Abstract
Sirtuins have been shown to regulate life-span in response to nutritional availability. We show here that levels of the mammalian sirtuin, SIRT6, increased upon nutrient deprivation in cultured cells, in mice after fasting, and in rats fed a calorie-restricted diet. The increase in SIRT6 levels is due to stabilization of SIRT6 protein, and not via an increase in SIRT6 transcription. In addition, p53 positively regulates SIRT6 protein levels under standard growth conditions but has no role in the nutrient-dependent regulation of SIRT6. These observations imply that at least two sirtuins are involved in regulation of life-span by nutrient availability.
Keywords: SIRT6, Sirtuin, Calorie restriction, Nutrient availability
1. Introduction
The sirtuins are highly conserved enzymes that utilize NAD+ to modify other proteins [1]. The founder member of the sirtuin family, yeast Sir2, displays both protein deacetylase and protein mono-ADP ribosyltransferase activities. However, despite their conservation from bacteria to humans, some sirtuins exhibit only one of these enzymatic activities [2].
Studies of several organisms indicate that sirtuins are pivotal in the regulation of longevity. Mutation of Saccharomyces cerevisiae Sir2 (ySir2) shortens yeast replicative life-span by 40%, whereas increasing the activity of sirtuins in S. cerevisiae, Caenorhabditis elegans and Drosophila melanogaster through either genetic or chemical means, extends life-span by at least 30% [3–6]. In addition, observations of yeast and drosophila suggested that sirtuins are required for mediating the beneficial effect of a calorie-restricted (CR) diet on life-span [7]. CR slows the rate of aging, delays the appearance of many age-related disorders and extends the maximum life-span of various organisms, including yeast, nematodes, drosophila and rodents [8–10]. However, the molecular mechanisms of CR are still poorly understood.
Of the seven mammalian ySir2 homologues, SIRT1 to 7, only SIRT1 was implicated to date in the CR response. SIRT1 is induced upon nutrient deprivation in vitro in a p53-dependent manner [11] and after long-term CR [12], and mice over-expressing SIRT1 exhibit some physiological properties similar to those of mice on a CR regimen [13]. Another mammalian sirtuin recently implicated in the regulation of aging is SIRT6, a nuclear protein that fails to deacetylate acetylated lysine in vitro, but instead catalyzes auto-ADP-ribosylation [2]. SIRT6-deficient mice are small, and by 2–3 weeks of age, develop abnormalities usually associated with aging [14]. These abnormalities include profound lymphopenia, loss of subcutaneous fat, lordokyphosis, severe metabolic defects, and, eventually, death at about 4 weeks. Notably, cells deficient in SIRT6 exhibit high levels of genomic instability that are likely due to defects in base excision repair (BER) [14]. Yet, the association between defects in BER and aging remains tenuous, as mutation of other BER factors has yet to be demonstrated to display a similar aging phenotype [15].
In order to determine if SIRT6 was regulated by nutrient levels and was involved in CR response in a manner similar to SIRT1, we studied the effects of nutrient depletion both in vivo in rodents and in vitro in tissue culture cells, and observed elevated SIRT6 levels in both model systems. We demonstrate that the elevated expression of SIRT6 protein is not due to increased SIRT6 transcription or translation, but rather to stabilization of SIRT6 protein. Moreover, studies using inhibitors indicate that the proteosome degradation pathway is largely responsible for regulating SIRT6 levels. Taken together, these results suggest the involvement of SIRT6 in the response to nutrient levels, and raise the possibility that part of the influence of nutrient availability on life-span is mediated by increasing SIRT6 stability.
