Abstract
Brain activity can be measured with several non-invasive neuroimaging modalities, but each modality has inherent limitations with respect to resolution, contrast and interpretability. It is hoped that multimodal integration will address these limitations by using the complementary features of already available data. However, purely statistical integration can prove problematic owing to the disparate signal sources. As an alternative, we propose here an advanced neural population model implemented on an anatomically sound cortical mesh with freely adjustable connectivity, which features proper signal expression through a realistic head model for the electroencephalogram (EEG), as well as a haemodynamic model for functional magnetic resonance imaging based on blood oxygen level dependent contrast (fMRI BOLD). It hence allows simultaneous and realistic predictions of EEG and fMRI BOLD from the same underlying model of neural activity. As proof of principle, we investigate here the influence on simulated brain activity of strengthening visual connectivity. In the future we plan to fit multimodal data with this neural population model. This promises novel, model-based insights into the brain's activity in sleep, rest and task conditions.
Keywords: neural population model, multimodal integration, simultaneous EEG and fMRI BOLD, brain dynamics, cortical connectivity, mean-field model
1. Introduction
Non-invasive recording of human brain activity has a long history, beginning with the electroencephalogram (EEG) [1]. The EEG remains prominent both in research and in clinical practice [2] owing to its excellent time resolution, which allows, for example, the tracking of evoked potentials. In the meantime, functional magnetic resonance imaging (fMRI) based on blood oxygen level dependent (BOLD) contrast has become a standard for researching cognition [3,4], largely because fMRI BOLD can locate brain activity with millimetre accuracy. However, progress in neuroimaging is slowing, owing to fundamental restrictions in acquisition methods. For example, volume conduction limits the spatial resolution of EEG to the centimetre range, whereas fMRI BOLD relies on vascular changes with latencies of half a second or more.
A promising way forward lies in combining already available neuroimaging modalities [5–9]. Each modality provides a particular and distinct representation of the brain's state via its specific signal sources. For example, EEG relies on electrical and fMRI BOLD on haemodynamic sources, which relate to different aspects of neural activity, as we shall see. Furthermore, data from different modalities often have complementary characteristics. This is the case for EEG and fMRI BOLD concerning spatiotemporal resolution, as discussed above. Finally, it is possible to record EEG and fMRI BOLD simultaneously [10]. This avoids the question whether data obtained in different sessions really refer to the same brain state. Hence EEG and fMRI BOLD present a convenient test case for developing multimodal approaches. An example for the ‘added value’ that simultaneous EEG/fMRI can provide is the connection of fMRI resting-state networks to EEG cortical microstates recently discovered by Britz et al. [11] and Musso et al. [12] (cf. the commentaries by Laufs [13] and Lehmann [14]).
There are three basic approaches to multimodal integration [15], which are all ‘model-based’ in some sense. When using converging evidence, a researcher combines data argumentatively against a backdrop of established expert opinion. Clearly, such an implicit ‘human mind’ model is powerful, but qualitative and idiosyncratic. Data fusion combines the various recorded data directly using statistical methods. This is quantitative and repeatable, but implies some model of relations between signal sources. Such relations may be less simple than commonly assumed (e.g. [16]). Computational modelling makes explicit prior knowledge and assumptions through the process of model specification. In principle, this allows the fully objective assessment of theory in terms of a model fit to data. In practice, realistic models are often too complex for a comprehensive validation or unequivocal falsification. Nevertheless, the explication of theory through a computational model generally allows more rigorous testing.
We present here a complete model chain from neural activity to detector signal, based on a neural population model (NPM). Pioneered by Wilson & Cowan [17] and others, such neural population modelling approaches have attracted much attention—see the recent reviews by Deco et al. [18] and Coombes [19]. They can successfully describe epileptic seizures [20–23], evoked potentials [24,25], cognitive activity [26,27], drug effects [28,29] and—of particular significance for this Theme Issue—sleep [30–33]. NPMs for the magnetoencephalogram [34,35] and fMRI BOLD [36] are popular, in particular, when considering network dynamics [37]. Here, we use a discretization of an anatomically folded cortex, with activity propagation instantiated not with approximate partial differential equations (PDEs) [38,39], but by explicitly keeping track of all signal delays. This allows the introduction of realistic cortical connectivity. Similar approaches have been developed by Sotero et al. [40] and Valdes-Sosa et al. [41].
Our NPM has been introduced previously in Bojak et al. [42]. Here, we add considerable technical detail necessary for implementing such a model. The paper is organized as follows. The following section explains how we extract the head model from structural MRI. Section 3 explains our NPM and signal expression. Section 4 presents new results for variations of specific connectivity strength. We then conclude with a discussion and outlook.
2. The head model
(a). Surfaces extracted from structural MRI
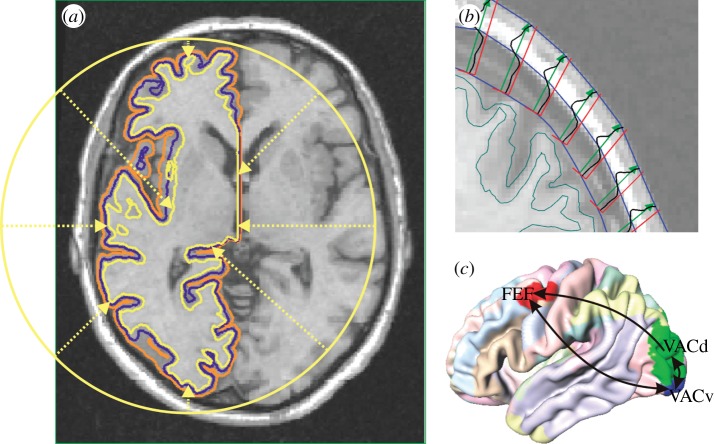
Surface approximations for the interfaces between grey matter (GM), cerebrospinal fluid (CSF) and white matter (WM) were obtained using the Civet software pipeline [43]. This involves a series of processing steps. Firstly, field non-uniformity artefacts are removed from T1-weighted structural images with the N3 algorithm [44]. T1-weighting is a standard MRI acquisition protocol that provides optimal intensity contrast between the tissue types of interest here. Secondly, the corrected images are normalized to stereotaxic space, and subsequently skull-stripped and classified into GM, WM and CSF. Thirdly, the GM/WM interface is constructed by deforming a spherical surface mesh subject to optimization constraints. Fourthly, Laplacian GM fields are computed, and CSF skeletons are constructed in deep sulci, as guides for an expansion of the GM/WM surface to the GM/CSF interface. The resulting meshes represent the borders of cortical GM, and we use an intermediate surface for modelling; see figure 1a.
Figure 1.
(a) Neural population model surface (blue) between the Civet interfaces of grey matter with white matter (yellow) and cerebrospinal fluid (orange). (b) Skull and scalp boundaries (blue) from intensity profiles (black) along outward vectors (green). (c) Visual connectivity used in this study. ‘Regional map’ areas are indicated by colours on an average cortical surface. FEF, frontal eye field; VACd and VACv, dorsal and ventral anterior visual cortex.
To estimate volume conduction for EEG predictions, it is also necessary to obtain skull and scalp surfaces. Using the same images, and the obtained cortical surfaces as a starting point, T1 intensities can be sampled along rays extending outwards until a voxel containing ‘air’ is found, indicating the edge of the scalp. In such a T1-weighted image, skull tissue produces little signal, and thus its inner and outer boundaries can be determined as the edges of a ‘dip’ along the ray. Figure 1b illustrates this method. More details will be provided in a forthcoming publication.
