Abstract
Autism spectrum disorders (ASD) represent a group of developmental disabilities with a strong genetic basis. The laboratory mouse is increasingly used as a model organism for ASD, and MGI, the Mouse Genome Informatics resource, is the primary model organism database for the laboratory mouse. MGI uses the Mammalian Phenotype (MP) ontology to describe mouse models of human diseases. Using bioinformatics tools including Phenologs, MouseNET, and the Ontological Discovery Environment, we tested data associated with MP terms to characterize new gene-phenotype associations related to ASD. Our integrative analysis using these tools identified numerous mouse genotypes that are likely to have previously uncharacterized autistic-like phenotypes. The genes implicated in these mouse models had considerable overlap with a set of over 300 genes recently associated with ASD due to small, rare copy number variation (Pinto D. et al, 2010). Prediction and characterization of autistic mutant mouse alleles assists researchers in studying the complex nature of ASD and provides a generalizable approach to candidate gene prioritization.
Keywords: Autism spectrum disorders, phenotype ontology, mouse disease models
1. Introduction
Autism spectrum disorders (ASD) are the fastest growing group of serious developmental disabilities in the United States, affecting one in every 110 children [1]. ASD is used to describe multiple disorders that are primarily characterized by deficits in socialization and communication, as well as restricted, stereotyped and repetitive patterns of behavior [2]. ASD, broadly defined, has a substantial genetic basis with a 92% concordance rate among monozygotic twins compared to 10% in dizygotic pairs [3]. Some cases are associated to genetic disorders such as Fragile X, to rare mutations in synaptic genes, or to risk loci identified from genome-wide association studies [4]. However, the genetic determinates and molecular mechanism of ASD remain largely unknown.
Until recently, the prevailing hypothesis for the cause of complex genetic diseases was that a combination of alleles that are relatively common in a population collectively act to manifest a disease phenotype; any one allele on its own having little phenotypic contribution [5]. By this notion, the common alleles contributing to disease should be identified in patient populations by performing genome wide association studies (GWAS). ASD has been subject to a number of GWAS, but to date only a few common alleles have been implicated and the majority of these have not been confirmed in follow up studies [6]. However, there is a growing appreciation that many complex human disorders are not the result of a combination of common alleles but, rather, they result from one or few rare variant alleles that have a large phenotypic contribution [7]. An important tenant of this model is that a large number of gene variants must reside within a population to explain the prevalence of disease. Support for this model in ASD comes from a recent study that implicates over 300 genes in autism by small (1 or 2 gene) copy number variants (CNV) found with a higher burden in 996 ASD patients [8].
With such a large number of genes being implicated in ASD, a bioinformatics and computational approach can usefully be employed to assess how these alleles can manifest in a common phenotype. In the last ten years, such scientists have taken advantage of the Gene Ontology (GO) to evaluate the characteristics of sets of genes [9]. The GO, like other formal ontologies, consists of a structured hierarchical controlled vocabulary whose use serves to standardize the representation of gene and gene product attributes. GO term enrichment experiments use the transitive closure power of the GO hierarchical structure to identifying biological processes that sets of genes are statistically more frequently associated with[10]. The authors of the CNV study, for example, performed such a “GO term enrichment” test that revealed biological processes which were not previously implicated in ASD including cellular proliferation and GTPase/Ras signaling [8]. While pointing out potential new avenues of investigation, there are limitations to this type of GO analysis since GO annotations describe gene function in normal biological processes and specifically do not describe pathological processes. To understanding why certain alleles associate with ASD, however, requires that the analysis include information about the associated disease phenotypes. For example, a common experimental approach is to subject a mouse strain carrying a targeted mutation of an ASD candidate gene to behavioral and neurological studies [11]. Such studies provide valuable insights into the human disorder, but as more genes are implicated in ASD, and more mouse models become available, researchers can be overwhelmed in just reviewing the literature.
