 is expressed at pMN in control NT, whereas GOF-JAG2 EP shows migratory Olig2+ cells. (c) Co-EP with human Jagged2 (GOF-JAG2) restores Olig2+ cells to pMN. (d and e) Sox9 is expressed throughout the VZ in control NT, whereas LOF-JAG2 electroporated NT shows depletion of Sox9+ cells at pMN and migratory Sox9+ cells. (f) Co-EP with GOF-JAG2 restores Sox9 expression in pMN. (g) Olig2+ migratory cells (red) co-expressed the OLP marker O4 (green). (h) Quantitative analysis of Olig2+ and Sox9+ migratory cells at 48 h PE after electroporation of the indicated DNAs. Numbers are shown as migratory marker expressing cell in each experimental condition. Bars correspond to the standard error (S.E.M.). (i–n) At 72 h PE of LOF-JAG2, Sox9 is extinguished from the VZ, and premature expression of the OLP markers Sox10 and PLPDM 20 are detected by in situ hybridisation. In control electroporated spinal cords, Sox10 and PLPDM20 are only expressed in the ventral roots. (o) Premature OLP generation as revealed by the real-time–PCR detection of a significant increase in the expression of PDGF receptor α and in the myelin-specific gene PLP at 48 h PE of LOF-JAG2. (p) Premature OLP generation as revealed by the increased activity of the MBP-Luc reporter 24 h PE of LOF-JAG2
Furthermore, the SoxE proteins have a prominent role in OLP generation. Sox9 is required for OLP specification and is expressed transiently in early migrating OLP. Sox8 and Sox10 are also expressed in early migrating OLPs, and Sox10 activates myelin genes as part of the terminal differentiation programme of oligodendrocytes.27, 28 Consistent with migratory cells being OLPs, knockdown of JAG2 resulted in the premature migration of Sox9+ cells (mean 28.57±5.73 cells/section, n=16) and the reduction of Sox9+ cells from the pMN. Again in a JAG2-LOF background, the number of Sox9+ migratory cells was reduced when NT was co-electroporated with the human JAG2 cDNA (GOF-JAG2) that was resistant to the short hairpin (rescue experiments; mean 7.2±2.1 migratory cells/section) further supporting the specificity of this phenotype (Figures 5d–f and h). Furthermore, premature migratory OLPs expressed Sox8 (not shown), Sox10 and the myelin-specific gene PLPDM20 (Figures 5i–n). Knockdown of JAG2 additionally increased levels of PDGFRa and PLP expression, as assessed by real-time PCR (Figure 5o), as well as the activity of the myelin basic protein (MBP) reporter as assessed by luciferase activity (Figure 5p). Altogether, these data indicate that knockdown of JAG2 activity results in the precocious generation of OLPs from the pMN domain.
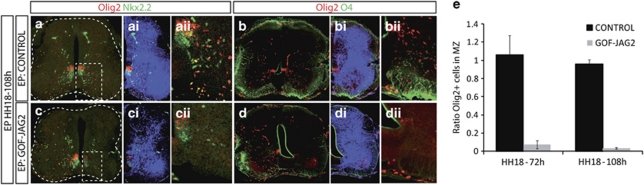
Converse GOF experiments allowed the maintenance of high levels of JAG2 expression at the time of OLP generation, when JAG2 expression is normally extinguished from pMN. This resulted in the loss of migratory OLP identified by double immunohistochemistry for Olig2+/Nkx2.2+ and Olig2+/O4+, at the stage at which migratory OLPs were abundant in the control contralateral side (Figures 6a–e).7, 29, 30 This is consistent with the finding that JAG2 has a negative role on Olig2 expression. Together these data supports the idea that MNs and OLPs are sequentially generated from a single progenitor and that JAG2 activity ensures progenitors do not switch from neurogenesis to gliogenesis prematurely.
Figure 6.