(b). Pruning the cortical mesh
The Civet mesh consists of 81 920 triangles with 40 962 vertices per hemisphere, but cortical folding can be represented faithfully with much fewer vertices. This is essential to reduce the NPM computation time, which scales linearly with the number of vertices. We will see below that ‘background connectivity’ scales roughly with the surface area, hence with the square of the number of vertices, and that longer edges between vertices reduce significantly the overall data transfer during the simulation. Thus, it is particularly advantageous to prune short edges. Furthermore, inspection of the Civet mesh shows that many small triangles are ‘wasted’ on relatively flat parts. In other places there appear unnatural ripples, typical evidence for ‘over-fitting’. These issues also are dealt with by removing short edges. Hence in each iteration of our algorithm, the shortest edge of the mesh is found. Subsequently, it is removed in a manner that we will detail next.
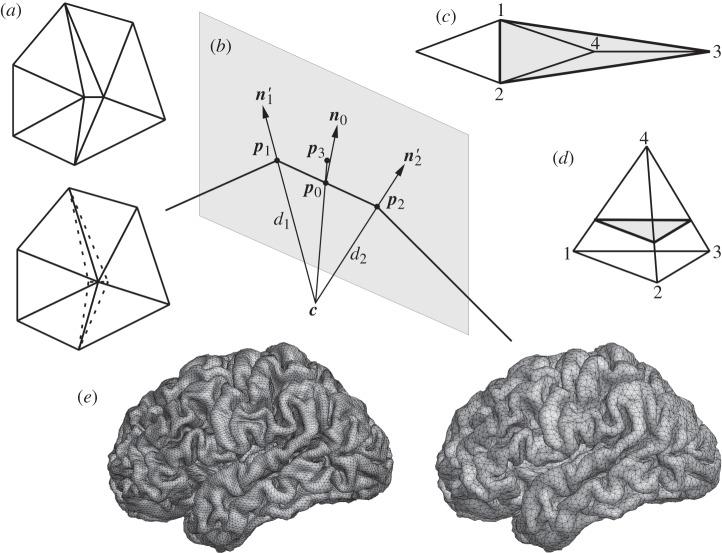
The two triangles that previously shared the shortest edge are removed, and their other two sides collapsed to a single edge terminating in a new vertex; see figure 2a. If the new vertex were to be positioned halfway on the former edge, the curvature of the cortex would be poorly preserved. Instead, the new vertex is positioned on a circle segment that approximates the surface (cf. figure 2b): p1 and p2 are the vertices to be removed, and n1 and n2 are the surface normals at these vertices, which are defined as the average over the area-weighted normals of the triangles they belong to. First 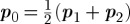 and
and 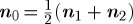 are computed. Next, n1 and n2 are projected onto the plane through p0 that is spanned by n0 and p1−p2, resulting in n1′ and n2′, respectively. Point c is the intersection of the lines through p1 with direction n1′ and through p2 with direction n2′. Then dk is defined as the distance between c and pk for k=1,2, and
are computed. Next, n1 and n2 are projected onto the plane through p0 that is spanned by n0 and p1−p2, resulting in n1′ and n2′, respectively. Point c is the intersection of the lines through p1 with direction n1′ and through p2 with direction n2′. Then dk is defined as the distance between c and pk for k=1,2, and 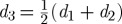 . Finally, p3 is defined on the line from c towards p0 at a distance d3 from c. The new vertex is placed at v=p0+b(p3−p0) with a fixed parameter 0≤b≤1, where b=1 would be perfect for a sphere and b=0 for a plane.
. Finally, p3 is defined on the line from c towards p0 at a distance d3 from c. The new vertex is placed at v=p0+b(p3−p0) with a fixed parameter 0≤b≤1, where b=1 would be perfect for a sphere and b=0 for a plane.
Figure 2.
(a) Before (top) and after (bottom) pruning. (b) The bold edge between p1 and p2 is replaced by a vertex between p0 and p3. (c) If edge 1–2 is removed, triangles 2–3–4 and 1–4–3 collapse. The shaded region is replaced by the 1–2–3 triangle; or (d) a lifted one. (e) Cortical surface before (left) and after (right) pruning to a minimum edge length of 2.5 mm.
The method fails for cases like edge 1–2 in figure 2c, since removing it would collapse two triangles onto each other. We refer to this as the ‘tetrahedron’, since the configuration resembles one if vertex 4 is not in the same plane, cf. figure 2d. Note that the base triangle 1–2–3 is empty, i.e. not itself part of the cortical surface. Tetrahedra are detected and removed by replacing them with single triangles; see figure 2d. We use a linear factor 0≤l<1, where l=0 indicates the edge base points 1, 2 or 3, and l=1 the peak 4. Thus, the tetrahedron is replaced by the base triangle for l=0 and with lifted ones for 0<l<1. While l>0 compensates better for loss of volume, it distorts the surrounding triangles. Figure 2e shows an original Civet mesh (left) pruned to a minimum edge length of 2.5 mm (right). We have used here b=1 and l=0. The pruned surface has 32 408 triangles with 17 208 vertices. The overall shape and surface area remain well preserved.
(c). Parcellation and specific connectivity
Tractography based on diffusion MRI is popular for determining connectivity [45], but has significant drawbacks. Firstly, it is biased towards short-range connections and has problems where tracts are densely packed. Secondly, it does not determine the direction of activity propagation. Thirdly, it cannot find precise termination points in the GM. Data obtained through histological tract tracing methods are free of these problems, but only available from animal studies. We use here a connectivity matrix for macaque monkey (available on request), based on tracer data in the CoCoMac database [46], together with a ‘regional map’ (RM). This RM is a parcellation of cortex, which is sufficiently generalized to accommodate anatomical homology across primate species and uses area names that are widely recognized and convenient to use [47]. Since the Civet pipeline is designed to obtain optimal correspondence between individual vertices of meshes extracted from different human brains, it is sufficient to map macaque brain regions to a Civet mesh once. The assignment of vertex number to equivalent brain region stays the same for all Civet meshes. The RM was manually delineated onto a template macaque cortical surface (F99-UA1) and, by using two landmark-based deformations included in the Caret software [48], first mapped to the human PALS-B12 surface and then to our Civet template surface. The resulting parcellation is shown in figure 1c, and is also available on request.
3. Neural activity model and signal expression
(a). The neural population model
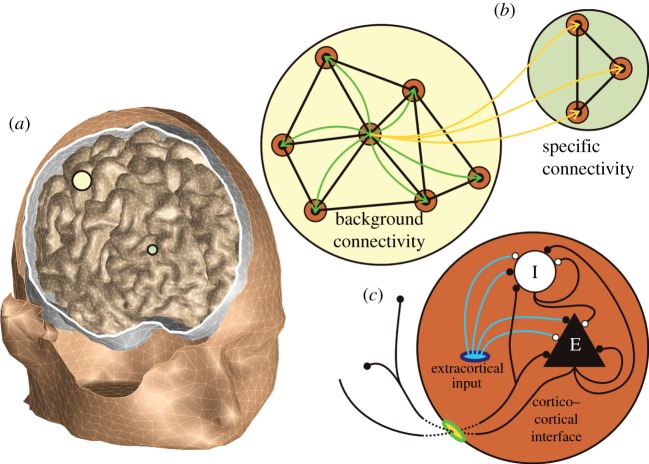
Our software is flexible concerning the employed model of neural activity. We compute here at each vertex the NPM of Liley et al. [49], as extended by Bojak & Liley [28]. It contains one excitatory (e) and one inhibitory (i) NPM neuron, which represent those populations of real neurons whose coherent activity dominates the macroscopic signal of interest. For example, one can speculate that the EEG is largely due to pyramidal layer V ‘output neurons’, which form long dendritic bundles [50] that can act as dipolar current sources. The NPM neurons are locally connected to each other ( ,
, 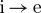 ) and to themselves (
) and to themselves ( ,
,  ); see figure 3c. NPM self-connections model real neurons of the same type connecting to each other.
); see figure 3c. NPM self-connections model real neurons of the same type connecting to each other.
Figure 3.
(a) Head model as extracted from structural magnetic resonance imaging. (b) For one vertex, the two kinds of long-range connectivity are illustrated. (c) The neural activity model with excitatory (black) and inhibitory (white) populations and connections.