Biocuration efforts, coupled to ontology development, make this literature more computable. The Mouse Genome Informatics (MGI) bioinformatics resource captures data about experiments conducted using the mouse as a model of human biology including those related to autism. MGI integrates genetic, genomic and phenotypic data for the laboratory mouse[12], and curates data using several different ontologies including the Mammalian Phenotype (MP) Ontology [13]. As of January 8, 2011, the MP contains over 7,800 terms that are used to describe the phenotypes of over 38,000 mouse genotypes. Mouse genotypes are also associated within MGI to the records in the Online Mendelian Inheritance in Man (OMIM) resource when a mouse is explicitly used as a model for a human disease.
The MP has been incorporated into several recently developed bioinformatics tools. 1)The Phenologs database, released in 2010 [14] identifies MP terms that are overrepresented among mouse genes that are orthologous to human genes associated with disease in OMIM. 2)The MouseNET system [15] supports prediction of functional assignments based on integration of genetic and genomic data. 3)The Ontological Discovery Environment (ODE) [16] integrates gene sets associated with phenotype terms using a variety of analysis tools such as Phenome Map analysis where a hierarchy of phenotypes is proposed based on their associated genes. And 4) ODE’s newest analytic module, Anchored Bicliques of Biomolecular Associates (ABBA) [16], finds genes that are functionally similar to an input set based on diverse evidence sources including empirical studies and curated annotations of mouse genes to GO and MP terms. We employed all of these tools in the analysis reported here.
Starting with genes clustered by MP terms, and utilizing informatics tools described above, we developed a method for discovering novel mouse models of autism. Using this new approach, we identified annotation gaps and doubled the number of mouse genotypes associated with autism within MGI. We defined a list of ASD candidate genes by finding overlap between human genes implicated in the CNV study and mouse orthologs that contribute to an autistic-like phenotype. This analysis has led to the prediction of new plausible mouse models of ASD for future experimental investigation.
2. Methods
2.1 Phenologs Database
The Phenologs database provides a mechanism to identify non-obvious equivalencies between mutant phenotypes in different species. The Phenologs database integrates data for a variety of human diseases represented in OMIM by compressing multiple variants of a disease together under one term. In the case of the Phenolog term “Autism”, multiple variants of ASDs were compressed to the singular term. By searching for “Autism” within the Phenologs database, a table displaying ASD-implicated human genes and their mouse orthologs is generated. MP terms that were significantly overrepresented among these mouse orthologs were ranked using a hypergeometric probability as described [14]. MP terms that had 2 or more orthologs among the human autism gene set, and had a p-value higher than p > 0. 2.05e-04, were selected for further analysis.
2.2 Phenome Map in the Ontological Discovery Environment (ODE)
The Ontological Discovery Environment (ODE) provides analysis of functional genomics data sets across species and experimental systems (http://ontologicaldiscovery.org/) [16]. The ODE database contains gene sets including those that consist of genes with alleles annotated to a MP term in the MGI database. These gene sets have the MP term as their identifier. Within ODE, a user is able to input gene sets that are centered on a specific phenotype or experimental result, retrieve publicly available sets, and analyze multiple gene sets using a variety of analysis tools. One analysis tool, Phenome Map [17], uses multi-way intersections of gene sets to create a hierarchy of gene-phenotype associations using a novel algorithm for biclique enumeration. We used Phenome Map to identify genes that are not associated with the OMIM term ‘Autism’ within MGI, but that appear to be good candidate genes based on functional similarity with autism-implicated genes that are in MGI. Mouse alleles found after review of the biomedical literature that were not yet associated with autism within MGI were submitted to the MGI curation pipeline.
2.3 MouseNET
MouseNET uses computational integration of diverse genetic, genomic, and phenotype data and Bayesian inference to predict functional relationships among mouse genes [15]. From a user-defined list of genes of interest, MouseNET generates a probabilistic network of functional relationships. The edges of the network represent statistical weighting of supporting evidence that includes protein-protein interactions, gene-phenotype association, and expression co-localization. The biological context of the network is provided by testing for enrichment of GO annotations associated with the individual genes in the predicted network. Associations among the mouse genes and disease terms for their orthologous counterparts are also provided by the resource. MouseNET can be used to identify potential novel disease gene candidates. MouseNET was used to predict new candidates genes related to Autism based on current data in the MGI system.