2. Results
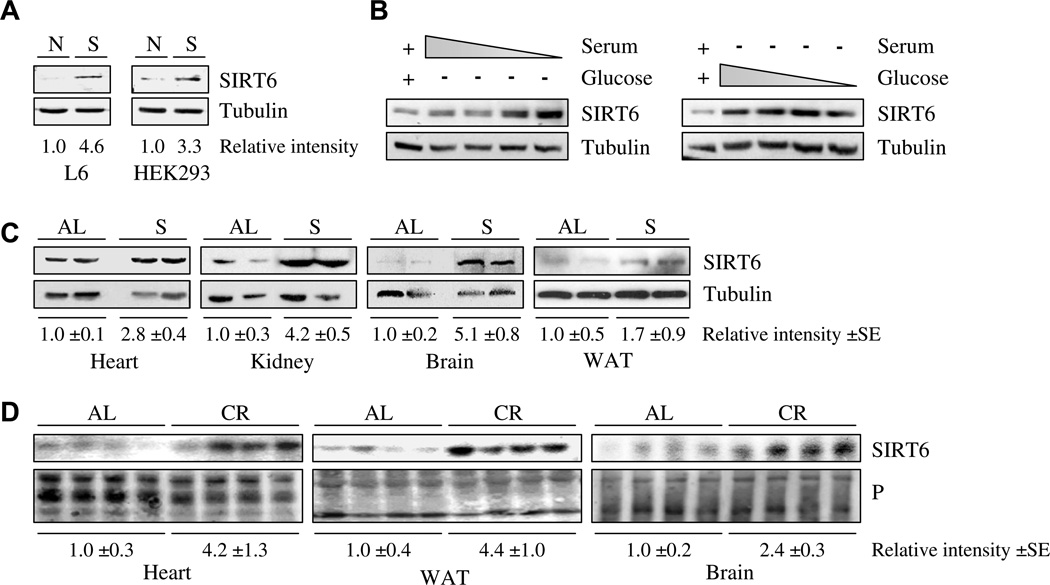
Nutrient availability was shown to regulate life-span, suggesting that proteins such as SIRT6, which are involved in aging and metabolism, might be regulated by nutrient levels. In order to investigate whether SIRT6 was involved in the response to nutrient levels, SIRT6 protein levels were monitored in mammalian models of nutrient deprivation. In human embryonic kidney 293 fibroblasts (HEK 293) and rat myoblast L6 cells, the level of SIRT6 was elevated after nutrient deprivation induced by the absence of serum and glucose (Fig. 1A). Under these experimental conditions, no significant change in cell morphology was detected after nutrient deprivation. These findings suggest that SIRT6 levels are regulated in vitro by nutrient deprivation.
Fig. 1.
SIRT6 is induced by nutrient deprivation: (A) SIRT6 protein levels in rat myoblast L6 or human embryonic kidney HEK293 cells grown in full medium [N], or starved [S]. (B) HEK293 cells were grown in serum free growth medium and decreased tenfold dilutions of glucose (right panel), or in glucose free growth medium and decreased tenfold dilutions of serum (left panel). Complete standard growth medium served as a control in each experiment (left lane). (C) Extracts of kidney, brain or WAT tissues from mice fed an ad libitum diet [AL] or mice fed for 24 h a water-only diet [S], were separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and probed with specific rabbit polyclonal antibodies against SIRT6. The data generated from two (out of five) representative mice from each treatment are shown; each lane represents an individual mouse. Tubulin levels served as a loading control. (D) Male Fisher 344 rats were fed either NIH-31 standard feed AL or CR diet until sacrifice at 12 or 24 months of age. Extracts of heart, WAT or brain tissues from AL and CR animals were separated by SDS–PAGE and probed with a specific rabbit polyclonal antibody against SIRT6. Equal amounts of protein extract were loaded onto each lane as measured by Bradford assay and by subsequent Ponceau protein staining of the membrane (P). Band intensity measurements were done using ImageJ analysis. Each lane represents a different rat.
To determine whether SIRT6 response to nutrient levels was dose-dependent or a threshold response, HEK 293 cells were grown in glucose free growth medium, with decreased concentrations of serum, or in serum free growth medium with decreased concentrations of glucose. As seen in Fig. 1B, SIRT6 levels were more sensitive to serum levels than to glucose levels. While the response to serum deficiency was robust and dose-dependent, SIRT6 response to glucose deficiency was moderate.
