Abstract
Murine L1Md-A5 retrotransposon is a redox-inducible element regulated by Nrf-2/JunD and E2F/Rb-binding sites within its promoter (5′-UTR). Because the human papillomavirus (HPV) oncoprotein E7 interacts with retinoblastoma (pRb) and members of the AP1 family, studies were conducted to examine functional interactions between HPV E7, pRb, and histone deacetylase 2 (HDAC2) in the regulation of L1Md-A5. Using a transient heterologous transcription system we found that HPV E7 alone, or in combination with HDAC2, disrupted pRb-mediated L1MdA-5 transactivation. HPV E7 also ablated the transcriptional response of L1Md-A5 to genotoxic stress, but did not interfere with basal activity. We conclude that HPV E7 associates with proteins involved in the assembly of macromolecular complexes that regulate antioxidant and E2F/Rb sites within L1MdA-5 to regulate biological activity
Keywords: 5′-untranslated region (5′-UTR), Benzo(a)pyrene (BaP), E7 viral oncoprotein, Long Interspersed Nuclear Element (LINE1 or L1), Retinoblastoma protein family
1. Introduction
Long interspersed nuclear elements (LINE-1 or L1) are genetic mobile retrotransposons that insert into the genome via a “copy-and-paste” mechanism using a self-encoded reverse transcriptase and RNA intermediates [1]. The full-length consensus human L1 (L1h) is a 6–7 kb DNA sequence [2, 3] that consists of a 903 bp 5′-untranslated region (5′-UTR) [4] containing a bidirectional RNA polymerase II internal promoter, followed by two open reading frames (ORF1 and ORF2), a 3′-UTR containing an AATAAA polyadenylation signal and a poly A tail [1]. Translation of the L1 mRNA leads to expression of ORF1p, a 40 kDa protein [4], with nucleic acid and protein-protein-binding capabilities [5], and ORF2p, a 150 kDa protein [6] with endonuclease [7] and reverse transcriptase activities [8]. Murine L1 (L1Md) retroelements are similar in function to L1h. In contrast to humans where only one active L1 family exists [9], structural differences within the 5′UTR region of murine L1 give rise to several lineages, namely, F, G, Tf, Gf and A [10]. The A-type 5′UTR is made of tandemly arranged 208 bp-long monomeric DNA repeats that vary in number [11]. The promoter strength is directly related to the monomer number within the 5′ UTR [11, 12].
Our laboratory has previously identified a novel retrotransposon, L1MdA5, as a target retroelement in the genotoxic stress response to benzo-a-pyrene (BaP), an environmental carcinogen [12]. Subsequent studies identified Nrf-2/JunD and E2F/Rb binding sites within the promoter that participate in transcriptional regulation of the L1 promoter [12, 13], and established a mathematical model of retrotransposon reactivation by BaP in heterologous systems [14]. Given that many of the proteins involved in transcriptional control of L1 interact with human papillomavirus (HPV) E6 and HPV E7 oncoproteins [15–17], we hypothesized that a functional link exists between stress and viral protein-regulated gene expression. Here we show that transient heterologous overexpression of Rb and HDAC2 in human cervical cancer cells (HeLa cells) as well as challenge with BaP transactivate the L1 promoter, and that forced overexpression of HPV E7 decreased the transcriptional response of the luciferase reporter gene to both Rb and carcinogen challenge.
2. Materials and Methods
2.1 Cell Culture
Human cervical cancer-derived HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco BRL, Carlsbad, CA) supplemented with 10% FBS and 1% penicillin-streptomycin antibiotics (Cellgro, Manassas, VA), and kept at 37 °C and 5% CO2.
