Abstract
Every research laboratory has a wealth of biomedical data locked up, which, if shared with other experts, could dramatically improve biomedical and healthcare research. With the PhysiomeSpace service, it is now possible with a few clicks to share with selected users biomedical data in an easy, controlled and safe way. The digital library service is managed using a client–server approach. The client application is used to import, fuse and enrich the data information according to the PhysiomeSpace resource ontology and upload/download the data to the library. The server services are hosted on the Biomed Town community portal, where through a web interface, the user can complete the metadata curation and share and/or publish the data resources. A search service capitalizes on the domain ontology and on the enrichment of metadata for each resource, providing a powerful discovery environment. Once the users have found the data resources they are interested in, they can add them to their basket, following a metaphor popular in e-commerce web sites. When all the necessary resources have been selected, the user can download the basket contents into the client application. The digital library service is now in beta and open to the biomedical research community.
Keywords: physiome, virtual physiological human, living human project, digital library, data sharing
1. Introduction
Data sharing is essential for the rapid translation of research results into knowledge, protocols, products and services to improve human health. Every hospital and research laboratory in the world has a wealth of biomedical data locked up somewhere, which, if shared with other experts, could dramatically improve the research practice itself, as well as the development of better biomedical products.
The data-repository problem has been governed for many years mainly by the genomics data sharing needs, but it is now increasing in all the medical and biomedical fields. In fact, data sharing is attracting the attention of the research community as is also demonstrated by the recent special of Nature (vol. 461, issue no. 7261, September 2009). This growth of interest on data sharing is also evident from the increasing number of papers with ‘data sharing’ as the keyword (resulting in about 400 references on PubMed) in different biomedical fields (Van Horn & Ball 2008; Fennema-Notestine 2009; Koike et al. 2009; Yuan et al. 2009), and from the support to data sharing given by funding agencies such as the National Institute of Health (http://grants.nih.gov/grants/guide/notice-files/NOT-OD-03-032.html). However, from the summary of the recent Toronto meeting (Toronto International Data Release Workshop Authors 2009) discussions, it is evident that, even if there is a desperate need for data sharing, this rarely happens mainly owing to the lack of appropriate tools to control the quality and access to data (Nelson 2009). To our knowledge, no tools are currently available that allow easy sharing and repository of public biomedical data of whatever type and format.
The living human project (LHP) is a grass-roots initiative aimed at developing an in silico model of the human neuro-musculoskeletal system able to predict how mechanical forces are exchanged internally and externally, from the whole body down to the protein level, consistently with the scope of the European virtual physiological human initiative. To pursue this very ambitious objective, it was necessary for large research communities to share highly heterogeneous collections of data and models through a repository fully integrated and directly accessible by any researcher in the world. Although inspired by the neuro-musculoskeletal research community, this problem is very general in nature, and its solution may significantly and positively affect research and its translation into clinical and industrial practices.
The living human digital library (http://www.livinghuman.org, FP6-2004-ICT-026932) project developed and deployed the resource-sharing infrastructure required by the LHP community and by many other similar groups involved with biomedical research and practice. The data-sharing service has been called PhysiomeSpace and it is described in the next sections with the following structure:
— the PhysiomeSpace architecture and general features,
— details on the client application,
— details on the server web interface, and
— ontology and metadata management.
At the end of the paper, some information on protection and privacy issues is also provided.
2. PhysiomeSpace service
PhysiomeSpace (http://www.physiomespace.com) is a data management and sharing service dedicated to biomedical data. In particular, it is designed to help biomedical researchers to share their data, but also to search for data owned by other users and download them after authorization.