The NPM consists of the following ordinary differential equations (ODEs):
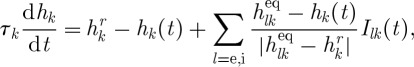 |
3.1 |
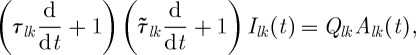 |
3.2 |
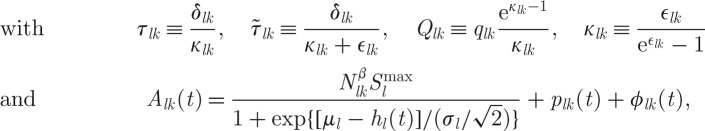 |
3.3 |
where l,k=e,i and κlk|ϵlk=0≡1 is continuous in ϵlk. The model parameters could vary from vertex to vertex, but we choose here uniformly those of [51] and set ϵlk=0 throughout.1 Equation (3.1) gives the response of the mean soma membrane potential hk to a sum of post-synaptic potentials (PSPs) Ilk. In the absence of input, hk decays exponentially to  with characteristic time τk. PSP impact is weighted, with a sign change at the Nernst potentials heqlk. Equation (3.2) responds to a pre-synaptic Dirac pulse Alk(t)=δ(t) with an alpha PSP
with characteristic time τk. PSP impact is weighted, with a sign change at the Nernst potentials heqlk. Equation (3.2) responds to a pre-synaptic Dirac pulse Alk(t)=δ(t) with an alpha PSP 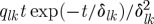 for ϵlk=0, and a bi-exponential PSP
for ϵlk=0, and a bi-exponential PSP 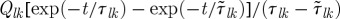 otherwise. Qlk is proportional to the total charge transferred, δlk is the rise time to maximum PSP amplitude, and ϵlk prolongs the characteristic PSP decay time.
otherwise. Qlk is proportional to the total charge transferred, δlk is the rise time to maximum PSP amplitude, and ϵlk prolongs the characteristic PSP decay time.
Equation (3.3) collects the sources for pre-synaptic spikes. The first term corresponds to the local firing rate, limited to 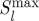 , multiplied by the number of local synapses
, multiplied by the number of local synapses 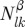 . The second term, plk, allows for extracortical input. Here, one could insert thalamocortical loops [52] or specific sensory inputs. We assume simply that extracortical input is noise-like in pee [28,49]. As a continuous average of neural signals, pee should be low-pass filtered. We use the spline variant of Catmull & Rom [53] with noise innovations as control points to construct such an input. This allows us to minimize the computational expense for random number generation and spline coefficient computation, which is important since the noise input is computed for every node independently. Catmull–Rom splines are unbiased, interpolating cubic splines with zero local tension and C1 (first derivative) continuity [54]. The following pseudo-code illustrates how one can interpolate pee at time steps s=0,1,2,…, where time t=s Δt, from the Gaussian white noise randn sampled every uth time step.
. The second term, plk, allows for extracortical input. Here, one could insert thalamocortical loops [52] or specific sensory inputs. We assume simply that extracortical input is noise-like in pee [28,49]. As a continuous average of neural signals, pee should be low-pass filtered. We use the spline variant of Catmull & Rom [53] with noise innovations as control points to construct such an input. This allows us to minimize the computational expense for random number generation and spline coefficient computation, which is important since the noise input is computed for every node independently. Catmull–Rom splines are unbiased, interpolating cubic splines with zero local tension and C1 (first derivative) continuity [54]. The following pseudo-code illustrates how one can interpolate pee at time steps s=0,1,2,…, where time t=s Δt, from the Gaussian white noise randn sampled every uth time step.
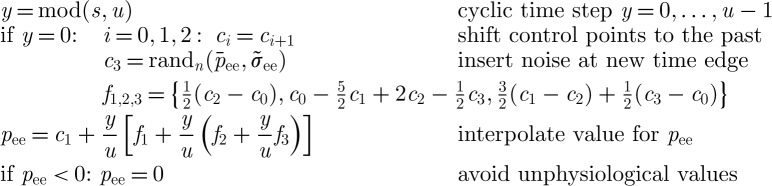
Setting up-sampling u=round[0.40449/(Δt fcut)] achieves an intended −3 dB frequency, chosen here as fcut=75 Hz. The spline output has mean 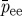 and standard deviation σee if
and standard deviation σee if 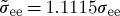 .
.
(b). Connectivity and propagation of activity
The third term of equation (3.3), ϕlk, supports long-range connectivity. We distinguish two kinds; cf. figure 3b. Background connectivity is roughly isotropic, homogeneous and diminishes exponentially with distance [55–57], typically leading to waves of cortical activity [58]. This type of connectivity has been commonly used, since it can be approximated by PDEs [38,39]. Background connections from vertices b to a particular vertex a are modelled here by
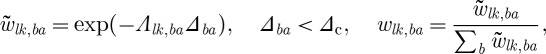 |
3.4 |
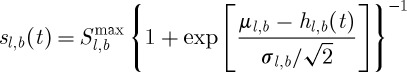 |
3.5 |
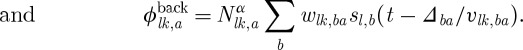 |
3.6 |
Background synaptic weights wlk,ba sum to one and are multiplied by 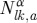 , the number of synapses formed at vertex a. The firing rates sl,b from vertices b arrive with delays Δba/vlk,ba, where Δba indicates the distance along the cortical surface and vlk,ba the conduction velocity; Δba is estimated as the shortest path through the cortical mesh. We consider here only distances up to a cut-off Δc, where
, the number of synapses formed at vertex a. The firing rates sl,b from vertices b arrive with delays Δba/vlk,ba, where Δba indicates the distance along the cortical surface and vlk,ba the conduction velocity; Δba is estimated as the shortest path through the cortical mesh. We consider here only distances up to a cut-off Δc, where 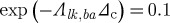 , in order to limit the number of connections to those with significant impact. Long-range connectivity is here considered as exclusively excitatory (
, in order to limit the number of connections to those with significant impact. Long-range connectivity is here considered as exclusively excitatory (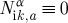 ), with characteristic decay (1/Λek,ba=2.5 cm) and cut-off (Δc≃5.76 cm) distances in the right range for the loss of coherence measured with subdural electrodes [59]. Note that other synaptic footprints can be introduced simply by changing the functional form of
), with characteristic decay (1/Λek,ba=2.5 cm) and cut-off (Δc≃5.76 cm) distances in the right range for the loss of coherence measured with subdural electrodes [59]. Note that other synaptic footprints can be introduced simply by changing the functional form of 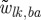 .
.
Specific connectivity is implemented by adding further synaptic weights 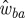 :
:
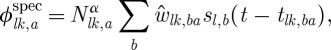 |
3.7 |
where the conduction delays tlk,ba must be given. We will explain our estimation method below. For example, a specific synaptic weight 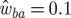 means that the added connectivity from vertex b to vertex a has 10% of the strength of the total background connectivity that a is receiving, since
means that the added connectivity from vertex b to vertex a has 10% of the strength of the total background connectivity that a is receiving, since 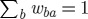 . Specific connectivity can accommodate arbitrary connections between brain regions, and typically will be constructed according to some experimental connectivity matrix. Since the conduction delays Δba/vlk,ba and tlk,ba are not known with great accuracy, we discretize both internally as multiples of the time step Δt=5×10−5 s. This makes delay bookkeeping much easier. The ϕlk,a represent averaged and hence continuous signals, for which we again use the spline approach detailed above. Let us consider fcut=1.6 kHz as sufficient, then up-sampling u=5 follows. Thus, we actually need to transmit ϕlk,a values only every uΔt=0.25 ms. However, the spline needs four control points spaced by three time steps, hence we can only spline delays greater than 3uΔt=0.75 ms. For vee,ba=3 m s−1, the minimum allowed distance is Δba=2.25 mm. This demonstrates the connection between data transfer and shortest edge length: if we reduce the latter, then we must lower the up-sampling and hence transmit ϕlk more often.