Persistent overexpression of Jagged2 inhibits OLP generation. Embryos EP at HH18 with Jagged2 (GOF-JAG2), or control empty vector (Control), were analysed at 4, 5 days after EP for oligodendrocyte generation. (a and c) Oligodendrocytes are identified as migratory cells departing from the pMN expressing Olig2 (red) and Nkx2.2 (green). Electroporated side is shown to the right, GFP (blue) shows transfected cells. Controls show numerous migratory OLPs. GOF-JAG inhibits OLP migration. (b and d) Oligodendrocytes are identified as migratory expressing Olig2 (red) and O4 (green). Electroporated side is shown to the right, GFP (blue) shows transfected cells. Controls show numerous migratory OLPs. GOF-JAG inhibits OLP migration. (e) Quantitative analysis of Olig2+ migratory cells at 72 h and 108 h after electroporation of the indicated DNAs. Numbers are shown as migratory marker expressing cell in each experimental condition. Bars correspond to the standard error (S.E.M.)
JAG2 controls OLP generation through the regulation of Hes5 expression
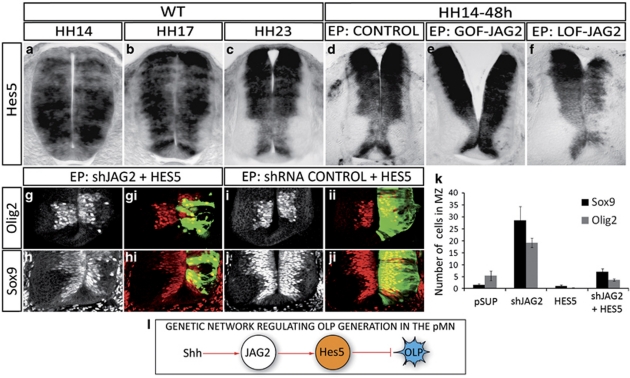
How might JAG2 prevent precocious OLP generation? Members of the Hes family of bHLH transcription factors are the best characterised Notch targets that function as effectors of the cell fate decisions mediated by Notch signalling. In the chick spinal cord, Hes5 is the only member expressed in the pMN domain (Figures 7a and b),31 and its expression is reduced from these progenitors as they switch from MN to OLP (Figure 7c). Moreover, mice lacking Hes5 generate excess oligodendrocytes.32 These data raised the possibility that Hes5 is the transcriptional effector that regulates the timing of OLP generation.
Figure 7.
Hes5 acts downstream of Jagged2 to prevent OLP generation. (a–c) In situ hybridisation of wild-type embryos shows that Hes5 is transiently expressed in pMN at the time of MN generation and extinguished after MN generation. (d) Control EP show transient expression of Jagged2 at the pMN. (e) Overexpression of Jagged2 (GOF-JAG2 48 h PE at HH14) maintains Hes5 expression at pMN. (f) Depletion of Jagged2 (LOF-JAG2 48 h PE of shJagged2 at HH14) inhibits Hes5 expression. (g–j) Hes5 overexpression retains Olig2+ and Sox9+ cells at pMN, even in the absence of Jagged2 function. Electroporated side is shown to the right, GFP (green) shows transfected cells. (k) Quantitative analysis of migratory Olig2+ and Sox9+ cells. Numbers are shown as total migratory cells on the electroporated side in each experimental condition. Bars correspond to the standard error (S.E.M.). (l) Summary of the genetic network operating for OLP generation
We tested whether JAG2 regulates Hes5 expression in the pMN. Reduction of JAG2 function caused the loss of Hes5 expression (Figures 7d and f). Moreover, even though Hes5 is broadly expressed, the loss of Hes5 was restricted to the pMN. Conversely, GOF experiments showed that overexpression of JAG2 resulted in the maintenance of high levels of Hes5 in the pMN domain (Figure 7e).
We next asked whether Hes5 activity could rescue the premature generation of OLP observed in the absence of JAG2. Indeed, in embryos in which JAG2 was depleted by shRNA, the forced expression of Hes5 significantly reduced the premature migration of OLPs (Olig2+ from 19.14±1.99 to 3.62±0.53 cells/section; Sox9+ from 28.57±5.73 to 7.09±1.28 cells/section, Figures 7g–k). Consequently, Olig2- and Sox9-expressing cells were maintained in the VZ of the pMN domain (Figures 7g and h). This supports the conclusion that Hes5 acts downstream of JAG2 to control the premature generation of OLPs.