2.2 Transient transcription assays
Hela Cells were plated at a density of 1.5×104 cells/well in 24-well plates. Transient transfections with 500 ng of pBASIC-luciferase or pL1Md-A5 Wild Type-luciferase reporter gene constructs [12] with or without 100 ng of HPV E7, and/or Rb or HDAC2 overexpression vectors, were performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Transient transfections were performed according to manufacturer’s specifications. Briefly, cells were plated in 24-well plates the day before transfection and maintained in growth medium without antibiotics. The growth medium was replaced with serum-free Opti-MEM medium the following day. DNA-Lipofectamine complexes were added to the cells and transfections allowed to continue for 6 hours. Next, the transfection medium was aspirated and 0.5 mL of fresh medium without antibiotics added to each well. Cells were allowed to recover for 18 hours before initiation of chemical treatments [18]. To correct for randomness during transfection, 10 ng of the Renilla luciferase reporter gene (pRL) was cotransfected in each assay. The total DNA load per transfection was kept constant at 800 ng with pBlueScript. Cells were lysed 36 hours after transfection and the luciferase activity measured using a Dual-Luciferase Reporter Assay System protocol (Promega, Madison, WI). Measurements were performed on a 20/20 Luminometer (Turner BioSystems, Sunnyvale, CA). Corrected Arbitrary Luciferase Units (ALU) were calculated as the ratio of activity of the pBASIC or the pL1Md-A5 luciferase reporter gene and the pRL activity. Each assay was performed in triplicate and at least two independent assays were performed for each experiment. The data shown are from one of the representative experiments and the error bars represent the standard deviation from triplicate assays.
2.3 Chemical treatments
24 hours after transfection, fresh medium containing 0.06% (V/V) dimethyl sulfoxide (DMSO) vehicle or 3 μM BaP was added to the cells for 16 hours, as previously reported [18]. At the concentrations used in our experiments DMSO did not activate cellular redox signaling. Cells were subsequently processed for luciferase activity quantification as described above.
2.4 Statistical analyses
Analyses were done either by student’s t test or ANOVA (p<0.05) followed by post-hoc tests.
3. Results
3.1 Rb protein and HPV E7 oncoprotein exert opposite effects on the mouse L1MdA5 retroelement promoter
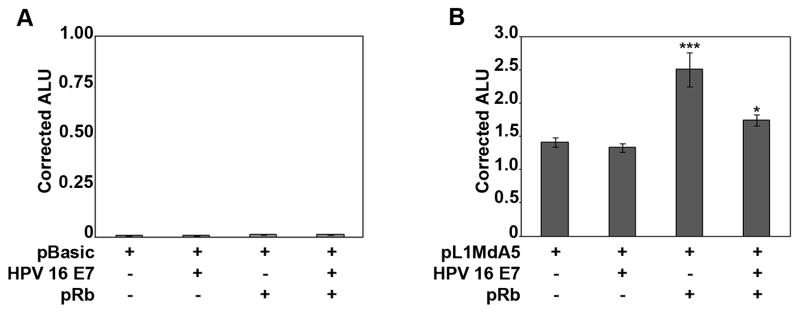
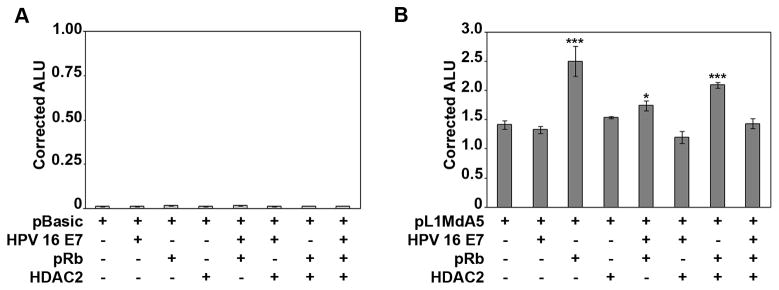
We previously identified L1MdA5 as a redox-inducible element [12] containing two functional electrophile response elements (EpRE)-like elements (Fig. 1A). We also have reported that E2F/pRb complexes bind to, and regulate, mouse L1 elements through mechanisms that involve HDAC proteins and epigenetic repression [13]. Since HPV E6 and HPV E7 oncoproteins interact with pRb and inactivate its cell cycle-related repressor function, we hypothesized that overexpression of HPV E7 oncoprotein alters L1 promoter activity. Thus, the effects of forced Rb and HPV E7 overexpression on L1MdA5 5′ UTR in vitro were investigated [12]. Transient transfections of HeLa cells with 500 ng of pBASIC-luciferase control or pL1Md-A5 Wild Type-luciferase