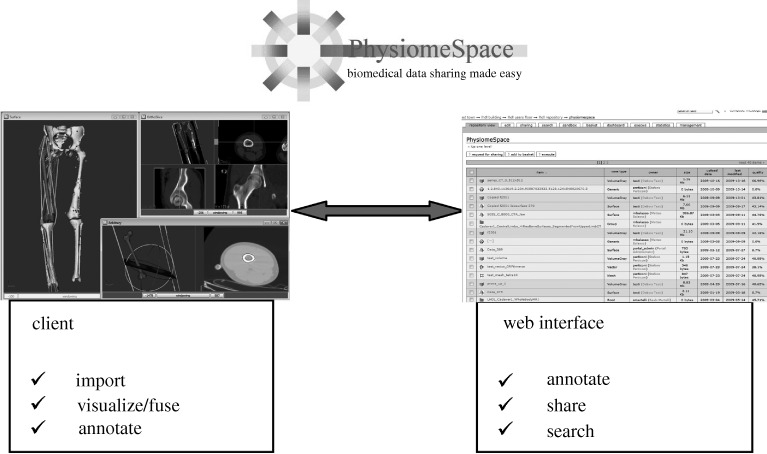
The service is composed of a client application (called PSLoader) that allows users to import almost any type of biomedical data, visualize and annotate the data, upload data to the repository and download the data already selected with the web interface. Then, the user can manage the uploaded data by accessing the repository via a web interface that allows the user to complete the data annotation, share the data with other PhysiomeSpace users, search for new data and select data for downloading into the local computer (figure 1).
Figure 1.
PhysiomeSpace user’s experience.
In particular, once the user has created the data collection within the PSLoader, the entire collection can be uploaded to the PhysiomeSpace servers with a single click, where it is stored in the user’s private space. The private space, called the sandbox, can then be accessed from the simple web interface, with which the user can add, remove, annotate data resources and assign to each resource a different set of access permissions. By default, all uploaded data are open only to the data owner, but at any time, the owner can choose with whom to share each dataset. After sharing, the data are moved from the personal sandbox to the repository where, even if the binary information are accessible only to those who have the permissions, the associated metadata are visible and searchable by all users. Thus, other PhysiomeSpace users can search every dataset. If the data is uploaded in the repository, but its owner has not made it available for sharing, the user can send a message requesting access. This gives the data owner the possibility to talk directly to anyone wishing to download the data before granting access.
Once a dataset is shared, the user can place it in the basket, ready to be downloaded from PSLoader. The dataset can then be exported in a long list of formats, and used with other specialized applications.
PhysiomeSpace data resources can be searched and browsed in various ways, relying on the fact that each data resource is annotated by a set of metadata defined according to a specialized ontology (details can be found in a later section). In addition, depending on the type of data, the user can choose additional sub-ontologies providing special concepts that are specific to a certain data generation modality, or that are related to a chosen way to describe what the dataset ‘represents’. A typical example is the anatomical description: anatomical sub-ontologies, such as the functional anatomy ontology (http://www.biomedtown.org/biomed_town/LHDL/Reception/ontologies/FAontology/) developed by the Université Libre de Bruxelles, can be used to describe in functional anatomy terms what the dataset represents. While the PSLoader automatically annotates a good part of the metadata of the main ontology, the user is still expected to do some manual curation. Each data resource stored on PhysiomeSpace has a quality index that scores the resources in terms of how extensive the annotation is. The idea is that better annotated datasets have higher scores and are likely to get more downloads.
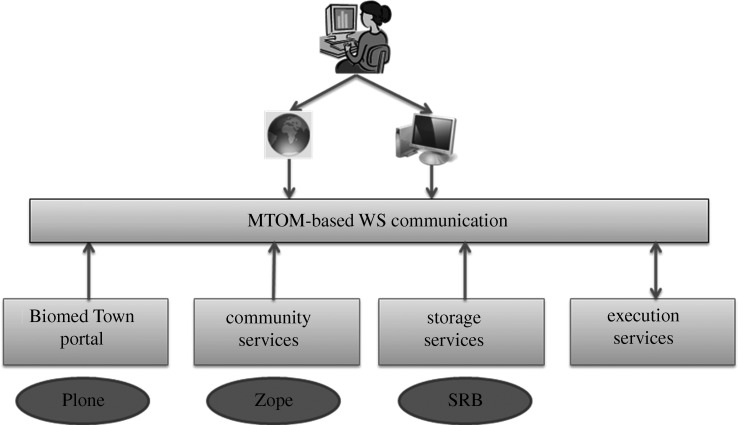
The PhysiomeSpace modules communicate with each other via web services (WSs) with the infrastructure schematized in figure 2.
Figure 2.
PhysiomeSpace architecture. MTOM is message transmission optimization mechanism, SRB is storage resource broker.