Discussion
In this study, we provide evidence that the generation of MN and OLP in the ventral spinal cord depends on the activity of the Notch ligand JAG2. The activity of JAG2 prevents the neighbouring Olig2-expressing progenitor from entering the differentiation pathway. In pMN progenitors in which the Notch pathway is activated by JAG2, MN differentiation programme is prevented, at least in part, by reducing the Olig2 protein levels. This preserves a pool of progenitors and, at the same time, maintains high levels of Hes5 expression, that in turn directly inhibit the oligodendrocyte generation during the neurogenic phase.
In the absence of JAG2, pMN progenitors enter the differentiation programme to generate first MNs and next, when Hes5 levels are sufficiently reduced, to generate oligodendrocytes. These data support a model in which MN and OLP arise sequentially from a common progenitor population residing in the pMN. JAG2 controls the timing of the switch from MN to OLP production by suppressing OLP generation during the neurogenic phase.
During the process of writing these results, two independent studies showed post-transcriptional modifications of Olig2 that changed the activity of Olig2, and thus the outcome of the Olig2+ progenitors in the pMN. Li et al.33 found a phosphorylation site in the bHLH domain of Olig2 that favoured the homodimerisation capability of Olig2, and thus its transcriptional activity. Mice carrying a mutant version of Olig2 (Olig2S147A) show the loss of progenitors at the pMN domain and the loss of differentiated MNs, a phenotype that is highly comparable with the overexpression of JAG2. This suggested that changes in the MN differentiation capacity of Olig2, that we reported in the presence of JAG2, might be the result of a similar post-trancriptional modification. Sun et al.34 found a triple serine phosphorylation motif at the N-terminal part of the Olig2 protein, which are required for the Olig2-mediated proliferation of neural progenitors, however, are dispensable for the Olig2-mediated specification of OLPs. Altogether, these data indicate a pivotal role played by post-transcriptional modifications of Olig2 in the outcome of the pMN progenitors, and our data indicate that JAG2 might be among the upstream signals regulating these modifications.
The generation of both MNs and OLPs requires Shh signalling. Olig2 expression depends on Shh and is required for the generation of both MNs, and OLPs, suggesting an explanation for the requirement for Shh in pMN derivatives. Here we provide evidence that, in addition to Olig2, the regulation of JAG2 expression by Shh signalling contributes to the generation of MNs and OLPs. Shh activity is required and sufficient for the restricted expression of JAG2 to the pMN domain. Moreover, the involvement of JAG2 in regulating the transition from MN to OLP production links the process of spatial patterning, which positions the pMN domain in the ventral neural tube, with temporal patterning that determines when each cell type is generated. Whether JAG2 expression is directly or indirectly regulated by Shh signalling, remains to be determined. Furthermore, the timely loss of JAG2 expression from the pMN domain appears as an important pre-requisite for OLP formation. Molecular mechanism operating directly to pause JAG2 expression might include transcriptional repression and/or epigenetic silencing, among many others, and understanding these mechanisms remains as an important unresolved question.
Whether the Notch pathway is required to maintain pools of undifferentiated progenitor cells or whether it has instructive roles in promoting specific cell fates has been the subject of debate. Our data indicate that Notch signalling, activated by JAG2, maintains pMN progenitors in an undifferentiated state but also provides an instructive signal that blocks the fate switch from MN to OLP. This suggests a dual role for Notch signalling and is consistent with other studies of glial development. The Notch effector Hes1 promotes Muller and radial glia identities,35 whereas Hes5 directly regulates expression of myelin genes.32 Moreover, Notch signalling has been implicated in the diversification of neuronal identities in the neural tube.24, 25 The observation that Notch ligands are expressed in discrete domains along the DV axis of the CNS is in line with the idea that Notch ligands have specific roles in the cell fate decisions of progenitors. Thus, our data support the idea that Notch ligands can serve both to prevent the untimely differentiation of neural progenitors and to generate diversity within a population of progenitors.21, 24, 25, 36
Our results suggest a model in which, early in the developing spinal cord, a common neuroglioblast progenitor is present in the pMN domain, and this has the capacity to generate MN and OLP.33, 34 During MN generation, Hes5 induced by JAG2 prevents premature OLP differentiation and maintains a proportion of these progenitors in an undifferentiated state. The switch to OLP generation depends on the downregulation of JAG2 expression, which facilitates the differentiation of the remaining progenitors and disinhibits OLP generation. When JAG2 activity is artificially depleted in vivo, the precocious generation of OLP is accompanied by a reduction in MN generation. This argues against a model in which separate OLP and MN precursors are present in the pMN, and suggests that in the absence of Notch activity, the progenitors of MN adopt a gliogenic identity. Nevertheless, our results cannot formally rule out the presence of separate lineage-restricted neuroblasts and glioblasts within the pMN that are derived from a single common precursor.37 In this case, JAG2 activity would act on these populations separately to inhibit differentiation and OLP generation. High-resolution analysis of the behaviour of individual pMN progenitors will be necessary to conclusively test this possibility. Notwithstanding this question, the data presented here demonstrate a novel role for JAG2 and Notch signalling in the temporal control of cell type identity in the vertebrate CNS.