reporter gene constructs [12, 18] (Fig. 1B), in the presence or absence of 100 ng of Rb or HPV E7 expression vectors, were performed. To correct for transfection artifacts, 30 ng of the Renilla luciferase reporter gene (pRL) were cotransfected in each assay. Thirty six hours after transfection, cells were lysed and luciferase activity measured. Transfection of the pBASIC vector alone, or in combination with Rb, HPV E7, or both, yielded low levels of corrected ALU, showing that the weak basal promoter activity of the parent vector was not affected by these proteins (Fig. 2A). In contrast, placement of the pL1Md-A5 5′ UTR sequence upstream of the luciferase reporter gene showed strong promoter activity, as evidenced by increased corrected ALU for the L1MdA5 promoter alone (0.012 vs. 1.4 ALU for pBASIC versus pL1Md-A5, respectively) (Fig. 2B). Expression of Rb increased pL1Md-A5 activity over the pBASIC vector (Fig. 2A), and over wild type controls (Fig. 2B), and this response was inhibited by HPV E7 viral oncoprotein (Fig. 2B). HPV E7 expression vector alone did not alter the corrected luciferase reporter activity of pL1Md-A5 (Fig. 2B). To further investigate the role of Rb in transactivation of L1MdA5, forced overexpression of HDAC2, a corepressor protein was examined [13, 19]. HeLa cells were cotransfected with pBASIC or pL1Md-A5 luciferase reporter vectors in the presence of 100 ng of Rb, HDAC2 or HPV E7 expression vectors, or their combination as indicated (Fig 3A). HDAC2 alone did not change corrected luciferase reporter activity (Fig 3B). When coexpressed with Rb, HDAC2 did not disrupt Rb-mediated transactivation of the L1Md promoter in vitro (Fig 3B). In contrast, coexpression of Rb, HDAC2 and HPV E7 led to strong inhibition of L1MdA5 transactivation, indicating that HPV E7 in association with HDAC2 blocks pRb effects. In all, these data suggest that inhibitory effects of HPV E7 are mediated through functional interactions with both pRb and HDAC2.
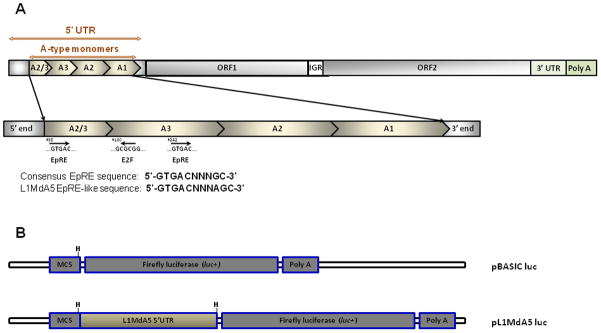
Fig. 1. The mouse L1MdA5 retroelement promoter contains several EpRE-containing monomers.
(A) Schematic structure and monomer organization for the L1MdA5 retrotransposon. A full-length L1MdA5 retroelement (top) is composed of a 5′ untranslated region (UTR) that contains three full-length A-type monomers (A3, A2, and A1) preceded by a 2/3 A-type monomer. Two open reading frames (ORF1 and ORF2), separated by a short intergenic region (IGR), encode the cis-acting proteins ORF1p and ORF2p respectively. The 3′ terminal region is composed of a 3′UTR followed by a poly-A tail. The location and orientation of putative EpRE and E2F DNA binding sites within the A-type monomer promoter are shown (bottom). (B) Simplified map of the luciferase reporter vectors used in the study. pGL3-BASIC (pBASIC) firefly luciferase (top) is a promoterless reporter vector used as the control in the study. The HindIII (H) restriction site within the vector multiple cloning site (MCS) was used to place the L1MdA5 promoter in front of the luciferase reporter gene (bottom). The vector also contains the simian virus 40 (SV 40) late poly adenylation signal (poly-A).
Fig. 2. Transactivation of L1 promoter activity by Rb is ablated by HPV E7 viral oncoprotein.