Next, we surveyed SIRT6 levels in mice fed an ad libitum (AL) diet and in mice after 24 h of fasting (supplemented with water only). SIRT6 was assessed in the following tissues: brain, spleen, muscle, heart, kidney, liver, white adipose (WAT) and testis. After 24 h of fasting, SIRT6 levels were elevated more than twofold in the brain, kidney and heart (Fig. 1C) but not in the other tissues tested. A moderate increase in SIRT6 levels was detected in WAT (Fig. 1C).
It is likely that an acute 24-h fast induces a range of physiological responses, some of which are also induced by long-term CR. Therefore, we examined SIRT6 levels in rats fed a CR diet for at least a year as compared to rats fed AL. In rats exposed to a CR diet, SIRT6 levels were elevated significantly in WAT, heart and brain tissues (Fig. 1D), and moderately in the kidney and liver (data not shown). Notably, CR had a more prominent effect on SIRT6 levels in WAT than 24-h fasting. Taken together, these data indicate that SIRT6 levels are regulated in vivo by short- and long-term nutrient limitation.
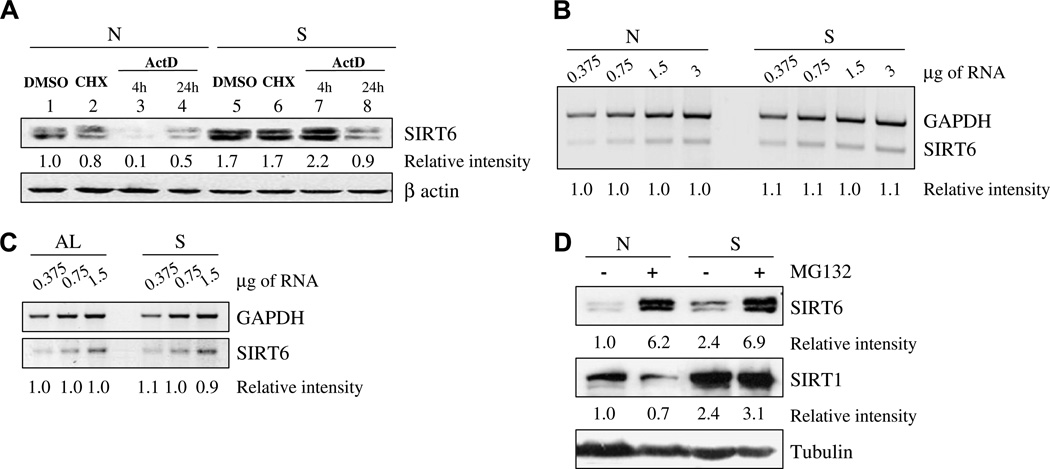
To address the mechanism by which SIRT6 levels increased, we examined its levels in HEK293 cells under normal growth and nutrient-deprived conditions in the presence and absence of various inhibitors. Treatment with the transcription inhibitor actinomycin D for the entire 24-h starvation period, but not when it was added for only the last 4 h of starvation, blocked the starvation-dependent increase in SIRT6 levels (Fig. 2A). No change in SIRT6 transcription was detected by RT-PCR analysis in nutrient-deprived HEK293 cells (Fig. 2B). Similarly, no change in SIRT6 mRNA levels was detected in the kidneys of starved mice in comparison to mice fed an AL diet (Fig. 2C), suggesting that actinomycin D inhibited the expression of another gene(s) affecting SIRT6 protein levels. Notably, under normal growth conditions, actinomycin D treatment for 4 h reduced SIRT6 protein levels by 90% (Fig. 2A). Treatment with the translational inhibitor, cyclohexamide, had no effect on SIRT6 levels under normal or starvation conditions (Fig. 2A). To examine if the proteasome was involved in regulating SIRT6 protein stability, we made use of the proteasome inhibitor MG-132. Under both normal and nutrient-depleted conditions, MG-132 treatment resulted in increased SIRT6 levels (Fig. 2D), whereas it had no significant effect on the protein levels of SIRT1, the other mammalian sirtuin reported to be induced by long-term CR [12] or nutrient depletion [11]. In summary, SIRT6 levels are regulated by the proteosome, and the increase in SIRT6 protein levels induced by nutrient depletion is due to an increase in SIRT6 protein stability, probably because of reduced proteasomal degradation of SIRT6.