Materials and Methods
Fluorescence-associated cell sorting (FACS) and microarray analysis
Plasmid DNA encoding either green fluorescence protein (GFP) alone, a deleted form of mouse patched lacking the second large extra cellular loop (mPtc1Δloop2) or an activated version of Gli3 (Gli3Act aa1402-4743) were inserted into pCAGGS-ires-EGFP vector as reported,23 for in vivo electroporation into HH11-12 stage embryos. Neural tubes were dissected out at 14 or 36 h later, and single-cell suspension was obtained by 10–15 min incubation on Tripsin-EDTA (Sigma-Aldrich, Madrid, Spain). GFP fluorescence was determined by flow cytometry using a MoFlo flow cytometer (DakoCytomation, Fort Collins, Colorado, USA). The resulting cell population, consisting of ∼90% of GFP+ cells, was used for total RNA extraction. GeneChip Chicken Genome Arrays (Affymetrix, Madrid, Spain) containing 32 773 chicken transcripts were used for the hybridisation of the samples. Unpaired analysis of the data, based on the comparison between the average values of the logRed and logGreen intensities, was performed using SOLAR System v2.0 (Alma Bioinformatics, Madrid, Spain). Results were filtered using unpaired t-tests using thresholds of ≥1.5-fold change and P-value ≤0.05.22
Mouse and chick embryos
Mice homozygous for the Shh-null allele were as published.6 Eggs from White Leghorn chickens were staged according to Hamburger and Hamilton (HH).38 For chick embryos electroporation, plasmid DNA was injected into the lumen of neural tubes and electroporation carried out using and Intracel Dual Pulse (TSS10) electroporator (Intracel, Royston, UK).
For BrdU labelling, 5 μg/μl BrdU was injected into the neural tubes at the indicated times before fixation. Transfected embryos were allowed to develop to the indicated stages, then dissected and processed for immunohistochemistry, in situ hybridisation or FACS sorting, and qPCR analysis.
Immunohistochemistry and in situ hybridisation
For immunohistochemistry, embryos were fixed 2–4 h at 4 °C in 4% PF in PB. For BrdU detection, sections were incubated in 2 N HCl for 30 min followed by 0.1 M Na2B4O7 (pH8.5) rinses further PBT rinses and anti-BrdU incubation. Immunostainings were performed following standard procedures on either vibratome or cryostat sections. Antibodies against the following proteins were used; GFP (Molecular Probes, Invitrogen, Barcelona, Spain), a unique β-tubulin (Tuj-1, Medpass International, Paris, France), phospho-histone 3 (p-H3, Upstate Biochemicals, Millipore, Madrid, Spain), Olig2,30 Pax2 (Zymed, Invitrogen), Sox9 (M Wegner), O4 (C Soula). Monoclonal antibodies to BrdU, MNR2, Nkx2.2, Islet1 and Lhx2.9 were obtained from the Developmental Studies Hybridoma Bank. Alexa488-, Alexa555- and Alexa633-conjugated anti-mouse or anti-rabbit antibodies (Molecular Probes) were used. After single or double staining, sections were mounted, photographed using a Leica (Leica Microsystems, Barcelona, Spain) SP5 Confocal microscope and processed with Adobe Photoshop CS5.