Transient transcription assays in HeLa cells with 500 ng of either the pBASIC-luciferase or the pL1Md-A5-luciferase reporter vectors and the forced overexpression of HPV E7 or pRb alone or in combination are shown. As an internal control for normalization, 10 ng of a second reporter vector, Renilla luciferase (pRL), were cotransfected within each experiment. (A) pBASIC-luciferase reporter activity corrected for pRL after cotransfection with 100 ng of empty vector control, HPV E7, Rb or HPV E7 and Rb together. (B) pL1Md-A5-luciferase reporter activity corrected for pRL after cotransfection with 100 ng of empty vector control, HPV E7, Rb or HPV E7 and Rb together. Cells were transfected for 6 hours, allowed to recover for 30 hours prior to luciferase activity measurements. Results are expressed as arbitrary luciferase units (ALU) and represent the mean ± S.D. of triplicate experiments after normalization to pRL. Statistic analyses were done using ANOVA (*, **, and *** indicate statistically significant differences, p<0.05, p<0.005 and p<0.0005 respectively). Each experiment was repeated at least 2 times.
Fig. 3. Transactivation of L1 promoter activity by Rb and HDAC2 is ablated by HPV E7 viral oncoprotein.
Transient transcription assays in HeLa cells with the pBASIC-luciferase or the pL1Md-A5-luciferase reporter vectors and the forced overexpression of HPV E7, pRb, or HDAC2 proteins alone or in combination are shown. As an internal control for normalization, 10 ng of pRL were cotransfected within each experiment. (A) pBASIC-luciferase reporter activity corrected for pRL after cotransfection with 100 ng of empty vector control, or expression vectors indicated in the figure label. (B) pL1Md-A5-luciferase reporter activity corrected for pRL after cotransfection with 100 ng of empty vector control, or expression vectors indicated in the figure label. Data were normalized as indicated in the Y axis. Cells were transfected for 6 hours, allowed to recover for 30 hours prior to luciferase activity measurements. Results are expressed as arbitrary luciferase units (ALU) and represent the mean ± S.D. of triplicate experiments after normalization to pRL. Statistic analyses were done using ANOVA (*, **, and *** indicate statistically significant differences, p<0.05, p<0.005 and p<0.0005 respectively). Each experiment was repeated at least 2 times.
3.2 HPV E7 ablates the response of the L1MdA5 reporter to genotoxic stress
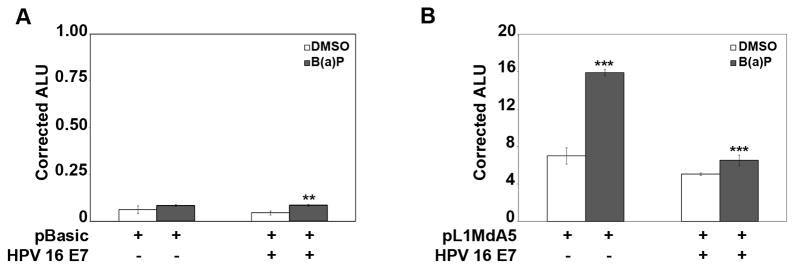
Since pRb is a frequent target of viral oncoproteins including adenovirus oncoprotein early region 1A (AdE1A) and HPV E7 [20, 21], and E2F/pRb complexes interact with human and mice L1 5′UTR regions [13], we hypothesized that HPV E7 overexpression alters the regulatory transactivation role for pRb on pL1Md-A5 under conditions of genotoxic stress. To this aim, we transiently transfected HeLa cells with pBASIC or pL1Md-A5 luciferase reporter vectors in the presence or absence of HPV E7 expression vector, followed by 3 μM BaP or control DMSO [18] (Fig 4A and 4B, respectively). Experiments conducted with pBASIC luciferase reporter alone showed low levels of corrected luciferase ALU for both DMSO and BaP treated cells (Fig. 4A). When cells were cotransfected with pBASIC and HPV E7 expression vector a similar result was obtained indicating that DMSO, BaP or HPV E7 did not influence the transcriptional activity of the control reporter vector. When cells were transiently transfected with pL1Md-A5 vector alone followed by exposure to 3 μM BaP, the corrected reporter gene ALU showed a robust response compared to control DMSO (Fig 4B). Importantly, BaP treatment elicited significant changes in the reactivation of the pL1Md-A5 reporter under all experimental conditions tested. DMSO alone increased reporter activity, a finding consistent with the redox-modulating effects of this solvent [12]. Forced overexpression of HPV E7 oncoprotein ablated the response of pL1Md-A5 reporter vector to genotoxic stress, but did not modify the response of the control pL1Md-A5 to DMSO (Fig. 4B). These data indicate that viral oncoproteins interfere with the cellular machinery responsible for L1 reactivation under conditions of genotoxic stress.