Fig. 2.
The increase in SIRT6 upon nutrient deprivation is due to an increase in its protein stability: (A) SDS–PAGE analysis of SIRT6 in HEK293 cells supplemented with normal levels of nutrients [N], or starved [S], in the presence or absence of the following agents: The translation inhibitor cyclohexamide (CHX) for 4 h; RNA transcription inhibitor actinomycin D (ActD) for 4 or 24 h; or dimethyl sulfoxide (DMSO), which served as a solvent control. (B) Semi-quantitative RT-PCR analysis of SIRT6 gene transcription of HEK293 cells under normal [N] or starvation [S] conditions. (C) Semi-quantitative RT-PCR analysis of SIRT6 gene transcription of kidney tissues from mice fed an ad libitum diet [AL], or mice fed for 24 h a water-only diet [S]. (D) SDS–PAGE analysis of SIRT1 and SIRT6 in HEK293 cells supplemented with normal levels of nutrients [N] or starved [S], in the presence or absence of a proteasomal degradation inhibitor, MG132. In each panel, the intensity of a given band relative to the relevant loading control (beta-actin or tubulin for proteins and GAPDH for RNA) is indicated below each lane. Band intensity measurements were done using ImageJ analysis. For RT-PCR, the RNA templates were serially diluted (the amount of RNA is indicated above the panel).
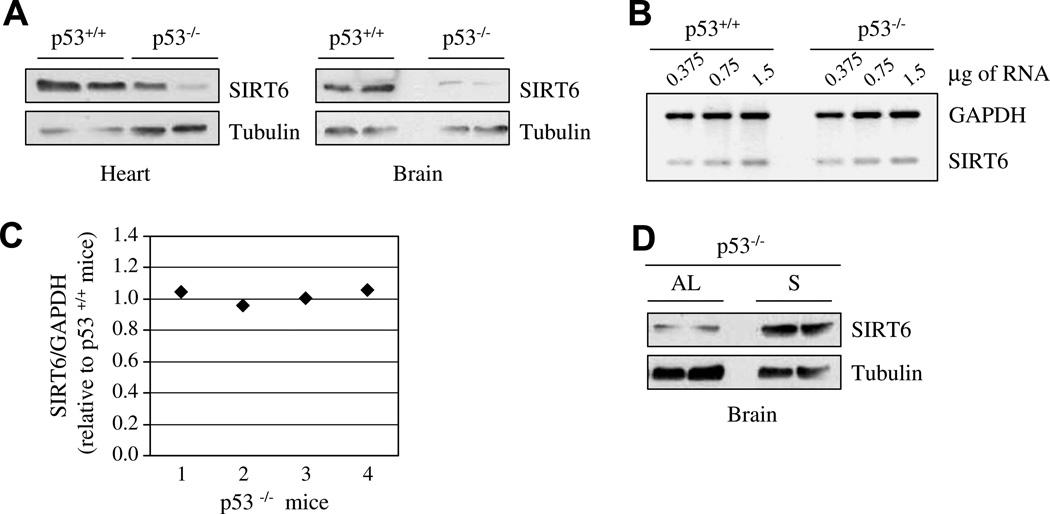
The tumor suppressor p53 was shown to regulate SIRT1 transcription levels in a manner dependent on nutrient availability [11]. Thus, the levels of SIRT6 might also be subject to p53 regulation. To test this possibility, SIRT6 protein levels were evaluated in brain and heart tissues of wild type or p53-deficient (p53−/−) mice, and were found to be lower in the absence of p53 (Fig. 3A). RT-PCR analysis with SIRT6-specific primers revealed similar SIRT6 mRNA levels in the brain of wild type and p53−/− mice (Fig. 3B and C). Thus, p53 regulates SIRT6 post-transcriptionally under normal growth conditions of AL diet. Surprisingly, even in p53-deficient mice, SIRT6 levels still increased significantly after 24-h of fasting (Fig. 3D). Taken together, our results demonstrate that p53 is a positive regulator of SIRT6 protein levels under normal growth conditions, but does not participate in the stabilization of SIRT6 under conditions of nutrient limitation.