(a). The client application: PSLOADER
PSLoader is a desktop application that is distributed for free to all PhysiomeSpace users. The software is developed based on the Multimod Application Framework (MAF; http://www.openmaf.org), an open-source framework for the rapid development of computer-aided medical applications (Viceconti et al. 2007). The C++ code is then extended with Python (http://www.python.org/) scripts to allow communication with the infrastructure WSs.
As a first step, the PSLoader allows the user to authenticate using a basic credentials-based mechanism (username and password) relying on a secure connection (hypertext transfer protocol over secure socket layer; HTTPS). Then, the application allows users to import biomedical data stored in a long list of digital formats (such as DICOM, STL, JPG, TIFF, ASCII, ANSYS, VTK and many more), and organize them in space using a hierarchical tree where the pose of one dataset is defined with respect to the parent one. The program also provides a long list of interactive views, designed to visualize whatever combination of data the user may have (i.e. orthoslice, arbitrary slice, isosurface). Then, a specialized operation is implemented to provide a metadata editor for the data curation. The editor interface allows the user to annotate the data according to any of the available ontologies. A good part of the metadata defined in the PhysiomeSpace resource ontology, which defines the core set of metadata, are generated automatically or extracted from the imported file (i.e. re-using the metadata already present in the DICOM files) by client application and are available for checking, while additional metadata are available for manual annotation, as described later in this paper. Once the operation is completed, the metadata information are associated with the binary object and uploaded into the repository.
In any moment, the user can then select one or more data and upload them into the repository sandbox. The upload operation preserves the information on the data hierarchy and linking so that the hierarchical tree can be regenerated in the download phase.
(b). The server: web interface
The web interface to access the PhysiomeSpace repository and services is hosted on Biomed Town (http://www.biomedtown.org), which is a Plone-based portal (http://www.plone.org) open to anyone with a professional interest in biomedical research.
As schematized in figure 2, the data-resources information are stored in extensible markup language (XML) in a ZopeDB (http://en.wikipedia.org/wiki/Zope_Object_Database) object database. The communication protocol used is XML-RPC (http://www.xmlrpc.com/), and SWIG (http://www.swig.org/) is used for wrapping gSOAP (http://www.cs.fsu.edu/~engelen/soap.html) clients for communication with storage WSs. The storage WSs use gSOAP as the communication protocol with a message transmission optimization mechanism (MTOM; http://www.w3.org/TR/soap12-mtom/) extension for large binary transmission, which allows an efficient transfer of large data objects by avoiding base 64 encoding, thus not increasing the dimension of the SOAP attachment. In order to allow the data to be distributed in different physical locations, the storage services rely on the storage resource broker middleware (http://en.wikipedia.org/wiki/Storage_Resource_Broker).
Within the web interface, the user can manage the loaded data by completing the metadata curation with an editor interface almost identical to that available on the client side. The curation can be completed and revised until the owner decides to share the data with other PhysiomeSpace users. Even after sharing the data, the owner can, at any moment, revise the assigned permissions by adding or removing any of the users. This, together with statistics on the download of each data, allows the user to have complete information on when and who accesses the owned data.
Once shared, the data can be downloaded by whoever has permission to, and searched by any other PhysiomeSpace user. Specific search mechanisms have been implemented and exposed to allow the users to look for the data in the most efficient way according to their specific needs. In particular, in addition to a simple search mechanism, an advanced search tool has been made available that allows the user to perform the search using the metadata fields available in the ontology and used to curate the data.
When a dataset of interest is found, the user can, with a single click, acquire permission from the data owner to download it and, once granted for access, download the file locally using the PSLoader.
(c). Ontology and metadata annotation
As already mentioned, the data resources shared in PhysiomeSpace are associated to a group of metadata: a coherent set of concepts that provides the information framework for searching and retrieving resources and the creation of complex workflows (i.e. traceability).