. Specific connectivity can accommodate arbitrary connections between brain regions, and typically will be constructed according to some experimental connectivity matrix. Since the conduction delays Δba/vlk,ba and tlk,ba are not known with great accuracy, we discretize both internally as multiples of the time step Δt=5×10−5 s. This makes delay bookkeeping much easier. The ϕlk,a represent averaged and hence continuous signals, for which we again use the spline approach detailed above. Let us consider fcut=1.6 kHz as sufficient, then up-sampling u=5 follows. Thus, we actually need to transmit ϕlk,a values only every uΔt=0.25 ms. However, the spline needs four control points spaced by three time steps, hence we can only spline delays greater than 3uΔt=0.75 ms. For vee,ba=3 m s−1, the minimum allowed distance is Δba=2.25 mm. This demonstrates the connection between data transfer and shortest edge length: if we reduce the latter, then we must lower the up-sampling and hence transmit ϕlk more often.
For parallel computation, we divide up the cortical mesh between compute nodes. To minimize the communication overheads, we compile data into chunks for transmission between nodes. Assume a compute node 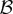 has been assigned vertices vb with b∈B. A different node
has been assigned vertices vb with b∈B. A different node 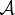 contains vertices va with a∈A. Call
contains vertices va with a∈A. Call 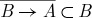 the list of vertices for which connection strengths wlk,ba or
the list of vertices for which connection strengths wlk,ba or 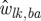 , or both, are non-zero. The compilation of values {sl,b(t)} with
, or both, are non-zero. The compilation of values {sl,b(t)} with  is what gets transmitted from
is what gets transmitted from 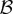 to
to 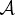 . To simplify our explanation of how node
. To simplify our explanation of how node 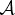 distributes the received data, we will assume that data are exchanged at every time step Δt. In reality, only the spline control points are sent. This leads to a significant reduction of traffic, here by a factor u=5. Every vertex has its own ‘delay buffer’ for accumulating inputs, with a size set by the maximum (discretized) delay of incoming connections. A pointer indicating ‘current time’ advances in this buffer and at its end cycles back to the beginning. Distributing {sl,b(t)} now simply reduces to adding
distributes the received data, we will assume that data are exchanged at every time step Δt. In reality, only the spline control points are sent. This leads to a significant reduction of traffic, here by a factor u=5. Every vertex has its own ‘delay buffer’ for accumulating inputs, with a size set by the maximum (discretized) delay of incoming connections. A pointer indicating ‘current time’ advances in this buffer and at its end cycles back to the beginning. Distributing {sl,b(t)} now simply reduces to adding 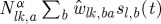 to the position tlk,ba/Δt steps ahead of the ‘current time’ pointer (modulo the buffer length), and likewise for background connections. When the ‘current time’ pointer has advanced to a new position in the buffer, the sum of equations (3.6) and (3.7) will have been built up there. The buffer entry is hence added to the system as the current input ϕlk,a, and then reset to zero, ready to accumulate input again.
to the position tlk,ba/Δt steps ahead of the ‘current time’ pointer (modulo the buffer length), and likewise for background connections. When the ‘current time’ pointer has advanced to a new position in the buffer, the sum of equations (3.6) and (3.7) will have been built up there. The buffer entry is hence added to the system as the current input ϕlk,a, and then reset to zero, ready to accumulate input again.
The RM does not provide an estimate for fibre length and hence time delays tlk,ba. To calculate an estimate, we assume that vertices are connected by the shortest possible route, which minimizes conduction delays in the brain. Our algorithm first determines all those straight lines between pairs of vertices, which are completely contained within the cortical volume. This yields a network of allowed paths. At this stage any pair of vertices is connected at least via the surface edges, but ‘shortcuts’ through the volume generally exist. The optimal route is computed by means of a standard ‘all pairs shortest path’ algorithm like Floyd–Warshall [60]; cf. figure 4a. Examples of routes so determined are shown in figure 4b. For route finding, we prune the cortical surface further to only several thousand vertices. This limits the computation time for, and final size of, the distance matrix. Vertices of the NPM surface are then matched by Euclidean proximity.
Figure 4.
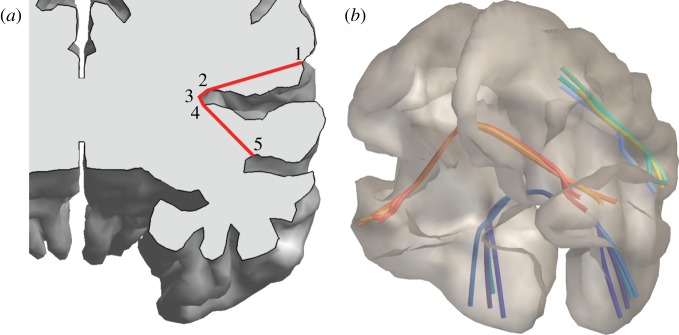
(a) Direct connections  ,
,  ,
,  and
and  within the cortical volume are concatenated to the shortest route
within the cortical volume are concatenated to the shortest route  . (b) Examples of shortest routes so determined.
. (b) Examples of shortest routes so determined.
(c). EEG and fMRI BOLD signal sources
The details of extracting EEG and fMRI BOLD signals from the NPM simulation have been described in our previous publication; see Bojak et al. [42] for details. Electric dipole strength is here assumed to be roughly proportional to the average excitatory membrane potential he [61]. A volume conductor model with three compartments was constructed, representing the scalp (conductivity 0.2 S m−1), the skull (0.03 S m−1 [62]) and the inside of the skull (0.2 S m−1). For piecewise homogeneous volume conductors such as this, the boundary element method can be used to compute the EEG transfer matrix. We assume that fMRI BOLD is driven by glutamate release [63,64]. The neural drive z is hence proportional to the sum feAee+fiAei of excitatory inputs only, cf. equation (3.3). Values fe≃0.85 and fi=1−fe represent the fraction of excitatory and inhibitory neurons, respectively. We implement a ‘balloon–windkessel’ model of haemodynamics at every vertex with four ODEs according to Friston et al. [65]. This predicts the local fMRI BOLD contrast y. For the results shown here, a baseline of resting activity is subtracted by hand.
(d). Comparing voxel data with surface simulations
The prediction of voxel values from vertex ones must be informed by anatomy: in the normal direction of the surface we assume here that fMRI BOLD is roughly uniform across the cortical mantle. It is however straightforward to introduce a gradient, e.g. to weigh contributions from the pial surface more strongly. The weighting factors are then determined as follows (cf. figure 5a): if we designate dij as the vector between a given vertex si and voxel centre point vj, then nij is its projection onto the surface normal at si; and tij is a tangential vector uniquely defined by being orthogonal to nij and coplanar with nij and dij. Weights are computed by the Gaussian dispersion
| 3.8 |
where 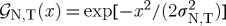 , and we use σN=2.0 mm and σT=1.66 mm. The latter equates the Gaussian full width at half-maximum with the average edge length of our cortical surface (3.9 mm). Finally, the projected fMRI BOLD contrast y(vj) is
, and we use σN=2.0 mm and σT=1.66 mm. The latter equates the Gaussian full width at half-maximum with the average edge length of our cortical surface (3.9 mm). Finally, the projected fMRI BOLD contrast y(vj) is
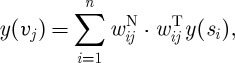 |
3.9 |
where n is the number of vertices in the surface mesh. A global normalization restores the original range of fMRI BOLD values. The same approach can be used as well to project fMRI BOLD voxel data onto surface vertices.
Figure 5.
