Abstract
The International Cancer Genome Consortium (ICGC) is a collaborative effort to characterize genomic abnormalities in 50 different cancer types. To make this data available, the ICGC has created the ICGC Data Portal. Powered by the BioMart software, the Data Portal allows each ICGC member institution to manage and maintain its own databases locally, while seamlessly presenting all the data in a single access point for users. The Data Portal currently contains data from 24 cancer projects, including ICGC, The Cancer Genome Atlas (TCGA), Johns Hopkins University, and the Tumor Sequencing Project. It consists of 3478 genomes and 13 cancer types and subtypes. Available open access data types include simple somatic mutations, copy number alterations, structural rearrangements, gene expression, microRNAs, DNA methylation and exon junctions. Additionally, simple germline variations are available as controlled access data. The Data Portal uses a web-based graphical user interface (GUI) to offer researchers multiple ways to quickly and easily search and analyze the available data. The web interface can assist in constructing complicated queries across multiple data sets. Several application programming interfaces are also available for programmatic access. Here we describe the organization, functionality, and capabilities of the ICGC Data Portal.
Database URL: http://dcc.icgc.org
Project description
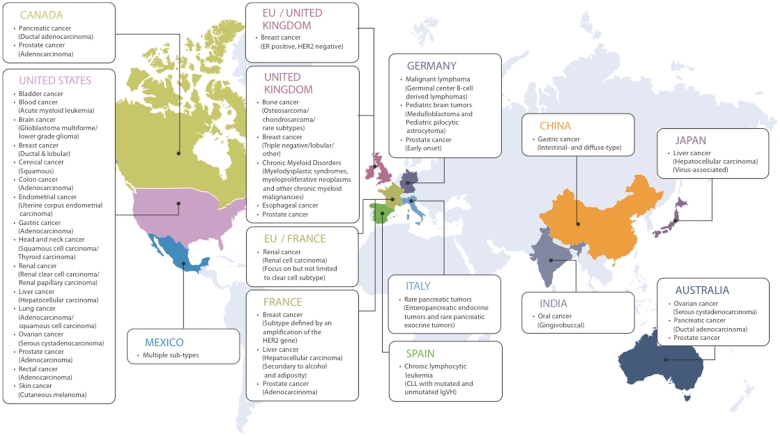
The International Cancer Genome Consortium (ICGC) is a multidisciplinary, multi-institutional collaborative effort aiming to systematically and comprehensively characterize somatic mutations in 50 different cancer types and subtypes (1). Five hundred tumor genomes, as well as matched normal control genomes for each cancer type, will be analyzed using high-throughput next-generation sequencing technologies to detect a wide range of somatic mutations, including single nucleotide mutations, small insertions/deletions, copy number alterations, translocations and other chromosomal structural rearrangements. Genome-wide methylation state analysis and whole-transcriptome sequencing have also been planned to provide additional molecular-level characterizations. To make the effort more scalable each member institution specializes in generating data for a particular tumor type. At the time of writing, 12 countries have joined the effort (Figure 1).
Figure 1.
International Cancer Genome Consortium (ICGC) projects (March 2011).
One of the major goals of ICGC is to rapidly bring these data to the cancer research community in order to accelerate studies on the discovery of cancer causes, to enhance the accuracy of diagnoses and to improve treatments. In order to achieve this task, the data generated by the consortium members have to be managed efficiently. Amongst the most important data management challenges faced by the consortium is the high complexity and heterogeneity of the data types involved, the necessity to link different data types, and the need to protect controlled data. Furthermore, the high volume of data and the distributed nature of the sources make traditional centralized approaches to data management impractical. Consequently, the ICGC has adopted federated data architecture to address their data management needs. The scalability of the system is improved by having each member institution store and process data locally; the data federation software then presents these separate sources as a single access point for remote data access.
BioMart, an open source data federation system (2), has been chosen as the ICGC data management platform. A number of features make BioMart an attractive solution for the ICGC. BioMart’s flexible data model makes it generally applicable to a wide range of biological data types and built-in query optimizations make it suitable for large data sets. In addition, BioMart supports an industry-standard security framework that is needed to provide secure access to the controlled data. Finally, a large number of existing expert-maintained public annotation databases can be readily federated, adding value to the interpretation of the ICGC experimental data.