For in situ hybridisation, embryos were fixed overnight at 4 °C in 4% PF in PB, rinsed and processed for whole-mount RNA in situ hybridisation following standard procedures using probes for chick Jagged2, Hes5, Sox8, Sox10 and PLPDM20 (from the chicken EST project, UK-HGMP RC). Mouse Jagged2 probe was used (JL de la Pompa). Hybridised embryos were developed either with NBT/BCIP, or with FAST-RED Chromogen (Roche Diagnostics, Barcelona, Spain) for fluorescent in situ, rinsed in PBT and vibratome sectioned and photographed on a Leica DMR microscope or a Leica SP5 confocal.
DNA constructs
A deleted form of mouse patched (mPtc1Δloop2), deleted forms of human Gli3 (Gli3Rep aa1–768 and Gli3Act aa1402–4743), human full-coding sequences for Jagged2 and Hes5 were cloned into pCAGGS-ires-GFP for in vivo GOF experiments. The full-length mouse Olig2 cDNA into pCDNA3.1 was also used.14
For in vivo LOF experiments, shRNA-based expression vectors for RNA interference were generated using the pSUPER RNAi system (OligoEngine Inc.). A pair of custom oligonucleotides that contained a unique 19-nt sequence derived from the chicken Jagged2 mRNAs were designed using the RNAi Design Tool (http://www.oligoengine.com). Forward and reverse 64-nt oligos (Sigma) were used for annealing and cloning into the pSUPER vector (Supplementary Figure 1a), according to the manufacturer's guidelines. Control shRNA was generated for a human HDAC sequence, cloned into the pSUPER vector and electroporated at same developmental stages for each experiment.
RT–quantitative real-time PCR
EGFP-containing plasmid DNAs were electroporated, and neural tubes dissected out 24–48 h later. Single-cell suspension was obtained by 10–15 min incubation on Tripsin-EDTA (Sigma). GFP-expressing cells were FACS sorted, and the resulting cell population was used for total RNA extraction. PCR reactions were carried out by triplicate using fixed amounts of template DNA.
Oligonucleotides were obtained from Qiagen (Madrid, Spain), (QuantiTec Primer Assays) for use in real-time RT–PCR with SYBR green detection. References for QuantiTect Primer Assay were as follows: GgJagged1 QT00598003, GgJagged2 QT01512749, GgPDGFRalpha QT00598969, GgPLP QT00598955. A standard curve was made for each amplicon by plotting the number of cycles at which the fluorescence crossed the threshold (crossing values) against increasing amounts of DNA template. All experimental values were normalised to those obtained for GAPDH.
Western blot
HH stage 14 embryos were electroporated with a Myc-tagged-Jagged2, a V5-tagged-Olig2 and pCAGGS-ires-RFP for transfection control. Neural tubes dissected 12 h PE were collected in PBS and directly lysed in 1 × SDS loading buffer (10% glycerol, 2% SDS, 100 mM DTT and 60 mM Tris-HCl pH 6.8) and the DNA disrupted by sonication. Selected samples were treated with calf intestine alkaline phosphatase for 30 min, normalised by total protein content, separated by SDS–PAGE gel electrophoresis and transferred to nitrocellulose membranes; blocked with 8% non-fat dry milk in TTBS (137 mM NaCl; 0.05 Tween 20 and 20 mM Tris HCl (pH 7.4)) and probed with in house produced monoclonal anti-Myc antibody (9E10) and polyclonal mouse anti-RFP or rabbit polyclonal anti-Olig2.30 The blots were developed using anti-rabbit or anti-mouse coupled to IRDye800CW or IRDye680LT and scanned with the Odyssey Infrared Imaging System (LI-COR Biotechnology, Ltd, Cambridge, UK).
Luciferase reporter assay
A transcriptional activity assay of JAG2 activity on the MBP reporter was performed in vivo. Chick embryos were electroporated with indicated DNAs or empty pCAGGS-GFP vector as control; together with a firefly luciferase reporter construct containing the MBP reporter (M Wegner) and a renilla luciferase reporter construct (Promega Biotech, Madrid, Spain) for normalisation. Embryos were harvested after 24 h incubation in ovo, and GFP-positive neural tubes were dissected and homogenised with a douncer in Passive Lysis Buffer on ice. Firefly and renilla luciferase activities were measured by the Dual Luciferase Reporter Assay System (Promega).