2.4 Anchored Bicliques of Biomolecular Associates (ABBA)
ODE’s Anchored Bicliques of Biomolecular Associates (ABBA) tool [16] is another analysis module that incorporates data and information from ODE, GO, and the MP to find genes that have similar characteristics based on discrete functional associations. When a list of genes is input into ABBA, the tool generates a list of all of the gene sets from within ODE’s database that contain a user determined number of the input genes. The tool then returns ranked list of other similar genes that are enriched among the same gene sets as the input genes. The user may designate a connectivity threshold for the number of gene sets that contain the predicted genes. This tool was used to identify genes that may be functionally similar to genes associated with autism within MGI’s database. For this study, the inputs used were: the eight genes associated with autism in MGI as of 8/2010, ODE genes sets that contain at least 2 genes from input list, genes for output list must be on at least 9 ODE gene sets. This list of genes was reviewed in the current scientific literature for any relevance to autism and any evidence was reported to MGI curators as necessary.
2.5 Generation of candidate ASD genes based on orthology to human ASD implicated genes and/or association with a neurological phenotype
A recent study led by the Autism Genome Project Consortium revealed rare copy number variation (CNV) as a determining risk factor for autism [8]. In the study, 996 autistic patients and their parents were genotyped to determine non-redundant CNVs in any individual experiencing the disorder. A list of 231 CNVs that affect one gene were identified along with 195 CNVs that affect two genes. Using MGI mouse/human orthology data, the lists of CNVs were compared with the gene lists from the ABBA and MouseNET analyses to determine overlap (Table 1). Probability of candidate gene overlap with ASD-associated CNVs due to random chance was determined by a Fisher’s exact test method described in Fury et al [18]. Genes identified by the ABBA and MouseNET tools were also manually screened for association with the MP terms “abnormal behavior”, “abnormal nervous system morphology” and “abnormal nervous system physiology” and are included on Table 1.
Table 1. Candidate ASD Genes based on Mouse Phenomics.
Genes from ABBA or MouseNET analysis that have orthologous with human genes implicated in ASD by CNV analysis and/or genes associated with an abnormal neurological phenotype. The first three columns represent ASD candidate genes identified by different stages of our analysis and the fourth column represents genes associated with an abnormal neurological phenotype in mice. The last column represents human orthologs implicated in ASD by CNV analysis as described in Pinto et al [8]
| Gene symbol |
MGI ID | Phenome Map |
MouseNET | ABBA | Neurological phenotype (mice) |
CNV implicated ASD |
|---|---|---|---|---|---|---|
| Anks1b | MGI:1924781 | x | x | |||
| Camk4 | MGI:88258 | x | x | x | ||
| Cask | MGI:1309489 | x | x | x | ||
| Chrna3 | MGI:87887 | x | x | x | ||
| Chrnb4 | MGI:87892 | x | x | x | ||
| Dsc3 | MGI:1194993 | x | x | |||
| Fgfr3 | MGI:95524 | x | x | x | ||
| Ghr | MGI:95708 | x | x | x | ||
| Plcb4 | MGI:107464 | x | x | x | x | |
| Thrb | MGI:98743 | x | x | x | ||
| Unc5c | MGI:1095412 | x | x | x | x | x |
| Amph | MGI:103574 | x | x | |||
| Apc | MGI:88039 | x | x | |||
| Cacna1a | MGI:109482 | x | x | x | x | |
| Cadps | MGI:1350922 | x | x | |||
| En1 | MGI:95389 | x | x | x | ||
| Foxp2 | MGI:2148705 | x | x | x | ||
| Gria2 | MGI:95809 | x | x | x | x | |
| Grin1 | MGI:95819 | x | x | x | ||
| Grin3b | MGI:2150393 | x | x | x | ||
| L1cam | MGI:96721 | x | x | x | ||
| Met | MGI:96969 | x | x | x | ||
| Mll1 | MGI:96995 | x | x | |||
| Pax3 | MGI:97487 | x | x | x | ||
| Pkd1 | MGI:97603 | x | x | x | ||
| Rora | MGI:104661 | x | x | x | ||
| Slc6a4 | MGI:96285 | x | x |
2.6 Statistical analysis
Precision (Pi), recall (Rho), and F-measure (f) were determined as described [19]. Briefly, precision measures the fraction of returned genes that are relevant to the search and recall measures the fraction of relevant genes that are returned by the search. F-measure represents the harmonic mean between precision and recall. The true positive gene set used was from a list of ASD-implicated genes chosen by a consortium of ASD researchers [8]. Probability of candidate gene overlap with ASD-associated CNVs due to chance was determined by a Fisher’s exact test method described in Fury et. al. [18]. Precision, recall and the F measure were calculated for each value of the rank threshold applied to ABBA results, and were compared to the distribution of measures estimated from 1000 draws of randomly chosen genes.