The architecture of the ICGC data management system has been modeled on the BioMart Central Portal (3, 4). For a number of years, the portal has been successfully providing a single point of access to a large number of biological databases distributed across the world. Each BioMart database federated in this portal is maintained independently, released and updated on its own schedule. Similarly, each ICGC member maintains their local BioMart database where clinical annotations and the data produced from genomic analyses are deposited. However, in order to make these data comparable across different cancer projects and create a unified ICGC data set, a mechanism for enforcing uniformity is required. Thus, the same set of data models, controlled vocabularies, ontologies and reference data sets are used in all of the ICGC member databases. Furthermore, a gene-centric data model adopted from Ensembl Mart (5) is used to reference experimental data to the same set of annotated genes.
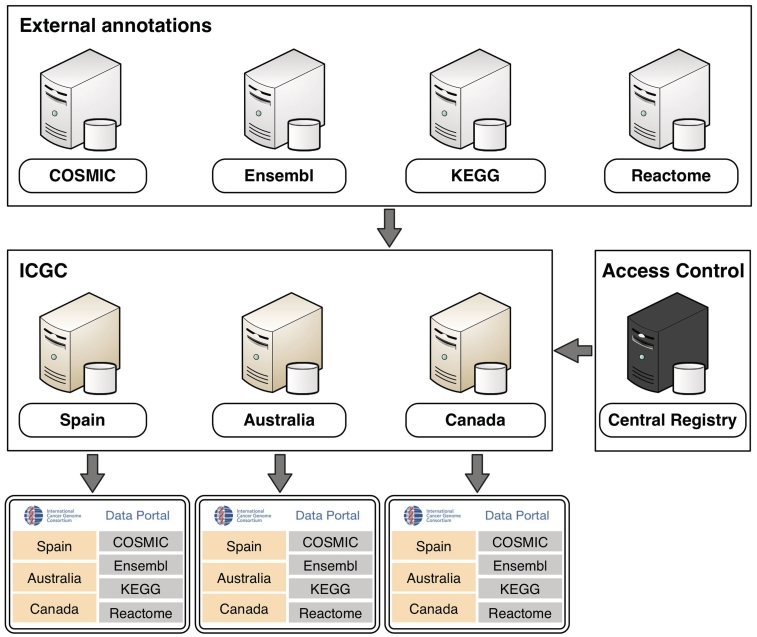
In order to link individual BioMart databases into a unified system, each of the ICGC member institutions maintains an instance of the BioMart server. These servers can each access data in three ways: they can access their own local databases directly; they can communicate with the other ICGC BioMart servers, in order to retrieve data from remote databases; and they can communicate with non-ICGC BioMart servers, such as COSMIC (6) and Reactome (7), to retrieve publicly-available annotation data. In this manner, the BioMart server maintained by each ICGC member acts as a fully featured ICGC data portal, providing unified access to all the consortium data. The users select a combination of different data sets and specify query criteria using a variety of graphical user interfaces and analysis tools that are available from the portal. Application programming interfaces (APIs) for Java, REST, SOAP and SPARQL are also available for programmatic access. Behind the scenes, BioMart software breaks down this query into smaller parts that are distributed to the remote data sources. The results are collected and compiled into a single unified results set that is presented to the user.
Some of the data in the portal has access restrictions in order to preserve patient privacy. To protect this sensitive data, communication between BioMart servers is secured by HTTPS and authentication is handled using the oAuth protocol. Access to controlled data is managed by the Central Registry server. When a user logs into an ICGC data portal server using his/her OpenID, the server will consult the central registry to ensure that the user is authorized to access the data (Figure 2).
Figure 2.
An overview of the ICGC Data Portal architecture.
Data content
At present the portal contains data from 24 cancer projects, consisting of 3478 genomes and 13 cancer types and subtypes. This includes the data generated from seven studies performed by five ICGC participating institutions located in four countries. The Data Portal also hosts data from other large-scale cancer genome projects including The Cancer Genome Atlas (TCGA) (8, 9), Tumor Sequencing Project (TSP)(10) and Johns Hopkins University (11–14). Open access data sets include (i) simple somatic mutations, (ii) copy number alterations, (iii) structural rearrangements, (iv) gene expression, (v) miRNA, (vi) DNA methylation and (vii) exon junctions. Secure access to controlled data sets, such as germline variations, is available to authorized users. A summary of data currently available on the Data Portal is shown in Table 1. In addition to cancer genomic data, the Data Portal also federates several public databases (Table 2). Currently this includes Ensembl genome database (5), Kyoto Encyclopedia of Genes and Genomes (KEGG) (15–17), Reactome (7), COSMIC (6), Pancreatic Expression Database (18) and the Breast Cancer Campaign Tissue Bank (19), and the number of resources is continuously growing.