The metadata are described in an extensible ontology composed of a master ontology and a series of sub-ontologies (http://www.biomedtown.org/biomed_town/LHDL/Reception/ontologies/). The master ontology (Physiome-Space resource ontology) is used to describe the resource types: data resources, datasets stored in the digital library; service resources that are user accessible WSs that can be used to process data resources directly in the repository, without downloading and uploading frequently; MAF resources that have a meaning only within MAF applications. There are also slots for documentation that provides extra documentation on the resource and access that defines the rules under which the users can access the resource. For each type of resource, specific metadata have been identified. For example, for the data-resource type, the following groups of metadata have been selected:
— data type: nature of the stored dataset (dimensionality, topology and time variance),
— size: dimension of the dataset (byte, entity count, etc.),
— dataset: resource uniform resource identifier (URI), checksum, upload date, etc.,
— ownership: details of the resource owner,
— traceability: details (who, when, what) on the event that created the resource and of all subsequent events that modified it,
— quality: multiple quality scores, obtained by the owner submitting the resource to different quality assurance (QA) services,
— source: slot for sub-ontologies that contains concepts specific to the source of the data (i.e. DICOM source includes all concepts derived from the medical-imaging sources and the relevant DICOM metadata), and
— representation: slot for sub-ontologies that describes what the datasets represent.
Sub-ontologies describing domain-specific concepts can be added to the master ontology. At present four sub-ontologies have been included (functional anatomy, microCT, DICOM and motion analysis), but more can be added depending on user needs.
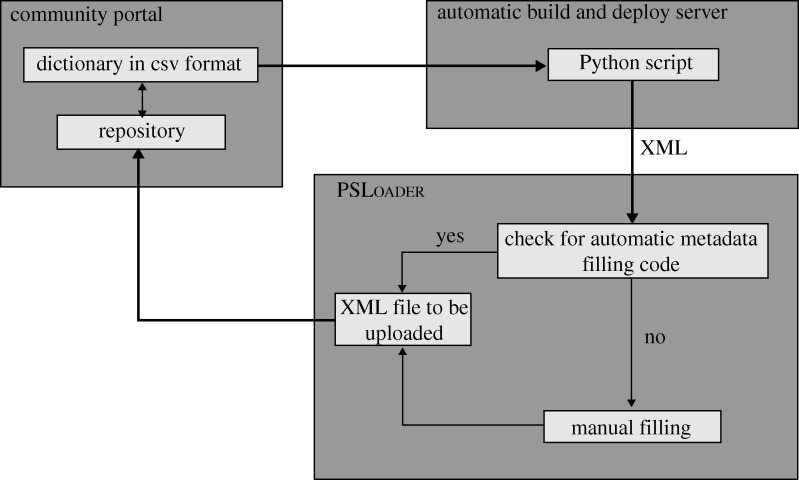
The ontologies are maintained by the domain experts and stored in a protected area with a revision control system active; then, the build and deploy server launches a Python script that automatically converts the script into an XML format for internal use by the application (figure 3).
Figure 3.
Ontology synchronization and metadata curation. CSV is comma-separated values.
To ascertain that, when uploaded, the data resource has the most updated version of the metadata if changes to the ontology are made, the PSLoader connects to the server when launched and checks if the ontology has changed. If the versions are different, it invites the user to download a new version of the client application.
The ontology also identifies some tags as ‘automatic’: these have to be filled automatically into the XML by the application. For every tag that comes from the downloaded ontology, a MAF class checks for the existence of the corresponding tag class for the calculation of its value. If the class exists, the value is calculated and written into the XML file, if the class does not exist, the tag is left for manual annotation, which can be done by the user both in the client application and into the web interface, as described earlier.
3. Data re-use, protection and privacy policy
During the beta testing phase, the service is open as a totally free service, with up to 1 Gb of storage and sharing space; extra space might be provided on a case-by-case basis when it is to be used for sharing resources of particular importance for the VPH/Physiome communities. The data are uploaded under complete responsibility of the users, and no guarantee is provided whatsoever for the continuity of the service, for the storage, the integrity and the preservation of the data stored; PhysiomeSpace is not an online backup service. The confidentiality of the data is protected only through the access limitations of the service, and in principle, the system administrators (SCS staff managing and developing the service) are in a position to access all data uploaded to the service.
In the future, through an advanced security mechanism based on double encryption keys (entirely automated in the PSLoader and thus transparent to the final user) that is currently under development, the data will leave the user computer only in an encrypted form, and only the data owner and those users to whom they grant access will have the key to decrypt the data; everyone else will be unable to see anything but the metadata, including the system managers. As well as adding inherent security to the service, this will make it legally possible to define a service licence agreement for PhysiomeSpace that transfers the entire responsibility of the upload and sharing of the data to the data owner. The service managers will not be allowed to see what the datasets are, and thus cannot be deemed responsible for eventual breaches of confidentiality, misuse and abuses associated with the uploading, sharing and selling of data resources.