Fig. 3.
p53 regulates SIRT6 levels: (A) Protein extracts of brain or heart tissues from wild type or p53−/− mice maintained on a normal diet were separated by SDS–PAGE. SIRT6 protein levels were measured with specific rabbit polyclonal antibody against SIRT6. (B) Representative semi-quantitative RT-PCR of SIRT6 RNA levels in the brain of wild type or p53−/− mice fed with AL diet. (C) The ratio between SIRT6 mRNA and GAPDH mRNA in each p53−/− mice versus wild type mice, as measured by densitometric analysis, was plotted and shown in a graph. Band intensity measurements were done using ImageJ analysis. (D) SIRT6 protein levels in the brain were also measured in p53−/− mice fed normally [AL], or with water only for 24 h [S].
3. Discussion
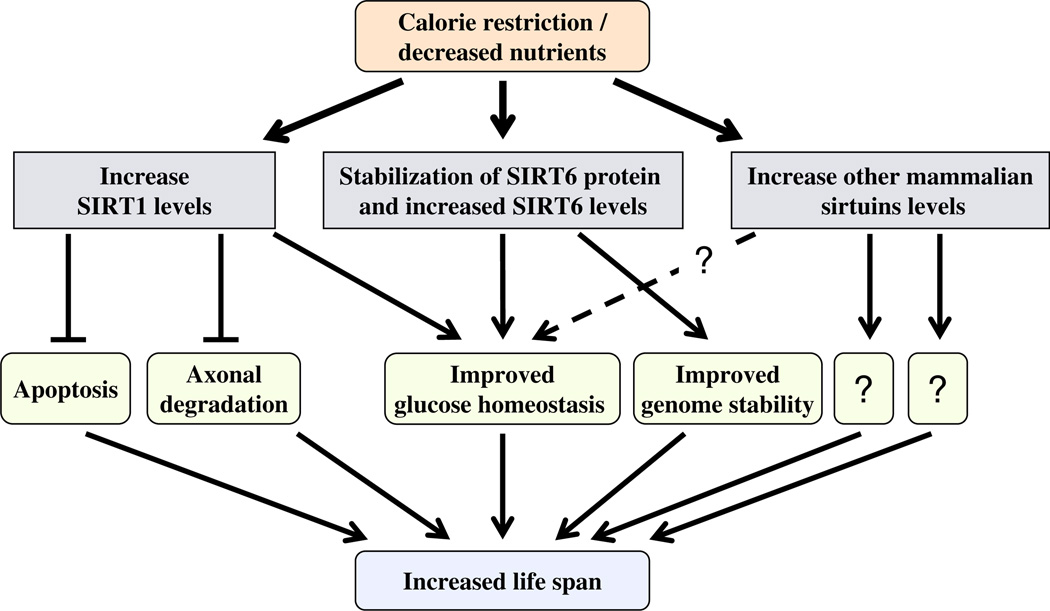
The yeast and fly sirtuins were demonstrated to control life-span and to influence the CR response [16]. However, the role of mammalian sirtuins in this process is still elusive. Here, we follow the effect of nutrient depletion and CR on SIRT6, a mammalian sirtuin whose absence promotes degenerative abnormalities associated with aging [14].We show that: (1) elevated SIRT6 levels are induced in multiple organisms by 24 h of nutrient depletion in vitro or in vivo, and by long-term CR; (2) SIRT6 induction is tissue specific, being most pronounced in the brain, kidney, heart and adipose tissues; (3) elevated SIRT6 levels after nutrient deprivation are due to increased SIRT6 protein stability, and SIRT6 levels were regulated significantly by proteasomal degradation; and (4) p53 positively regulates SIRT6 protein levels. Taken together, these results suggest that SIRT6 protein levels are regulated by nutrient levels, and that one way by which nutrient limitation acts to extend life-span is by increasing SIRT6 levels. This increase has the potential to increase SIRT6 activity, resulting in proper glucose homeostasis and genome stability (Fig. 4).