Table 1.
The summary of data available on the ICGC Data Portal divided by cancer project
| Source | Cancer project | Data set |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Simple mutations | Copy number alterations | Structural rearrangements | Gene expression | miRNA expression | Exon junctions | DNA methylation | Germline variations | ||
| ICGC | Breast carcinoma (WTSI, UK) | • | |||||||
| Liver cancer (NCC, JP) | • | • | |||||||
| Liver cancer (RIKEN, JP) | • | • | |||||||
| Malignant melanoma (WTSI, UK) | • | • | |||||||
| Pancreatic cancer (OICR, CA) | • | • | • | ||||||
| Pancreatic cancer (QCMG, AU) | • | • | • | • | • | • | • | ||
| Small cell lung carcinoma (WTSI, UK) | • | • | |||||||
| TCGA | Acute myeloid leukemia | • | |||||||
| Breast invasive carcinoma | • | • | |||||||
| Colon adenocarcinoma | • | • | |||||||
| GlioblastomaMultiforme | • | • | • | ||||||
| Kidney renal clear cell carcinoma | • | • | |||||||
| Kidney renal papillary cell carcinoma | • | ||||||||
| Lung adenocarcinoma | • | • | |||||||
| Lung squamous cell carcinoma | • | • | |||||||
| Ovarian serous cystadenocarcinoma | • | • | • | ||||||
| Rectum adenocarcinoma | • | • | |||||||
| Stomach adenocarcinoma | • | ||||||||
| Uterine corpus endometrioid carcinoma | • | ||||||||
| Other | Breast cancer (JHU, US) | • | |||||||
| Colorectal cancer (JHU, US) | • | ||||||||
| GlioblastomaMultiforme (JHU, US) | • | • | |||||||
| Lung adenocarcinoma (TSP, US) | • | ||||||||
| Pancreatic cancer (JHU, US) | • | • | |||||||
Table 2.
Public databases federated with the ICGC Data Portal
| Source | URL | Description of contents |
|---|---|---|
| Ensembl | www.ensembl.org | Genome annotation |
| Reactome | www.reactome.org | Pathway annotation |
| KEGG | www.genome.jp/kegg | Pathway annotation |
| COSMIC | www.sanger.ac.uk/genetics/CGP/cosmic/ | Somatic mutations in cancer |
| Pancreatic Expression Database | www.pancreasexpression.org | Pancreatic cancer expression data |
| Breast Cancer Campaign Tissue Bank | www.breastcancertissuebank.org/bio-informatics.php | Breast cancer expression data |
Query examples
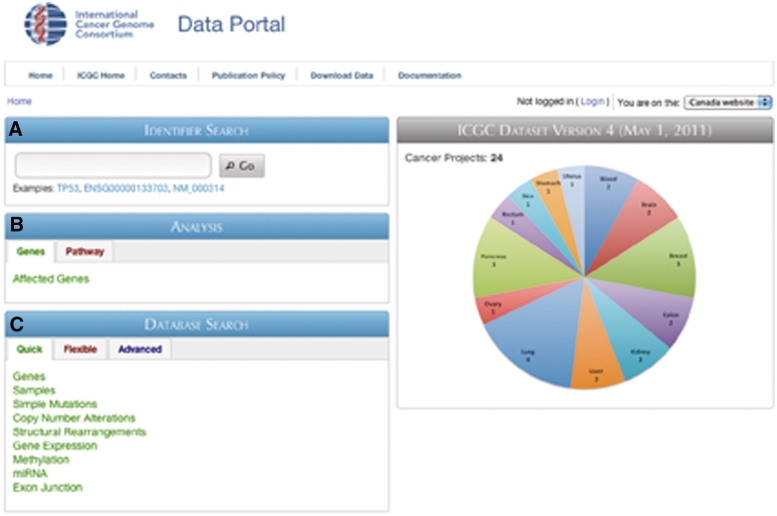
The Data Portal provides three major interactive entry points: Identifier Search, Analysis and Database Search (Figure 3). Identifier search (Figure 3A) lets users input identifiers that are commonly used in public annotation databases (e.g. HGNC gene symbol, Ensembl ID, RefSeq ID, UniProt ID and other accessions) and returns links to the corresponding Gene Report page, which displays basic gene description, pathway annotation, mutations found in the COSMIC database, as well as pancreas and breast cancer expression data (Figure 4). In addition, the Gene Report displays a summary of mutation frequencies in the selected gene across all cancer projects (Figure 4D). Records in the gene report (i.e. genomic coordinates, pathway name, publication) are linked to appropriate resources when available, allowing users to easily retrieve additional information on their data of interest.