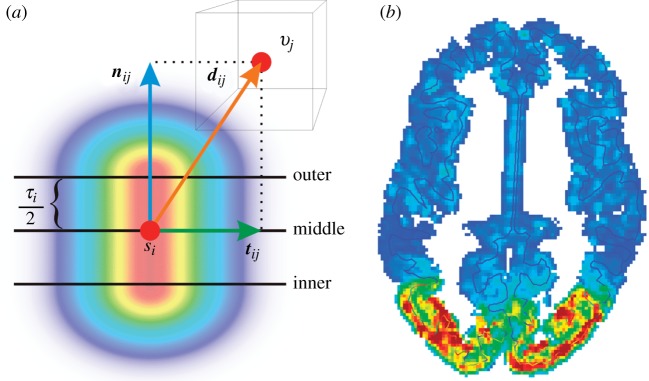
(a) Projection vectors for vertex si and voxel vj, and resulting weights from high (red) to low (blue). (b) Vertex to voxel projection of fMRI BOLD in horizontal section.
4. Results
The CoCoMac matrix provides basic information about connectivity strength by assigning values 0 (absent), 1 (weak), 2 (moderate) and 3 (strong) based on the reported histochemical staining. Here, we explore the relationship between CoCoMac's anatomical connectivity strength and the effective (causal) connectivity of our model, gauge the strength of specific versus background connectivity, and test the functional visibility of a cortical network consisting of the dorsal (VACd) and ventral (VACv) anterior visual cortex and the frontal eye fields (FEF). As can be seen in figure 1c, these areas form a triangle of connections, all with ‘moderate’ CoCoMac strength. We also include contra-lateral connectivity from every vertex to the vertex nearest to its mirror position in the other hemisphere. For the sake of simplicity, we employ only one universal 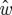 value for all specific connections. Varying this connection strength parameter will provide a basic test for how significant current experimental and theoretical uncertainties concerning (effective) connectivity are for model predictions of brain dynamics.
value for all specific connections. Varying this connection strength parameter will provide a basic test for how significant current experimental and theoretical uncertainties concerning (effective) connectivity are for model predictions of brain dynamics.
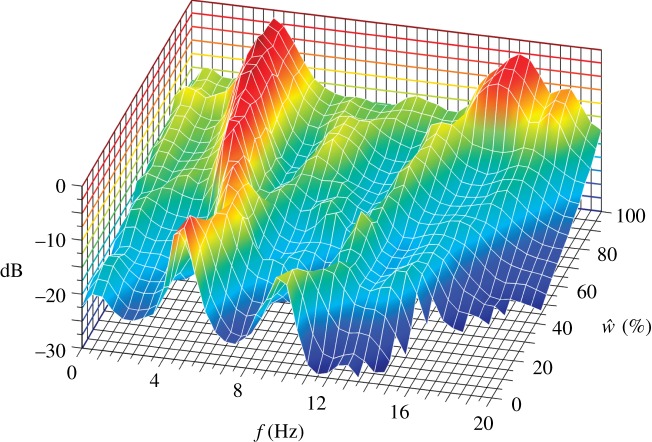
We have simulated 10 s of brain time for 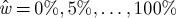 , i.e. from no specific connectivity to one as strong as the background connectivity. The only input here is the noise shaped as described above, hence all dynamical features at frequencies well below fcut=75 Hz emerge from the system. The power spectral density of the excitatory mean soma potential he (simply called ‘ECoG’ henceforth) was computed for the last 4 s of brain time (2 s Hann window, 50% overlap), avoiding initial transients and averaged over all vertices. The result is shown in figure 6: a strong peak is evident in the delta/theta band, which drifts from 5 to 4 Hz for increasing
, i.e. from no specific connectivity to one as strong as the background connectivity. The only input here is the noise shaped as described above, hence all dynamical features at frequencies well below fcut=75 Hz emerge from the system. The power spectral density of the excitatory mean soma potential he (simply called ‘ECoG’ henceforth) was computed for the last 4 s of brain time (2 s Hann window, 50% overlap), avoiding initial transients and averaged over all vertices. The result is shown in figure 6: a strong peak is evident in the delta/theta band, which drifts from 5 to 4 Hz for increasing 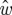 . There is also a weak alpha peak at 9.5 Hz, which weakens further as it drifts to higher frequencies. Strikingly, upon moving from
. There is also a weak alpha peak at 9.5 Hz, which weakens further as it drifts to higher frequencies. Strikingly, upon moving from 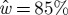 to
to 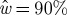 , suddenly a strong beta peak appears at around 16 Hz, which for
, suddenly a strong beta peak appears at around 16 Hz, which for 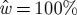 reaches three-quarters of the maximum power of the slow oscillation. The power of the slow oscillation itself by then has more than doubled. All the described features of the power spectra are due to self-sustaining oscillations, i.e. can be elicited also without the noise drive. We have tested that these results are stable for the simple (forward Euler) integration scheme used here by repeating the calculations with a five times smaller time step.
reaches three-quarters of the maximum power of the slow oscillation. The power of the slow oscillation itself by then has more than doubled. All the described features of the power spectra are due to self-sustaining oscillations, i.e. can be elicited also without the noise drive. We have tested that these results are stable for the simple (forward Euler) integration scheme used here by repeating the calculations with a five times smaller time step.
Figure 6.
Dependence of the power spectral density of the ECoG on the strength of specific connectivity 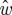 . The power is shown in dB relative to the largest overall value.
. The power is shown in dB relative to the largest overall value.
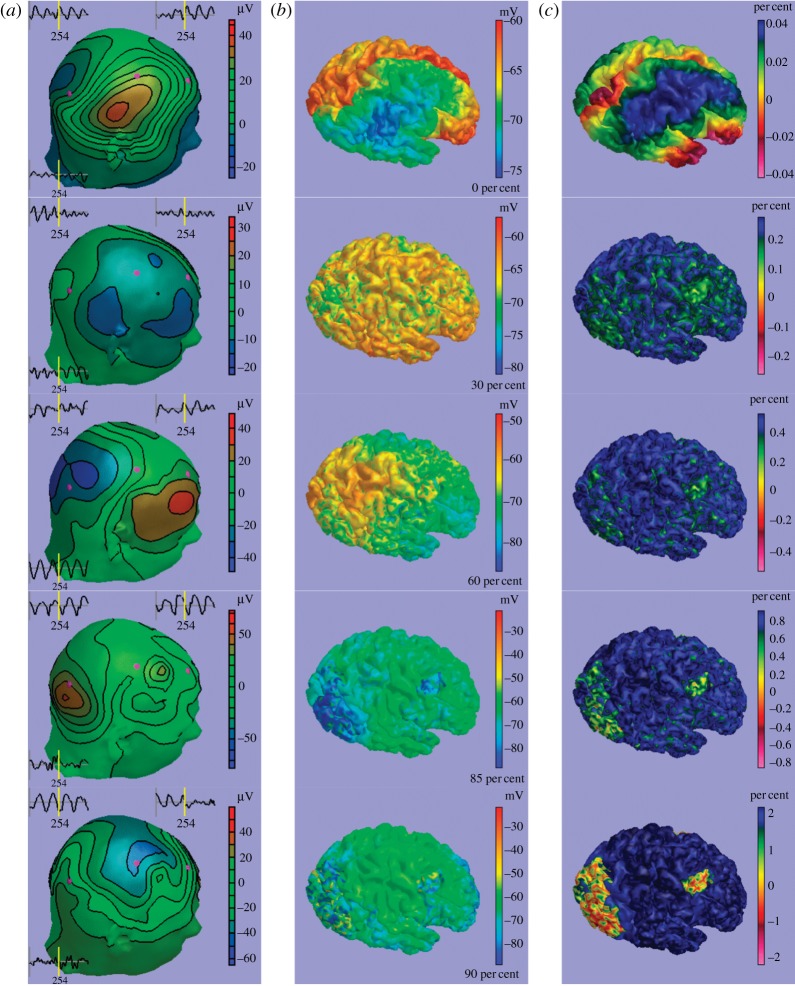
In figure 7, we compare activity patterns for (a) scalp EEG, (b) ECoG and (c) fMRI BOLD for 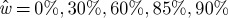 , from top to bottom. In the EEG column, we also see recordings from three ‘electrodes’ indicated in purple, which show the last second of the simulation. Without specific connectivity, large waves of activity dominate. fMRI BOLD contrast gets sufficiently strong to be detected over experimental noise only between 30 and 60 per cent, and VACd, VACv and FEF also become somewhat visible there. The transition from 85 to 90 per cent appears less dramatic in the activity snapshots than in the power spectrum. However, for 90 per cent and higher we can easily identify the regions of our chosen cortical network, in particular with fMRI BOLD contrast. In summary, our variation of
, from top to bottom. In the EEG column, we also see recordings from three ‘electrodes’ indicated in purple, which show the last second of the simulation. Without specific connectivity, large waves of activity dominate. fMRI BOLD contrast gets sufficiently strong to be detected over experimental noise only between 30 and 60 per cent, and VACd, VACv and FEF also become somewhat visible there. The transition from 85 to 90 per cent appears less dramatic in the activity snapshots than in the power spectrum. However, for 90 per cent and higher we can easily identify the regions of our chosen cortical network, in particular with fMRI BOLD contrast. In summary, our variation of 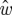 identified large parameter regions for which brain dynamics would appear qualitatively unchanged in simultaneous EEG/fMRI (30–85% and greater than 90%, respectively), but with the possibility for a transition between these regions induced by a small parameter change (from 85% to 90%).