Fig. 4.
Combinatorial regulation of sirtuin levels under decreased nutrient growth conditions. In response to CR or nutrient deprivation, the level of various sirtuins (SIRT1, SIRT6 and possibly others) increased in different tissues. As a result, sirtuin-dependent pathways are affected and the combinatorial outcome is expressed as the beneficial effect of CR on age-related pathologies.
The observation that CR regulates at least two mammalian sirtuins, affirms the critical role of sirtuins in the regulation of aging, despite differences in their enzymatic functions and substrates. Furthermore, our results suggest that the beneficial effect of CR on aging is likely to represent a combinatorial outcome mediated by several sirtuins in various organs (Fig. 4). For example, in rats fed a CR diet, SIRT1 [12] and SIRT6 levels increase in the brain, kidney and WAT, whereas only SIRT6 but not SIRT1 levels increase in the heart. Thus, we propose that in order to delineate the key regulators of life-span, one must search for master factors that are common to the regulation of different sirtuins. One promising candidate is the nutrient level regulated nicotinamide phosphoribosyl-transferase (Nampt), which recycles nicotinamide in the NAD+ de novo cycle, and was shown to regulate SIRT1 activity [17] and exhibit increased expression upon nutrient depletion [18].
We could not detect significant changes in SIRT6 transcription after nutrient deprivation in tissue culture (Fig. 2B) or in starved mice (Fig. 2C). Thus, our results indicate that SIRT6 is regulated mainly at the level of protein stability. Similarly, in HEK 293 cells, we could not detect changes in SIRT1 mRNA upon nutrient depletion, suggesting that this mode of regulation is common to several members of the mammalian sirtuin family (data not shown). Nevertheless, we found that treatment with the transcription inhibitor actinomycin D blocked the starvation-dependent increase in SIRT6 protein. These results suggest the existence of another protein(s) that is regulated at the level of transcription in response to nutrient deprivation and influences SIRT6 protein levels. This putative protein may either bind and stabilize SIRT6, or modify it post-translationally under nutrient-depleted conditions. This function is absent under normal conditions, and SIRT6 half life would be relatively short. Indeed, treatment for 4 h with actinomycin D reduced the levels of SIRT6 by 90% under normal conditions but had no effect under starvation (Fig. 2A). Inhibition of proteosomal degradation by MG-132 significantly increased SIRT6 but not SIRT1 levels to an extent similar to that observed after nutrient depletion. Therefore, we suggest that nutrient deprivation might exert its effect on SIRT6 levels by regulating the labeling of SIRT6 by poly-ubiquitylation.
We observed that, under normal conditions, inhibition of transcription reduced SIRT6 levels dramatically (Fig. 2A), suggesting that SIRT6 protein has a short half life. Thus, the main challenge of the cell under nutrient depletion is to stabilize SIRT6 protein rather than increase the rate of its transcription. Indeed, our results strongly support such a mode of mechanism. This scenario is reminiscent of the mode of regulation documented for another critical mediator of cellular response to its environment, namely p53. It would be of great interest to follow whether a similar mechanism stabilized SIRT6 upon the induction of other pathways in which SIRT6 was demonstrated to be involved, such as the base excision repair of DNA.
p53 exhibited opposite effects on SIRT1 and SIRT6 levels. Compared to wild type mice, p53−/− mice exhibited higher SIRT1 levels [11] but lower SIRT6 levels (Fig. 3D). SIRT6 levels still increased in p53−/− mice after fasting. Thus, although p53 positively regulated the levels of SIRT6 under normal growth conditions, p53 could not have been the main protein which stabilized SIRT6 protein upon nutrient deprivation. Why would p53 have positive effects on SIRT6? Mice with a SIRT6 knockout demonstrate a spectrum of phenotypes, including an increase in lymphocyte apoptosis, metabolic defects and deficiency in base excision repair (BER) of DNA lesions. Thus, p53 might induce SIRT6 protein levels as part of the involvement of p53 in the BER pathway [11]. Further analysis is required to determine whether p53 stabilizes SIRT6 directly by physical interaction with SIRT6 or via its effect on a third, unknown protein.