2.7 Integration of Bioinformatics tools
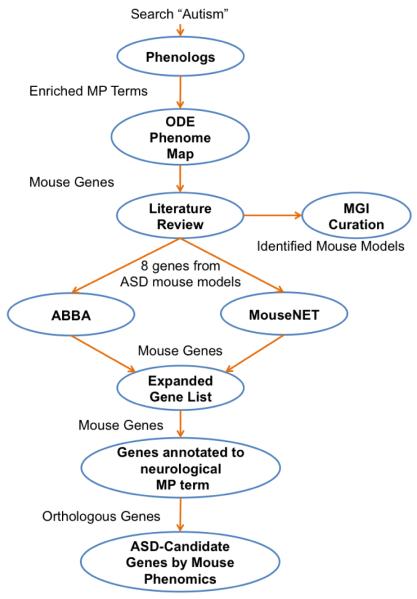
A workflow was established among the bioinformatics tools that utilize MP annotations as depicted in Figure 1. In summary, a manual input of “Autism” generated a list of associated MP terms from the Phenologs database. A “project” was then manually created within the ODE environment by selecting the relevant MP terms from the ODE database. Mouse genes were identified from a Phenome Map analysis and subjected to a thorough literature review. Genes with alleles from 8 definitive mouse ASD models were then separately used as inputs into the ABBA and MouseNET tools. Results were then integrated by comparing candidate genes to orthologous human genes implicated by the CNV analysis and/or genes associated with an abnormal mouse neurological phenotype.
Figure 1. Phenome analysis workflow.
The analysis starts with a keyword search in the Phenolog database and then proceeds by using the output from the previous tool as an input to the next. ODE= Ontological Discovery Environment, MGI= Mouse Genome Informatics, ABBA= Anchored Bicliques of Biomolecular Associates analysis.
3. Results
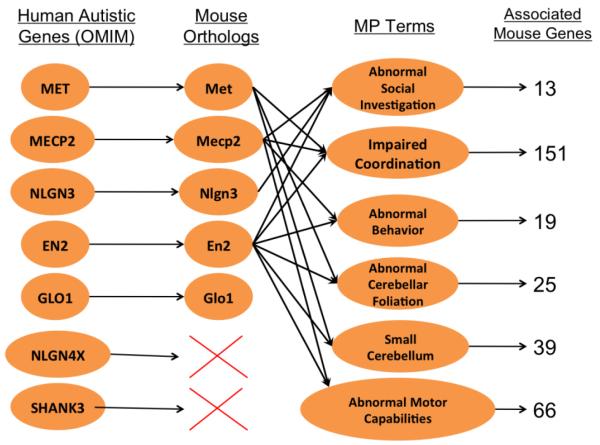
Using the Phenologs tool to find MP terms that are overrepresented with the five mouse orthologs of human genes implicated in ASD, six such MP terms were identified (Figure 2). The six MP terms are “abnormal social investigation”, “impaired coordination”, “abnormal behavior”, “abnormal cerebellar foliation”, “small cerebellum”, and “abnormal motor/capabilities/coordination movement.” Many of these same MP terms were found among MGI’s annotations of mouse models of autism confirming the validity of the Phenolog findings.
Figure 2. Phenologs of ASD-implicated genes.
A depiction of the process using the Phenologs tool to determine the MP terms most associated with disease-related genes.