Figure 3.
Screenshot of the ICGC Data Portal home page. Three main entry points are available: (A) Identifier search, (B) Analysis and (C) Database search.
Figure 4.
Gene Report for KRAS includes: (A) Gene annotation data from Ensembl, Pathway annotation from (B) KEGG and (C) Reactome, (D) Summary of mutation frequencies in each cancer project, (E) Mutation data from COSMIC, Expression data from (F) Pancreatic Expression Database and (G) Breast Cancer Campaign Tissue Bank (BCCTB). Different sections of this page come from federated BioMart sources.
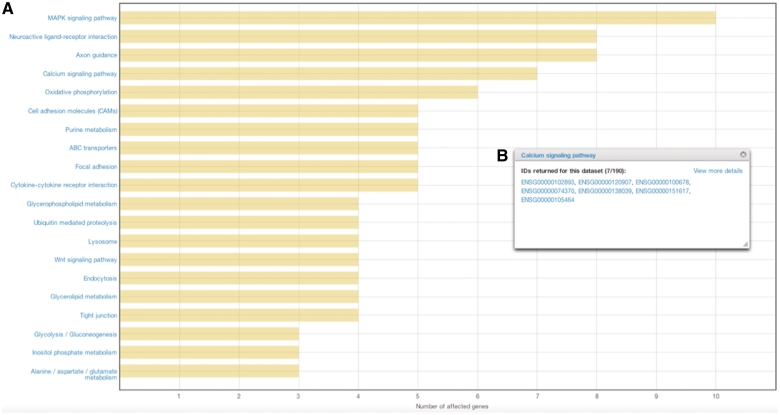
To help with the interpretation of cancer data, Gene and Pathway analysis tools are available in the Analysis section of the Data Portal (Figure 3B). These tools enable users to view the most commonly affected genes or pathways in one or more cancer projects. Results are presented in an easy-to-follow chart that can be exported as an image file, and the numerical data can also be downloaded for further processing (Figure 5).
Figure 5.
Results from affected pathways analysis for breast cancer (JHU, US). (A) Affected pathways are shown in a chart, with bars representing the number of affected genes in each pathway. (B) By clicking on the bar, users are able to view and download the genes that were mutated in each pathway.
Using the Database Search entry point (Figure 3C) users can interactively query the database by several data types: genes, samples, simple mutations, copy number alterations, structural rearrangements, gene expression, miRNA, DNA methylation and exon junctions. Three interfaces are available: Quick, Flexible and Advanced. The Quick interface contains a pre-selected set of the most commonly used filters such as gene type, mutation type and chromosome, and outputs a fixed set of attributes. The Flexible interface contains additional filters and a selection of attributes, allowing the user to choose which data are displayed in the output. The Advanced interface contains the complete set of filters and attributes, including technological platforms used for sequencing, and clinical parameters such as patient gender and tumor histopathology.
To demonstrate the utility of the ICGC Data Portal we present several queries that can be performed using different query interfaces.