3.1. Perspective
We show that SIRT6, similar to SIRT1, is influenced by CR and nutrient availability. SIRT1 and SIRT6 possess different enzymatic activities and are induced in an overlapping subset of organs. These findings raise the possibility that, in mammals, several sirtuins mediate the beneficial effects of CR on life span in a combinatorial manner. Hence, a systematic approach is required when studying the role of sirtuins in aging and CR. Furthermore, we propose that in order to develop small molecules which could mimic the ability of CR to prolong healthy life-span, one should search for master regulators with the ability to promote the activities of multiple sirtuins.
4. Materials and methods
4.1. Cell culture
All cell lines were maintained as previously described [12]. For in vitro nutrient starvation, the cells were grown without serum and glucose for 18–24 h.
4.2. Rodents
Male C57BL mice were either fed AL, or given only water for 24 h (starvation) prior to analysis. Male Fisher 344 rats were grown for 12 months on an AL diet or 60% (CR) food supply, as previously described [12]. Protein extraction from the tissues was done as described previously [12]. All experiments were approved by the Institutional Animal Care and Use Committee.
4.3. Antibodies and western blot
Whole cell extracts were prepared using lysis buffer (50 mM Tris [pH 8], 1% NP-40, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT and 1X EDTA-free protease inhibitor cocktail [Roche Diagnostics]). The following antibodies were used: Mouse monoclonal anti-α-actin (A4700 Sigma), mouse monoclonal anti-β-tubulin (12G10 Hybridoma Bank, University of Iowa), rabbit polyclonal anti-SIRT1 (07-131 Millipore), rabbit polyclonal anti-SIRT6 (kindly provided by Sigma–Aldrich Israel, Ltd.), and rabbit polyclonal anti-SIRT6 antibody (Supp. Fig. 1).
4.4. Primers
For PCR analysis, the following SIRT6 specific primers were used: S6 Fwd - 5’ CCA AGT TCG ACA CCA CCT TT 3’ and S6 Rev - 5’ CGG ACG TAC TGC GTC TTA CA 3’.
4.5. Inhibition of transcription or translation
For inhibition of transcription, the culture medium was supplemented with actinomycin D (5 µg/ml) for the entire 24 h or for the last 4 h of the experiment. For inhibition of translation, the medium was supplemented with cyclohexamide (50 µg/ml) 4 h before harvesting the cells.
Supplementary Material
Acknowledgements
We thank Doron Ginsberg (Bar-Ilan University, Israel) and members of the Cohen lab for helpful comments on the manuscript; Fred Alt (HMS) for the SIRT6−/− mice; Moshe Oren (Weizmann Institute, Israel) for the p53−/− mice; and Izumi Horikawa (NIH) for pcDNA-DEST40-SIRT6. This study was supported by grants from the Israeli Academy of Sciences, German–Israeli Foundation, Binational US–Israel Foundation, Israel Cancer Association, Koret Foundation, and the Israel Cancer Research Foundation. H.C. is funded by the Alon Foundation.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2008.01.019.
References
- 1.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 2.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes. Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 5.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 6.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 Shows no substrate specificity in vitro. J. Biol. Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 8.Weindruch R. Caloric restriction, gene expression, and aging. Alzheimer Dis. Assoc. Disord. 2003;17 Suppl. 2:S58–S59. doi: 10.1097/00002093-200304002-00008. [DOI] [PubMed] [Google Scholar]
- 9.Lane MA, Black A, Handy A, Tilmont EM, Ingram DK, Roth GS. Caloric restriction in primates. Ann. NY Acad. Sci. 2001;928:287–295. doi: 10.1111/j.1749-6632.2001.tb05658.x. [DOI] [PubMed] [Google Scholar]
- 10.Weindruch R. The retardation of aging by caloric restriction: studies in rodents and primates. Toxicol. Pathol. 1996;24:742–745. doi: 10.1177/019262339602400618. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 12.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 13.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 14.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 15.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 16.Guarente L. Calorie restriction and SIR2 genes – towards a mechanism. Mech. Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyl-transferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.