Quantifications and statistical analysis
Cell counting was made on 10–20 different sections of at least four different embryos after each experimental condition from confocal images. Quantitative data were expressed as mean±S.E.M.
Acknowledgments
We thank Susana Usieto and Mónica Pons for invaluable research assistance. For DNAs and antibodies we thank Dr D Rowitch, Dr M Wegner, Dr C Soula, Dr JL de la Pompa. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD, and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. Work in the EM lab is supported by Grant BFU2010-18959. Work in the SP lab in supported by Grant BFU-2008-02424BFI. Work in the JB lab is supported by MRC.
Glossary
- Shh
Sonic hedgehog
- pMN
motor neuron progenitor
- MN
motor neuron
- OLP
oligodendrocyte progenitor
- DV
dorsal–ventral
- GOF
gain-of-function
- LOF
loss-of-function
- VZ
ventricular zone
- TZ
transition zone
- JAG
Jagged
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Bazan
Supplementary Material
References
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, et al. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Martí E, Bumcrot DA, Takada R, McMahon AP. Requirement of 19K form of Sonic hedgehog for induction of distinct ventral cell types in CNS explants. Nature. 1995;375:322–325. doi: 10.1038/375322a0. [DOI] [PubMed] [Google Scholar]
- Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, Beachy PA, et al. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Soula C, Danesin C, Kan P, Grob M, Poncet C, Cochard P. Distinct sites of origin of oligodendrocytes and somatic motorneuons in the chick spinal cord: oligodendrocytes arise from Nkx2.2-expressing progenitors by a Shh-dependent mechanism. Development. 2001;128:1369–1379. doi: 10.1242/dev.128.8.1369. [DOI] [PubMed] [Google Scholar]
- Oh S, Huang X, Chiang C. Specific requirements of sonic hedgehog signalling during oligodendrocyte development. Dev Dyn. 2005;234:489–496. doi: 10.1002/dvdy.20422. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson D. The bHLH transcription factors OLIG1 and OLIG2 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic-helix-loop-helix factor Olig2 is essential for the development of motor neuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2003;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Myat A, Henrique D, Ish-Horowicz D, Lewis J. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol. 1996;174:233–247. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- Marklund U, Hansson EM, Sundström E, de Angelis MH, Przemeck GK, Lendahl U, et al. Domain-specific control of neurogenesis achieved through patterned regulation of Notch ligand expression. Development. 2010;137:437–445. doi: 10.1242/dev.036806. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, et al. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Park HC, Appel B. Delta-Notch signalling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- Cruz C, Ribes V, Kutejova E, Cayuso J, Lawson V, Norris D, et al. Foxj1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development. 2010;137:4271–4282. doi: 10.1242/dev.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuso J, Ulloa F, Cox B, Briscoe J, Martí E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, et al. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134:3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha FS, Santos-Lopes S, Gossler A, Henrique D. Dll1 and Dll4 function sequentially in the retina and pV2 domian of the spinal cord to regulate neurogenesis and creat cell diversity. Dev Biol. 2009;328:54–65. doi: 10.1016/j.ydbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Hollyday M, Hamburguer V. An autorradiographic study of the formation of the lateral motor neuron column in the chick spinal cord. Brain Res. 1977;132:197–208. doi: 10.1016/0006-8993(77)90416-4. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk DI, Kaing S, Stiles CD, et al. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol. 2001;11:1413–1420. doi: 10.1016/s0960-9822(01)00441-9. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ. The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron. 2001;31:791–807. doi: 10.1016/s0896-6273(01)00414-7. [DOI] [PubMed] [Google Scholar]
- Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;281:318–333. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, et al. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4822–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Paes de Faria J, Andrew P, Nitarska J, Richardson WD. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron. 2011;69:918–929. doi: 10.1016/j.neuron.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, et al. Phosphorylation state of olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Yeo SY, Chitnis AB. Jagged-mediated Notch signalling maintains proliferating neural progenitors and regulates cell diversity in the ventral spinal cord. Proc Natl Acad Sci. 2007;104:5913–5918. doi: 10.1073/pnas.0607062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi M. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.