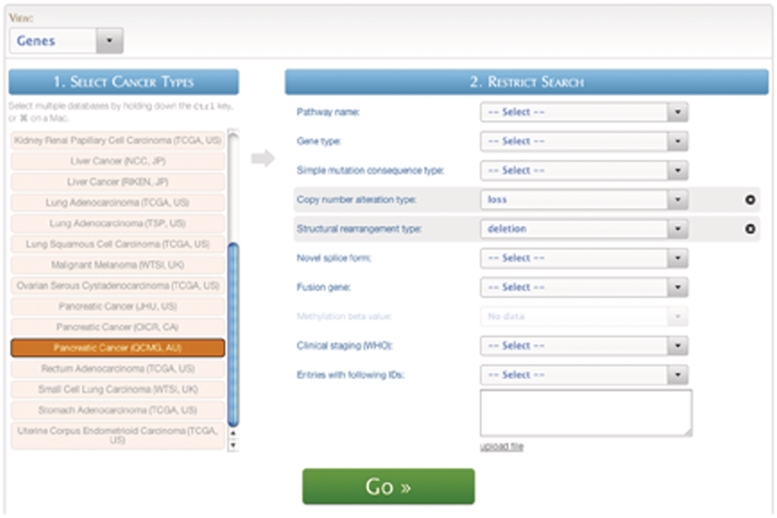
Query #1 (Quick Search): ‘Search for genes affected by copy number loss and also detected as deletion from structural rearrangement analysis.’ (Figure 6)
Figure 6.
A screenshot of the Quickquery interface.
| Data sets | Cancer projects | Filters | Attributes |
|---|---|---|---|
| Genes | Pancreatic Cancer (QCMG, AU) | Rearrangement type: deletion | Ensembl Gene ID |
| Copy number alteration type: loss | Gene symbol | ||
| Gene description |
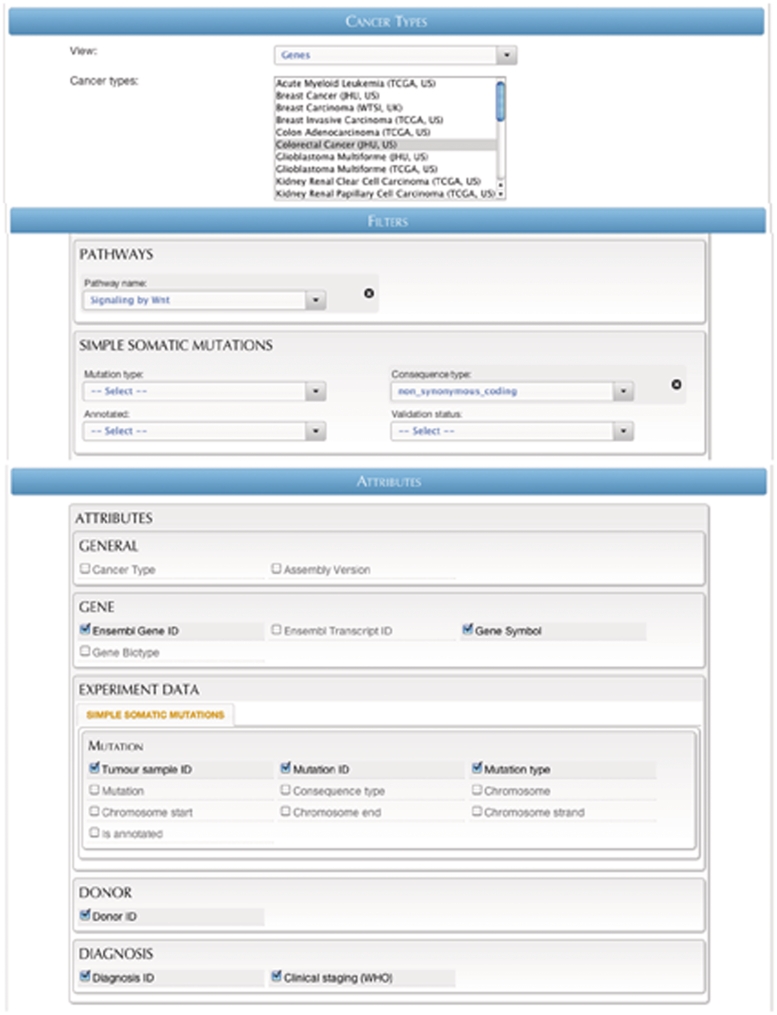
Query #2 (Flexible Search): ‘Retrieve clinical staging data for colorectal cancer patients with non-synonymous simple mutations in genes that are involved in WNT signaling pathway.’ (Figure 7)
Figure 7.