Fig. 4. BaP induction of L1 promoter activity is ablated by HPV E7 viral oncoprotein.
Transient transcription assays in HeLa cells with the pBASIC-luciferase (A), or the pL1Md-A5-luciferase (B), reporter vectors showing the effects of 0.06% DMSO or 3 μM BaP with or without HPV E7 oncoprotein forced overexpression. Cells were transfected for 6 hours, allowed to recover for 18 hours and chemically treated for 16 hours. Results are expressed as arbitrary luciferase units (ALU) and represent the mean ± S.D. of triplicate experiments after normalization to pRL. Statistic analyses were done using ANOVA (*, **, and *** indicate statistically significant differences, p<0.05, p<0.005 and p<0.0005 respectively). Each experiment was repeated at least 2 times.
4. Discussion
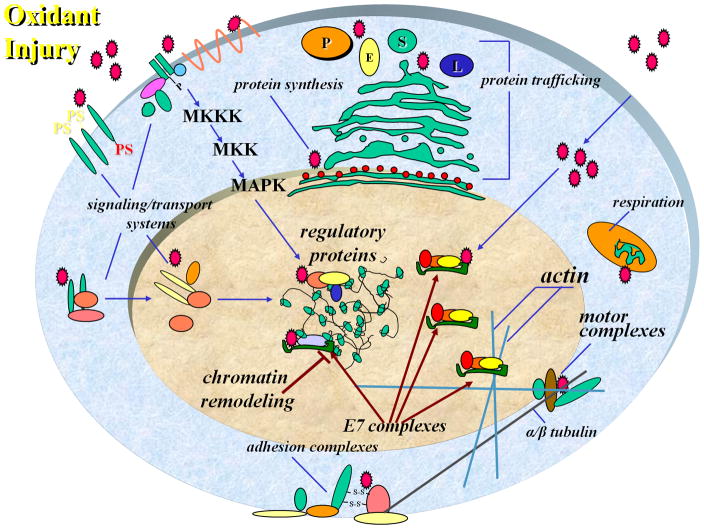
HPV E6 and HPV E7 proteins target a diverse variety of host cellular proteins involved in cytoplasmic and nuclear protein complex formation and regulation of cellular signaling (Fig 5). Evidence is presented here implicating viral oncoprotein E7 in heterologous downregulation of mouse L1MdA5 promoter transactivation, and this effect is mediated through interactions with Rb. This interpretation is consistent with the ability of viral oncoproteins including, AdE1A, simian virus 40 large T antigen, and HPV E6 and HPV E7, to compete with endogenous cellular proteins for binding to Rb proteins [15]. HPV E7 overexpression ablated the L1 transactivation exerted by both Rb and BaP, a response that may involve interference with, and modulation of, macromolecular complex assembly on the L1 promoter. pRb is known to interact with CNC-bZIP proteins, and that this interaction leads to pRb-mediated transactivation of c-Jun target genes [16]. Transient transfection assays in NIH3T3 cells have shown that coexpression of pRb and c-Jun transactivates AP-1 binding sites within the collagenase promoter [16]. Recent evidence also points to a dual role for pRb as a repressor as well as coactivator in the cellular response to toxic injury [22, 23]. Interestingly, pRb activity in those models seems to directly regulate proteins involved in L1 retroelement reactivation [22, 24, 25]. As such, the positive regulation of L1 seen upon forced expression of Rb may be linked to modulation of ARE-binding proteins that regulate L1 expression, and these interactions may be repressed by HPV E7. Viral oncoproteins also block redox-regulated transcription, as demonstrated previously in studies showing that AdE1A interferes with ARE signaling in both murine and human cells [26].