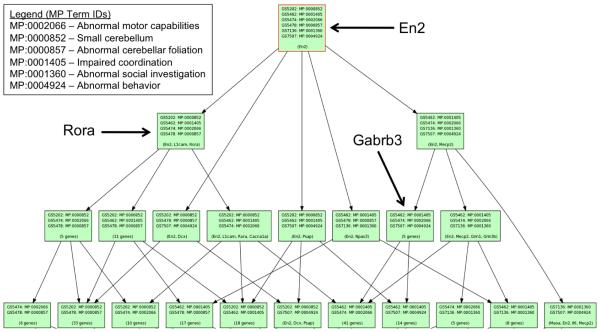
We then integrated these results with tools available in the Ontological Discovery Environment by taking the six MP terms from the Phenologs analysis and performing a Phenome Map analysis. This function creates a hierarchical tree that shows groupings of all genes annotated to these terms based on their annotations to different combinations of the input MP terms (Figure 3). Genes associated with two or more MP terms were examined for association with ASD in both the MGI database and in the scientific literature. For example, the only gene with annotations to all six MP terms, En2, was already annotated to autism in MGI. However, a mouse genotype containing Gabrb3 was not yet annotated to autism in MGI although we identified research clearly using it as a model (Figure 3) [20]. This process led to the addition of three mutant alleles that have experimental evidence for a linkage with autism within MGI. These are Gabrb3, Ehmt1, and Nrcam. One additional gene, Pten, was also identified as a strong candidate but lacked an explicit statement from researchers saying this was an appropriate ASD model.
Figure 3. Phenome Map.
Mapping of the six MP term-created gene sets generated by within ODE. The top of the map shows the one gene that has MP annotations to all six MP. Below this root node, different derivatives of MP terms and their associated genes are shown. Red box indicates a root node. See text for discussion of the three examples.
Of particular interest were genes from this analysis that lack mouse literature linking the gene to autism related phenotypes but whose human orthologs have been implicated in ASD. For example, mutations in the Rora gene have been linked to abnormal coordination and abnormal cerebellum development in mice but had not been considered a model for autism simply because the human gene was not implicated in the disease. However, it was recently demonstrated that idiopathic autistic patients have decreased expression of the RORA protein due to differences in the methylation state of the gene suggestive that RORA plays a role in the disease phenotype [21].
Intrigued by these findings, we compared the 426 genes implicated in autism by the CNV study to mouse genes that have similar functional and phenotypic similarity to known mouse ASD genes. We took advantage of two tools. The first was MouseNET. MouseNET creates a functional network based on an inputted list of genes. By entering into the MouseNET the eight genes associated with ASD in the MGI database (Cadps2, En2, Gabrb3, Gstm1, Nlgn3, Pten, Ehmt1, and Nrcam), a ranked list of genes from a functional network was generated. A ranked list containing 40 potential genes was identified by a scoring mechanism that incorporated the similarity between genes. Literature review of the genes identified by MouseNet found a subset of eight human genes associated with autism. These genes are CADPS, PAX3, DMD, MLL1, PKD1, AMPH, CACNA1A, and APC. However, the mouse orthologs for these genes were not recorded as models for autism due to a lack of experimental evidence. One additional gene, Unc5c was found to overlap with the CNV list.
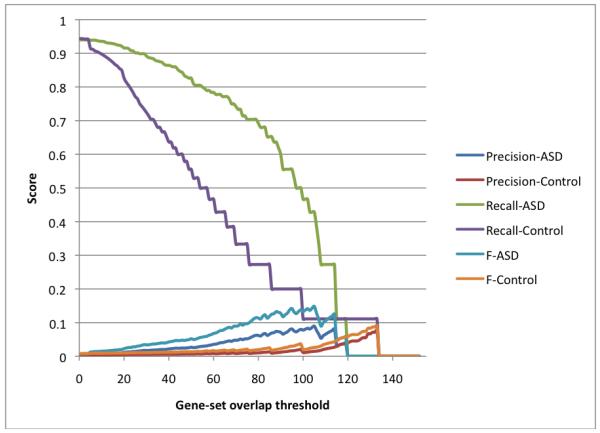
We also analyzed the autism-implicated gene set with the ABBA tool to find similar genes based on functional and phenotypic similarity. ABBA incorporates data from the ODE database to generate a list of candidate genes with similar characteristics to an input set. To assess this approach, we first measured precision and recall against a true positive list of 133 genes definitively linked to people affected with ASD as determined by a consortium of ASD researchers [8]. Precision and recall scores were calculated over a range of gene set overlap thresholds (figure 4). As expected, there is less recall and improved precision as stringency is increased. We compared these results to 1000 randomly drawn ranked gene lists and plotted the precision and recall at a 95% confidence level. Our ABBA analysis performs significantly better than chance on recall but not precision at very high thresholds, and for lower thresholds, consistently performs above chance. This analysis shows that our approach performs well despite scarcity in existing knowledge of genetic components of ASD. We expect as more genes are implicated in ASD and are added to the true-positive list, precision of our analysis will improve.