identified large parameter regions for which brain dynamics would appear qualitatively unchanged in simultaneous EEG/fMRI (30–85% and greater than 90%, respectively), but with the possibility for a transition between these regions induced by a small parameter change (from 85% to 90%).
Figure 7.
Predictions of (a) scalp EEG, (b) ECoG and (c) fMRI BOLD for different strengths of specific connectivity: 0% (top), 30%, 60%, 85% and 90% (bottom). See also the video in the electronic supplementary material.
5. Conclusions
In the present work and in Bojak et al. [42], we have outlined a complete simulation pipeline for forward predictions of simultaneous EEG and fMRI BOLD. It is based on a well-known NPM formalism [28,49], but uses a cortical tessellation with correct anatomical geometry in a realistic head model. Our software features a method for tracking conduction delays that allows the implementation of arbitrary corticocortical connectivity. We have explained here our pruning procedure for surface meshes, Catmull–Rom splining in noise generation and (multi-parallel) communication, the implementation of specific connectivity and finally the projection of surface fMRI BOLD predictions to voxel data. Furthermore, we have investigated the effects of strengthening specific connectivity. We found that a dynamical regime of dominant slow oscillations that allow the detection of functional networks with fMRI BOLD was qualitatively stable over a large parametric range (specific connectivity strength 30–85% relative to that of the homogeneous and isotropic ‘background connectivity’). However, a further small parameter increase (by 5%) would then result in the sudden appearance of additional strong beta oscillations. Consequently, for the time being it appears necessary to adjust connectivity strength carefully with respect to the resulting dynamics, since it is not guaranteed that predictions will be qualitatively similar for two different, but reasonable, parameter choices. We speculate that this difficulty generalizes beyond our current model, and suggest that any dynamical implementation of a connectivity matrix has to be accompanied by a careful exploration of the used connectivity strength.
With respect to frequency content, our simulations may already resemble stage II sleep. However, since our model oscillations are of purely cortical origin, lacking both realistic thalamic input and sleep cycle modulation, the current setup provides no direct insight into the mechanisms of sleep. Importantly, however, the NPM and connectivity can be easily modified or exchanged. Hence the sleep NPMs mentioned in §1 (cf. also Robinson et al. [66] in this Theme Issue) could be integrated with our software to allow realistic predictions of the activity of the sleeping brain as observed with (simultaneous) EEG and fMRI BOLD.
Footnotes
The paper provides γlk and Γlk at ϵlk=0, where δlk=1/γlk and 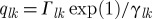 .
.
References
- 1.Berger H.1929Über das Elektrenkephalogramm des Menschen Arch. Psychiatr. Nervenkr. 87527–570. 10.1007/BF01797193 (doi:10.1007/BF01797193) [DOI] [Google Scholar]
- 2.Niedermeyer E., Lopes da Silva F. H., editors. Electroencephalography: basic principles, clinical applications, and related fields. 5th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2005. [Google Scholar]
- 3.Norris D. G.2006Principles of magnetic resonance assessment of brain function J. Magn. Reson. Imag. 23794–807. 10.1002/jmri.20587 (doi:10.1002/jmri.20587) [DOI] [PubMed] [Google Scholar]
- 4.Matthews P. M., Honey G. D., Bullmore E. T.2006Applications of fMRI in translational medicine and clinical practice Nat. Rev. Neurosci. 7732–744. 10.1038/nrn1929 (doi:10.1038/nrn1929) [DOI] [PubMed] [Google Scholar]
- 5.Stufflebeam S. M., Rosen B. R.2007Mapping cognitive function Neuroimag. Clin. N. Am. 17469–484. 10.1016/j.nic.2007.07.005 (doi:10.1016/j.nic.2007.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibasaki H.2008Human brain mapping: hemodynamic response and electrophysiology Clin. Neurophysiol. 119731–743. 10.1016/j.clinph.2007.10.026 (doi:10.1016/j.clinph.2007.10.026) [DOI] [PubMed] [Google Scholar]
- 7.Mulert C., Pogarell O., Hegerl U. Simultaneous EEG–fMRI: perspectives in psychiatry. Clin. EEG Neurosci. 2008;39:61–64. doi: 10.1177/155005940803900207. [DOI] [PubMed] [Google Scholar]
- 8.Blinowska K., Müller-Putz G., Kaiser V., Astolfi L., Vanderperren K., Van Huffel S., Lemieux L.2009Multimodal imaging of human brain activity: rational, biophysical aspects and modes of integration Comput. Intell. Neurosci. 2009813607. 10.1155/2009/813607 (doi:10.1155/2009/813607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman W. J., Ahlfors S. P., Menon V.2009Combining fMRI with EEG and MEG in order to relate patterns of brain activity to cognition Int. J. Psychophysiol. 7343–52. 10.1016/j.ijpsycho.2008.12.019 (doi:10.1016/j.ijpsycho.2008.12.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laufs H., Daunizeau J., Carmichael D. W., Kleinschmidt A. K.2008Recent advances in recording electrophysiological data simultaneously with magnetic resonance imaging NeuroImage 40515–528. 10.1016/j.neuroimage.2007.11.039 (doi:10.1016/j.neuroimage.2007.11.039) [DOI] [PubMed] [Google Scholar]
- 11.Britz J., Van De Ville D., Michel C. M.2010BOLD correlates of EEG topography reveal rapid resting-state network dynamics NeuroImage 521162–1170. 10.1016/j.neuroimage.2010.02.052 (doi:10.1016/j.neuroimage.2010.02.052) [DOI] [PubMed] [Google Scholar]
- 12.Musso F., Brinkmeyer J., Mobascher A., Warbrick T., Winterer G.2010Spontaneous brain activity and EEG microstates. A novel EEG/fMRI analysis approach to explore resting-state networks NeuroImage 521149–1161. 10.1016/j.neuroimage.2010.01.093 (doi:10.1016/j.neuroimage.2010.01.093) [DOI] [PubMed] [Google Scholar]
- 13.Laufs H.2010Multimodal analysis of resting state cortical activity: what does EEG add to our knowledge of resting state BOLD networks? NeuroImage 521171–1172. 10.1016/j.neuroimage.2010.05.034 (doi:10.1016/j.neuroimage.2010.05.034) [DOI] [PubMed] [Google Scholar]
- 14.Lehmann D.2010Multimodal analysis of resting state cortical activity: what does fMRI add to our knowledge of microstates in resting state EEG activity? NeuroImage 521173–1174. 10.1016/j.neuroimage.2010.05.033 (doi:10.1016/j.neuroimage.2010.05.033) [DOI] [PubMed] [Google Scholar]