Fig. 5. Hypothetical mechanisms for the role of HPV E7 in control of the cellular response to redox-mediated injury and redox-dependent L1 reactivation.
Redox stress alters both cell signaling and transcription programs. Injury might lead either directly or indirectly to pRb/HDAC-dependent repression of factors required for LINE-1 silencing thus allowing for its reactivation. Cells expressing HPV E7 viral oncoprotein inactivate the Rb/HDAC effect, likely through direct interaction with Rb, thus eliminating the redox-mediated retroelement reactivation.
We reported previously that mouse cells lacking the retinoblastoma family of proteins pRb/p105, p107, and pRb2/p130 show increased L1MdA5 transcription, a response that involved impaired recruitment of HDACs to the L1 promoter and changes in epigenetic silencing marks. Thus, the finding that forced Rb expression enhanced L1MdA5 transcription was unexpected and likely indicative of the complexity of Rb interactions and functions within the cell. In vivo Rb proteins not only interact with other DNA binding proteins but also play key roles in heterochromatin formation and recruitment of DNA methyltransferases (DNMTs) and HDACs that repress transcription of E2F-responsive promoters [27–29]. These complex interactions are governed by both DNA sequence and chromatin microenvironment and therefore, not captured in transient transfection experiments. Such relationships indicate that the cellular response observed in our studies likely reflect elements of the cellular response governed by DNA-protein interactions. For instance, transient transfection of the rat α-actin gene into L8 rat myoblasts is associated with fast and active demethylation [30]. In fact, the lack of proper nucleosomal and chromatin organization may render DNA methylation unresponsive as an epigenetic silencing mark [31]. Finally, DNA methylation is known to provide docking sites for methyl binding proteins that enhance repression of repetitive elements.
In summary, evidence is presented here that HPV E7 alone, or in combination with HDAC2, disrupt Rb-mediated transactivation of the L1 promoter. HPV E7 alone also ablated the transcriptional response of L1Md-A5-luciferase reporter gene to genotoxic stress, but did not interfere with basal activity. Thus, HPV E7 associates with proteins involved in the assembly of macromolecular complexes that regulate antioxidant and E2F/Rb sites within L1 5′UTR to regulate biological activity. The degree to which these interactions regulate L1 biology in vivo provides fertile ground for future investigations.
Highlights.
HPV E7 viral oncoprotein ablates L1MdA5 reactivation following genotoxic stress.
HPV E7 disrupts retinoblastoma-mediated L1MdA5 promoter transactivation.
The basal and carcinogen-induced activities of L1MdA5 promoter are independent of Rb repressor function.
HPV E7 disrupts assembly of protein complexes regulating L1MdA5.
Acknowledgments
FUNDING: This work was supported in part by National Institutes of Health (NIH, USA) [Grants 5R01 ES004849; ARRA GB090603A1, Kentucky Lung Cancer Research Program, and P30 ES014443 to K.S.R.]
The pRb expression vector was a kind gift of Dr. Douglas C. Dean, (Department of Ophthalmology and Visual Sciences, University of Louisville), the HPV 16 E7 expression vector was a gift from Dr. Robert Mitchell (J.G. Brown Cancer Center, University of Louisville), the HDAC2 expression vector was a kind gift of Dr. Ed Seto (H. Lee Moffitt Cancer Center & Research Institute).
Abbreviations
- 5′-UTR
5′-untranslated region
- AdE1A
adenovirus oncoprotein early region 1A
- BaP
Benzo(a)pyrene
- CNC-bZIP
Cap n′ collar-basic leucine zipper protein family
- DNMT
DNA methyltransferase
- DMSO
dimethyl sulfoxide
- EpRE-like
electrophile-like response element
- HDAC
histone deacetylase
- HMT
histone methyltransferase
- HPV
Human papillomavirus
- LINE1 or L1
Long Interspersed Nuclear Element
- MEF
mouse embryo fibroblast
- ORF
Open reading frame
- Rb
retinoblastoma
Footnotes
COMPETING INTEREST: The authors declare no potential competing interests.