Figure 4. Recall and Precision analysis.
Recall and precision scores were calculated for the ABBA analysis across a range of gene-set overlaps. Control sets represent the 95% percentile of 1000 randomly drawn gene lists of the same size as each thresholded ABBA result.
By inputting the eight autism-implicated genes into our ABBA analysis, 349 candidate genes were identified. Eleven orthologous genes was found to overlap with the CNV set including: Unc5c, Dsc3, Ghr, Cask, Fgfr3, Camk4, Chrna3, Anks1b, Chrnb4, Plcb4, and Thrb. Assuming 25,000 protein coding genes, the probability of having 11 or more orthologous genes overlap is p=0.0069. Investigation of the ABBA candidate genes within MGI showed 27 had neurological phenotypes (see Table 1) especially mutant alleles to Unc5c and Plcb4 that expressed neurological morphology and behavioral phenotypes potentially indicative of ASD. Besides the CNV analysis, neither gene has been associated with ASD before.
4. DISCUSSION
With this project, we discover potential mouse models of autism by employing bioinformatics tools that access and integrate MGI phenotype data. As a result, four additional mutant mouse alleles are now associated with autism within the MGI database. In addition, a list of 27 candidate genes that are potentially relevant to ASD has been identified. These findings may lead to additional mouse models of autism by highlighting those mutant mouse strains that should be examined for autistic-like behavior. To validate our approach, we have begun experimental study of candidate strains using new mouse behavioral tests for autism like phenotypes [22]. Behavioral tests include time spent investigating stranger mice [23], exploration of a novel environment, and scoring of repetitive behaviors such as grooming.
The workflow developed in this project is applicable to finding models for any human disease where a single allele has a large contribution to the disease phenotype. For example, of the 300+ genes implicated in the study linking small CNVs to autism, only 3% overlap with genes previously linked to autism or other genes associated with intellect-disabilities. The large number of unknown genes confounds evaluation by pattern recognition algorithms in finding additional candidate genes while the number of known genes is large enough to make reviewing the literature burdensome. The advantage of our approach is that it quickly highlights mouse strains containing highly penetrant alleles that have phenotypic characteristics relevant to the disease in question, making integrated use of both high-throughput genomic studies and individual gene characterizations. With inclusion of additional information on the temporal and spatial information known about a given gene’s expression in the rich datasets becoming available from projects like the Allen Brain Atlas [24], we may be able to relate subsets of the phenotype characteristics to time and place of gene expression in the developing brain. This is an avenue of analysis we are pursing to better understand how the alleles of so many different genes lead to the ASD phenotype.
While we have attempted an analysis of recall and precision of our approach, it is critical to consider the existing state of knowledge underlying true positive and true negative results. The possibility of unbiased whole genome experimentation is a recent development and as such, even the most well characterized disease processes have relatively sparse associations to genes. Therefore, we expect serious downward biased estimates of precision. By intent, the focus of the human component of the Phenologs database is on complex disorders whose genetic components are not completely known. Furthermore, the Phenologs database uses annotations from 2008 and is not updated. This impacts both the human genes associated with a human disease and the number of mouse orthologs associated with an MP term. For example, three independent studies published in 2008 find the CNTNAP2 is linked to an increased familial risk of ASD but this gene is not included in Phenolog’s ASD gene set [25-27]. Likewise, 18 genes are associated with the MP term “abnormal social investigation” in the Phenologs database based on the criteria of having an allele annotated to this term on a non-complex genetic background; as of February 2011, there are 32 genes that meet this criteria. A third limitation that impacts both Phenologs and our initial ODE analyses is that full transitive closure was not made of the MP annotations. Curators annotating data to an ontology make associations to the most appropriate granular term; annotations are meant to be associated to all the ancestral terms in the ontology hierarchy. Not performing transitive closure leads to an under-representation of genes associated with higher level MP terms, except when multiple genotypes at the same locus are annotated to various levels of the hierarchy. In light of these issues, we are working on our own MP term enrichment based on orthologous OMIM gene associations, and ODE now features full-transitive closure of MP annotations.