A screenshot of the Flexiblequery interface.
| Data sets | Cancer projects | Filters | Attributes |
|---|---|---|---|
| Genes | Colorectal cancer (JHU, US) | Pathway: Signaling by Wnt | Tumor sample ID |
| Consequence type: non_synonymous_ coding | Mutation ID | ||
| Mutation type | |||
| Donor ID | |||
| Diagnosis ID | |||
| Clinical staging (WHO) | |||
| Ensembl Gene ID | |||
| Gene Symbol |
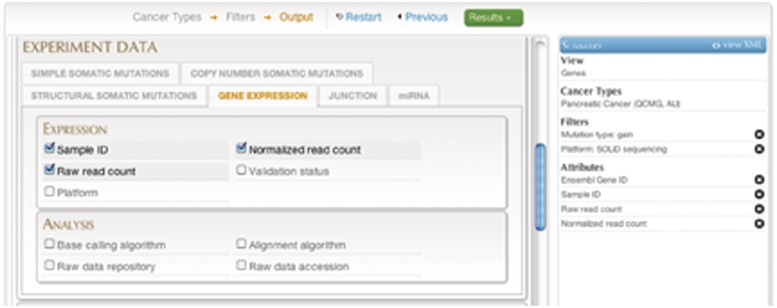
Query #3 (Advanced Search): ‘In pancreatic cancer data set, retrieve all RNA-seq expression data for genes that are affected by copy number gains.’ (Figure 8)
Figure 8.
A screenshot of the Advancedquery interface.
| Data sets | Cancer projects | Filters | Attributes |
|---|---|---|---|
| Genes | Pancreatic Cancer (QCMG, AU) | Copy number alteration type: gain | Ensembl Gene ID |
| Platform: SOLiD sequencing | Gene Expression: Sample ID | ||
| Normalized read count | |||
| Raw read count |
Conclusion and future directions
The ICGC Data Portal is the first project to successfully federate large amounts of cancer genomics data and rich annotation data in a single access point. It presents a scalable approach, not only in the traditional sense of parallelizing data processing and storage, but also in a more general sense of outsourcing the external annotation expertise, by federating annotations from independently maintained databases. This approach has proved to be successful in addressing ICGC data management needs and can be useful for similar, large-scale collaborative projects.
The ICGC Data Portal will continue to expand by adding more data, both from within the project in the form of new cancer genomics data from ICGC members, and by integrating other public annotation databases to further the depth of analysis possible through the data portal. Additionally, new tools will be developed to increase the flexibility and utility of the system. This includes tools for ICGC deployers, to further streamline the data processing, transformation and loading processes, and tools for users, to add new methods of data visualization and analysis to the portal.
Funding
Ontario Institute for Cancer Research and the Ontario Ministry for Research and Innovation. Funding for open access charge: XXX.
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank members of the International Cancer Genome Consortium who have provided data, use cases, and/or input into the ICGC Data Portal. The authors are also grateful to the programmers and bioinformaticians who have contributed to the BioMart project over the course of its development.
References
- 1.Hudson TJ, Anderson W, Artez A, et al. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Haider S, Guberman JM, et al. BioMart: a data federation framework for large collaborative projects. Database. 2011;((This issue)) doi: 10.1093/database/bar038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guberman JM, Arnaiz O, Baran J, et al. BioMart central portal: an open database network for the biological community. Database. 2011;((This issue)) doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haider S, Ballester B, Smedley D, et al. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinsella RJ, Kahari A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across the taxonomic space. Database. 2011;((This issue)) doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shepherd R, Forbes SA, Beare D, et al. Data mining using the catalogue of somatic mutations in cancer BioMart (COSMICMart) Database. 2011;((This issue)) doi: 10.1093/database/bar018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haw R, Croft D, Yung C, et al. The reactome BioMart. Database. 2011;((This issue)) doi: 10.1093/database/bar031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. The Cancer Genome Atlas. http://cancergenome.nih.gov/
- 9.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Hattori M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutts RJ, Emanuela G, Lemoine NR, et al. Using BioMart as a framework to manage and query pancreatic cancer data. Database. 2011;((This issue)) doi: 10.1093/database/bar024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Breast Cancer Campaign Tissue Bank. http://breastcancertissuebank.org/bio-informatics.php.