4. Discussions
The PhysiomeSpace repository service described in this paper is a powerful and simple tool that allows researchers to share their data in a controlled and safe way. The complete control on who can access the shared data should allow the biomedical researcher to be more confident in sharing of their data and to overcome the suspicion so far encountered by sharing services.
The PhysiomeSpace service has been implemented based on state-of-the-art technologies and relies on an architecture that makes its expansion easy with the integration of other libraries or services such as the cryptography library (i.e. Cripto++) or commercial data store services (i.e. Amazon S3).
In principle, we could allow the resource owners to expose their data with multiple licence agreements, i.e. not-for-profit and for-profit, and put a price tag on access of the data resource only when a for-profit licence is requested. The mechanism will also make it possible to sell the resources for very small prices (e.g. less than €10). This could create a single marketplace for both research and industry, where data are shared with different policies, under different user licences and with difference prices.
Another dimension that can be expanded is that of the QA services. Currently, each data resource is automatically scanned at the upload stage, and a quality score is generated according to the completeness of the metadata curation. But of course this is only one of the infinite QA criteria we could elaborate as a community. In general, we can imagine a plethora of QA services, managed by single researchers, institutions and governmental agencies or scientific societies. The simpler criteria could be designed to be fully automated, while the more complex would involve a workflow with a person in the middle, including peer review. At upload, each resource owner can choose to have their resource assessed by one or more of these services, and receive a score for the resource according to the criteria of that particular QA service.
While currently PhysiomeSpace can only be accessed via the PSLoader, the service supports a complete service application programming interface (API) exposed via standard WSs, and thus it would be quite simple to grant access to the service and also to other data-management or data-processing clients or event to include the PhysiomeSpace storage into automatic execution workflows, where multi-scale coupled models are executed on different computers and the intermediate data are stored in PhysiomeSpace.
However, it is clear that any future development is conditioned by the success this first beta service will have. In this sense, we invite the entire Physiome research community to start using the service as soon as possible and to report to us any request for improvement that might emerge.
Acknowledgements
This work has been partially funded by the ICT for Health Unit of the European Commission in the living human digital library project (FP6-2004-IST-4-026932) and in the VPHOP project (FP7-2008-ICT-224635).
Footnotes
One contribution of 13 to a Theme Issue ‘The virtual physiological human: computer simulation for integrative biomedicine II’.
References
- Fennema-Notestine C. Enabling public data sharing: encouraging scientific discovery and education. Methods Mol. Biol. 2009;569:25–32. doi: 10.1007/978-1-59745-524-4_2. ( ) [DOI] [PubMed] [Google Scholar]
- Koike A., Nishida N., Inoue I., Tsuji S., Tokunaga K. Genome-wide association database developed in the Japanese Integrated Database Project. J. Hum. Genet. 2009;54:543–546. doi: 10.1038/jhg.2009.68. ( ) [DOI] [PubMed] [Google Scholar]
- Nelson B. Data sharing: empty archives. Nature. 2009;461:160–163. doi: 10.1038/461160a. ( ) [DOI] [PubMed] [Google Scholar]
- Toronto International Data Release Workshop Authors. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. ( ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn J. D., Ball C. A. Domain-specific data sharing in neuroscience: what do we have to learn from each other? Neuroinformatics. 2008;6:117–121. doi: 10.1007/s12021-008-9019-9. ( ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viceconti M., Zannoni C., Testi D., Petrone M., Perticoni S., Quadrani P., Taddei F., Imboden S., Clapworthy G. The multimod application framework: a rapid application development tool for computer aided medicine. Comput. Methods Programs Biomed. 2007;85:138–151. doi: 10.1016/j.cmpb.2006.09.010. ( ) [DOI] [PubMed] [Google Scholar]
- Yuan S., Wei D., Xu W., Shen W.2009Web-based sharing of electrocardiogram: a framework for information publishing Conf. Proc. IEEE Eng. Med. Biol. Soc. 11671–1674 [DOI] [PubMed] [Google Scholar]