Authorship
D.E.M-D. participated in experimental design, data acquisition, manuscript drafting and final approval of submitted version. K.S.R. participated in manuscript drafting, editing, and critical review and approval of final version of submitted document
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High Frequency Retrotransposition in Cultured Mammalian Cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 2.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardies SC, Martin SL, Voliva CF, Hutchison CA, Edgell MH. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Molecular Biology and Evolution. 1986;3:109–125. doi: 10.1093/oxfordjournals.molbev.a040386. [DOI] [PubMed] [Google Scholar]
- 4.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. MCB.00332-07 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. Journal of Molecular Biology. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 6.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH. A potential role for the nucleolus in L1 retrotransposition. Human Molecular Genetics. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 7.Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 8.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 9.Farley AH, Luning Prak ET, Kazazian HH. More active human L1 retrotransposons produce longer insertions. Nucleic Acids Research. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardies SC, Wang L, Zhou L, Zhao Y, Casavant NC, Huang S. LINE-1 (L1) Lineages in the Mouse. Molecular Biology and Evolution. 2000;17:616–628. doi: 10.1093/oxfordjournals.molbev.a026340. [DOI] [PubMed] [Google Scholar]
- 11.Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison CA., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu KP, Ramos KS. Redox Regulation of a Novel L1Md-A2 Retrotransposon in Vascular Smooth Muscle Cells. Journal of Biological Chemistry. 2003;278:28201–28209. doi: 10.1074/jbc.M303888200. [DOI] [PubMed] [Google Scholar]
- 13.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res. 2009;665:20–28. doi: 10.1016/j.mrfmmm.2009.02.011. S0027-5107(09)00083-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rempala GA, Ramos KS, Kalbfleisch T, Teneng I. Validation of a Mathematical Model of Gene Transcription in Aggregated Cellular Systems: Application to L1 Retrotransposition. Journal of Computational Biology. 2007;14:339–349. doi: 10.1089/cmb.2006.0125. [DOI] [PubMed] [Google Scholar]
- 15.Chellappan S, Kraus VB, Kroger B, Munger K, Howley PM, Phelps WC, Nevins JR. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci U S A. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nead MA, Baglia LA, Antinore MJ, Ludlow JW, McCance DJ. Rb binds c-Jun and activates transcription. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antinore MJ, Birrer MJ, Patel D, Nader L, McCance DJ. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- 18.Teneng I, Stribinskis V, Ramos KS. Context-specific regulation of LINE-1. Genes Cells. 2007;12:1101–1110. doi: 10.1111/j.1365-2443.2007.01117.x. GTC1117 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21:2711–2716. doi: 10.1101/gad.1590607. 21/21/2711 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Marmorstein R. When viral oncoprotein meets tumor suppressor: a structural view. Genes Dev. 2006;20:2332–2337. doi: 10.1101/gad.1471706. 20/17/2332 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Huang G, Elferink CJ. Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol Pharmacol. 2005;67:88–96. doi: 10.1124/mol.104.002410. mol.104.002410 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Puga A, Barnes SJ, Dalton TP, Chang C, Knudsen ES, Maier MA. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem. 2000;275:2943–2950. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- 24.Watabe Y, Nazuka N, Tezuka M, Shimba S. Aryl hydrocarbon receptor functions as a potent coactivator of E2F1-dependent trascription activity. Biol Pharm Bull. 2010;33:389–397. doi: 10.1248/bpb.33.389. JST.JSTAGE/bpb/33.389 [pii] [DOI] [PubMed] [Google Scholar]
- 25.Ge NL, Elferink CJ. A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- 26.Chen YH, Ramos KS. A CCAAT/enhancer-binding protein site within antioxidant/electrophile response element along with CREB-binding protein participate in the negative regulation of rat GST-Ya gene in vascular smooth muscle cells. J Biol Chem. 2000;275:27366–27376. doi: 10.1074/jbc.M000405200. M000405200 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 28.Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. 117/11/2173 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Paroush Z, Keshet I, Yisraeli J, Cedar H. Dynamics of demethylation and activation of the alpha-actin gene in myoblasts. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-e. [pii] [DOI] [PubMed] [Google Scholar]
- 31.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]