4.1 Conclusions
We have developed an informatic approach to find mouse alleles that contribute to an autistic phenotype that have yet to be described as mouse models of ASD in the literature. We believe our approach will aid researchers looking to prioritize mouse models to any given disease that has a large number of implicated genes.
6. Acknowledgements
This work was supported by the National Human Genome Research Institute grant HG00330 (CB, CC, JB, TFM), the National Institute on Alcohol Abuse and Alcoholism grant AA18776 (EC, JJ), and the Jackson Laboratory Summer Student Program funded in party by the Horace W. Goldsmith Foundation (CC). The funding sources had no involvement in study design, collection, or analysis of data.
Abbreviations
- ASD
autism spectrum disorders
- MGI
the Mouse Genome Informatics
- MP
Mammalian Phenotype ontology
- CNV
copy number variants
- OMIM
Online Mendelian Inheritance in Man
- ODE
Ontological Discovery Environment
- ABBA
Anchored Bicliques of Biomolecular Associates
Footnotes
5. Conflict of interest The authors state that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- [1].Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ. 2009;58:1–20. [PubMed] [Google Scholar]
- [2].Levy SE, Mandell DS, Schultz RT. Autism. Lancet. 2009;374:1627–1638. doi: 10.1016/S0140-6736(09)61376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:e472–486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- [4].Kumar RA, Christian SL. Genetics of autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:188–197. doi: 10.1007/s11910-009-0029-2. [DOI] [PubMed] [Google Scholar]
- [5].Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- [6].Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- [7].McClellan J, King M. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- [8].Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, et al. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moy SS, Nadler JJ. Advances in behavioral genetics: mouse models of autism. Mol. Psychiatry. 2008;13:4–26. doi: 10.1038/sj.mp.4002082. [DOI] [PubMed] [Google Scholar]
- [12].Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Smith CL, Eppig JT. The Mammalian Phenotype Ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guan Y, Myers CL, Lu R, Lemischka IR, Bult CJ, Troyanskaya OG. A genomewide functional network for the laboratory mouse. PLoS Comput. Biol. 2008;4:e1000165. doi: 10.1371/journal.pcbi.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baker EJ, Jay JJ, Philip VM, Zhang Y, Li Z, Kirova R, et al. Ontological Discovery Environment: a system for integrating gene-phenotype associations. Genomics. 2009;94:377–387. doi: 10.1016/j.ygeno.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang Y, Chesler E, Langston M. On Finding Bicliques in Bipartite Graphs: a Novel Algorithm with Application to the Integration of Diverse Biological Data Types. Hawaii International Conference on System Sciences. 2008:473. doi: 10.1186/1471-2105-15-110. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- [19].Rijsbergen C. Information retrieval. 2.ed Butterworths; London: 1979. [Google Scholar]
- [20].DeLorey TM, Sahbaie P, Hashemi E, Homanics GE, Clark JD. Gabrb3 gene deficient mice exhibit impaired social and exploratory behaviors, deficits in non-selective attention and hypoplasia of cerebellar vermal lobules: a potential model of autism spectrum disorder. Behav. Brain Res. 2008;187:207–220. doi: 10.1016/j.bbr.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. Faseb J. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ricceri L, Moles A, Crawley J. Behavioral phenotyping of mouse models of neurodevelopmental disorders: relevant social behavior patterns across the life span. Behav. Brain Res. 2007;176:40–52. doi: 10.1016/j.bbr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- [23].Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- [24].Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat. Rev. Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- [25].Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]