- 15.Horwitz B. L., Poeppel D.2002How can EEG/MEG and fMRI/PET data be combined? Hum. Brain Mapp. 171–3. 10.1002/hbm.10057 (doi:10.1002/hbm.10057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou W., Nissilä I., Radhakrishnan H., Boas D. A., Hämäläinen M. S., Franceschini M. A.2009Study of neurovascular coupling in humans via simultaneous magnetoencephalography and diffuse optical imaging acquisition NeuroImage 46624–632. 10.1016/j.neuroimage.2009.03.008 (doi:10.1016/j.neuroimage.2009.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson H. R., Cowan J. D.1972Excitatory and inhibitory interactions in localized populations of model neuron Biophys. J. 121–24. 10.1016/S0006-3495(72)86068-5 (doi:10.1016/S0006-3495(72)86068-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deco G. R., Jirsa V. K., Robinson P. A., Breakspear M., Friston K. J.2008The dynamic brain: from spiking neurons to neural masses and cortical fields PLoS Comput. Biol. 4e1000092. 10.1371/journal.pcbi.1000092 (doi:10.1371/journal.pcbi.1000092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coombes S.2010Large-scale neural dynamics: simple and complex NeuroImage 52731–739. 10.1016/j.neuroimage.2010.01.045 (doi:10.1016/j.neuroimage.2010.01.045) [DOI] [PubMed] [Google Scholar]
- 20.Suffczyński P., Lopes da Silva F. H., Parra J., Velis D. N., Kalitzin S. N. Epileptic transitions: model predictions and experimental validation. J. Clin. Neurophysiol. 2005;22:288–299. [PubMed] [Google Scholar]
- 21.Liley D. T. J., Bojak I. Understanding the transition to seizure by modeling the epileptiform activity of general anesthetic agents. J. Clin. Neurophysiol. 2005;22:300–313. [PubMed] [Google Scholar]
- 22.Marten F., Rodrigues S., Benjamin O., Richardson M. P., Terry J. R.2009Onset of polyspike complexes in a mean-field model of human electroencephalography and its application to absence epilepsy Phil. Trans. R. Soc. A 3671145–1161. 10.1098/rsta.2008.0255 (doi:10.1098/rsta.2008.0255) [DOI] [PubMed] [Google Scholar]
- 23.Molaee-Ardekani B., Benquet P., Bartolomei F., Wendling F.2010Computational modeling of high-frequency oscillations at the onset of neocortical partial seizures: from ‘altered structure’ to ‘dysfunction’ NeuroImage 521109–1122. 10.1016/j.neuroimage.2009.12.049 (doi:10.1016/j.neuroimage.2009.12.049) [DOI] [PubMed] [Google Scholar]
- 24.Jansen B. H., Rit V. G.1995Electroencephalogram and visual evoked potential generation in a mathematical model of coupled cortical columns Biol. Cybern. 73357–366. 10.1007/BF00199471 (doi:10.1007/BF00199471) [DOI] [PubMed] [Google Scholar]
- 25.Rennie C. J., Robinson P. A., Wright J. J.2002Unified neurophysical model of EEG spectra and evoked potentials Biol. Cybern. 86457–471. 10.1007/s00422-002-0310-9 (doi:10.1007/s00422-002-0310-9) [DOI] [PubMed] [Google Scholar]
- 26.Zavaglia M., Astolfi L., Babiloni F., Ursino M.2006A neural mass model for the simulation of cortical activity estimated from high resolution EEG during cognitive or motor tasks J. Neurosci. Meth. 157317–329. 10.1016/j.jneumeth.2006.04.022 (doi:10.1016/j.jneumeth.2006.04.022) [DOI] [PubMed] [Google Scholar]
- 27.Bojak I., Liley D. T. J.2007Self-organized 40 Hz synchronization in a physiological theory of EEG Neurocomputing 702085–2090. 10.1016/j.neucom.2006.10.087 (doi:10.1016/j.neucom.2006.10.087) [DOI] [Google Scholar]
- 28.Bojak I., Liley D. T. J.2005Modeling the effects of anesthesia on the electroencephalogram Phys. Rev. E 71041902. 10.1103/PhysRevE.71.041902 (doi:10.1103/PhysRevE.71.041902) [DOI] [PubMed] [Google Scholar]
- 29.Rowe D. L., Robinson P. A., Gordon E.2005Stimulant drug action in attention deficit hyperactivity disorder (ADHD): inference of neurophysiological mechanisms via quantitative modelling Clin. Neurophysiol. 116324–335. 10.1016/j.clinph.2004.08.001 (doi:10.1016/j.clinph.2004.08.001) [DOI] [PubMed] [Google Scholar]
- 30.Steyn-Ross D. A., Steyn-Ross M. L., Sleigh J. W., Wilson M. T., Gillies I. P., Wright J. J.2005The sleep cycle modelled as a cortical phase transition J. Biol. Phys. 31547–569. 10.1007/s10867-005-1285-2 (doi:10.1007/s10867-005-1285-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comte J. C., Schatzman M., Ravassard P., Luppi P. H., Salin P. A.2006A three states sleep–waking model Chaos Solitons Fractals 29808–815. 10.1016/j.chaos.2005.03.054 (doi:10.1016/j.chaos.2005.03.054) [DOI] [Google Scholar]
- 32.Phillips A. J. K., Robinson P. A.2007A quantitative model of sleep–wake dynamics based on the physiology of the brainstem ascending arousal system J. Biol. Rhythms 22167–179. 10.1177/0748730406297512 (doi:10.1177/0748730406297512) [DOI] [PubMed] [Google Scholar]
- 33.Rempe M. J., Best J., Terman D.2010A mathematical model of the sleep/wake cycle J. Math. Biol. 60615–644. 10.1007/s00285-009-0276-5 (doi:10.1007/s00285-009-0276-5) [DOI] [PubMed] [Google Scholar]
- 34.Jirsa V. K., Jantzen K. J., Fuchs A., Kelso J. A. S.2002Spatiotemporal forward solution of the EEG and MEG using network modeling IEEE Trans. Med. Imag. 21493–504. 10.1109/TMI.2002.1009385 (doi:10.1109/TMI.2002.1009385) [DOI] [PubMed] [Google Scholar]
- 35.David O., Friston K. J.2003A neural mass model for MEG/EEG: coupling and neuronal dynamics NeuroImage 201743–1755. 10.1016/j.neuroimage.2003.07.015 (doi:10.1016/j.neuroimage.2003.07.015) [DOI] [PubMed] [Google Scholar]
- 36.Ghosh A., Rho Y. A., McIntosh A. R., Kötter R., Jirsa V. K.2008Noise during rest enables the exploration of the brain's dynamic repertoire PLoS Comput. Biol. 4e1000196. 10.1371/journal.pcbi.1000196 (doi:10.1371/journal.pcbi.1000196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deco G. R., Jirsa V. K., McIntosh A. R.2011Emerging concepts for the dynamical organization of resting-state activity in the brain Nat. Rev. Neurosci. 1243–56. 10.1038/nrn2961 (doi:10.1038/nrn2961) [DOI] [PubMed] [Google Scholar]
- 38.Jirsa V. K., Haken H.1996Field theory of electromagnetic brain activity Phys. Rev. Lett. 77960–963. 10.1103/PhysRevLett.77.960 (doi:10.1103/PhysRevLett.77.960) [DOI] [PubMed] [Google Scholar]
- 39.Bojak I., Liley D. T. J.2010Axonal velocity distributions in neural field equations PLoS Comput. Biol. 6e1000653. 10.1371/journal.pcbi.1000653 (doi:10.1371/journal.pcbi.1000653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sotero R. C., Trujillo-Barreto N. J., Iturria-Medina Y., Carbonell F., Jimenez J. C.2007Realistically coupled neural mass models can generate EEG rhythms Neural Comput. 19478–512. 10.1162/neco.2007.19.2.478 (doi:10.1162/neco.2007.19.2.478) [DOI] [PubMed] [Google Scholar]
- 41.Valdes-Sosa P. A., Sánchez-Bornot J. M., Sotero R. C., Iturria-Medina Y., Alemán-Gómez Y., Bosch-Bayard J., Carbonell F., Ozaki T.2009Model driven EEG/fMRI fusion of brain oscillations Hum. Brain Mapp. 302701–2721. 10.1002/hbm.20704 (doi:10.1002/hbm.20704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bojak I., Oostendorp T. F., Reid A. T., Kötter R.2010Connecting mean field models of neural activity to EEG and fMRI data Brain Topogr. 23139–149. 10.1007/s10548-010-0140-3 (doi:10.1007/s10548-010-0140-3) [DOI] [PubMed] [Google Scholar]
- 43.Kim J. S., Singh V., Lee J. K., Lerch J., Ad-Dab'bagh Y., MacDonald D. J., Lee J. M., Kim S. I., Evans A. C.2005Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification NeuroImage 27210–221. 10.1016/j.neuroimage.2005.03.036 (doi:10.1016/j.neuroimage.2005.03.036) [DOI] [PubMed] [Google Scholar]
- 44.Sled J. G., Zijdenbos A. P., Evans A. C.1998A nonparametric method for automatic correction of intensity nonuniformity in MRI data IEEE Trans. Med. Imag. 1787–97. 10.1109/42.668698 (doi:10.1109/42.668698) [DOI] [PubMed] [Google Scholar]
- 45.Hagmann P., Cammoun L., Gigandet X., Meuli R. A., Honey C. J., Wedeen V. J., Sporns O.2008Mapping the structural core of human cerebral cortex PLoS Biol. 6e159. 10.1371/journal.pbio.0060159 (doi:10.1371/journal.pbio.0060159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephan K. E., Kamper L., Bozkurt A., Burns G. A. P. C., Young M. P., Kötter R.2001Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac) Phil. Trans. R. Soc. B 3561159–1186. 10.1098/rstb.2001.0908 (doi:10.1098/rstb.2001.0908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kötter R., Wanke E.2005Mapping brains without coordinates Phil. Trans. R. Soc. B 360751–766. 10.1098/rstb.2005.1625 (doi:10.1098/rstb.2005.1625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Essen D. C.2005A population-average, landmark- and surface-based (PALS) atlas of human cerebral cortex NeuroImage 28635–662. 10.1016/j.neuroimage.2005.06.058 (doi:10.1016/j.neuroimage.2005.06.058) [DOI] [PubMed] [Google Scholar]
- 49.Liley D. T. J., Cadusch P. J., Dafilis M. P. A spatially continuous mean field theory of electrocortical activity. Network: Comput. Neural Syst. 2002;13:67–113. [PubMed] [Google Scholar]
- 50.Rockland K. S., Ichinohe N.2004Some thoughts on cortical minicolumns Exp. Brain Res. 158265–277. 10.1007/s00221-004-2024-9 (doi:10.1007/s00221-004-2024-9) [DOI] [PubMed] [Google Scholar]
- 51.Bojak I., Liley D. T. J., Cadusch P. J., Cheng K.2004Electrorhythmogenesis and anaesthesia in a physiological mean field theory Neurocomputing 58–601197–1202. 10.1016/j.neucom.2004.01.185 (doi:10.1016/j.neucom.2004.01.185) [DOI] [Google Scholar]
- 52.Robinson P. A., Rennie C. J., Wright J. J.1997Propagation and stability of waves of electrical activity in the cerebral cortex Phys. Rev. E 56826–840. 10.1103/PhysRevE.56.826 (doi:10.1103/PhysRevE.56.826) [DOI] [Google Scholar]
- 53.Catmull E. E., Rom R. A class of local interpolating splines. In: Barnhill R. E., Riesenfeld R. F., editors. Computer aided geometric design. New York, NY: Academic Press; 1974. pp. 317–326. [Google Scholar]
- 54.Kochanek D. H. U., Bartels R. H.1984Interpolating splines with local tension, continuity, and bias control ACM SIGGRAPH Comput. Graph. 1833–41. 10.1145/964965.808575 (doi:10.1145/964965.808575) [DOI] [Google Scholar]
- 55.Liley D. T. J., Wright J. J. Intracortical connectivity of pyramidal and stellate cells: estimates of synaptic densities and coupling symmetry. Network: Comput. Neural Syst. 1994;5:175–189. [Google Scholar]
- 56.Hellwig B.2000A quantitative analysis of the local connectivity between pyramidal neurons in layers 2/3 of the rat visual cortex Biol. Cybern. 82111–121. 10.1007/PL00007964 (doi:10.1007/PL00007964) [DOI] [PubMed] [Google Scholar]
- 57.Kaiser M., Hilgetag C. C., van Ooyen A.2009A simple rule for axon outgrowth and synaptic competition generates realistic connection lengths and filling fractions Cereb. Cortex 193001–3010. 10.1093/cercor/bhp071 (doi:10.1093/cercor/bhp071) [DOI] [PubMed] [Google Scholar]
- 58.Wu J.-Y., Huang X. Y., Zhang C.2008Propagating waves of activity in the neocortex: what they are, what they do Neuroscientist 14487–502. 10.1177/1073858408317066 (doi:10.1177/1073858408317066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bullock T. H., McClune M. C., Achimowicz J. Z., Iragui-Madoz V. J., Duckrow R. B., Spencer S. S.1995EEG coherence has structure in the millimeter domain: subdural and hippocampal recordings from epileptic patients Electroencephalogr. Clin. Neurophysiol. 95161–177. 10.1016/0013-4694(95)93347-A (doi:10.1016/0013-4694(95)93347-A) [DOI] [PubMed] [Google Scholar]
- 60.van Dam P. M., van Oosterom A.2003Atrial excitation assuming uniform propagation J. Cardiovasc. Electrophysiol. 14S166–S171. 10.1046/j.1540.8167.90307.x (doi:10.1046/j.1540.8167.90307.x) [DOI] [PubMed] [Google Scholar]
- 61.Nunez P. L. Electric fields of the brain: the neurophysics of EEG. 1st edn. New York, NY: Oxford University Press; 1981. [Google Scholar]
- 62.Oostendorp T. F., Delbeke J., Stegeman D. F.2000The conductivity of the human skull: results of in vivo and in vitro measurements IEEE Trans. Biomed. Eng. 471487–1493. 10.1109/TBME.2000.880100 (doi:10.1109/TBME.2000.880100) [DOI] [PubMed] [Google Scholar]
- 63.Petzold G. C., Albeanu D. F., Sato T. F., Murthy V. N.2008Coupling of neural activity to blood flow in olfactory glomeruli is mediated by astrocytic pathways Neuron 58897–910. 10.1016/j.neuron.2008.04.029 (doi:10.1016/j.neuron.2008.04.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giaume C., Koulakoff A., Roux L., Holcman D., Rouach N.2010Astroglial networks: a step further in neuroglial and gliovascular interactions Nat. Rev. Neurosci. 1187–99. 10.1038/nrn2757 (doi:10.1038/nrn2757) [DOI] [PubMed] [Google Scholar]
- 65.Friston K. J., Harrison L. M., Penny W. D.2003Dynamic causal modelling NeuroImage 191273–1302. 10.1016/S1053-8119(03)00202-7 (doi:10.1016/S1053-8119(03)00202-7) [DOI] [PubMed] [Google Scholar]
- 66.Robinson P. A., Phillips A. J. K., Fulcher B. D., Puckeridge M., Roberts J. A.2011Quantitative modelling of sleep dynamics Phil. Trans. R. Soc. A 3693840–3854. 10.1098/rsta.2011.0120 (doi:10.1098/rsta.2011.0120) [DOI] [PubMed] [Google Scholar]