Abstract
Dengue has become a global public health problem and a sensitive diagnostic test for early phase detection can be life saving. An internally controlled, generic real-time PCR was developed and validated by testing serial dilutions of a DENV positive control RNA in the presence of a fixed amount of IC with results showing a good linearity (R 2 = 0.9967) and a LOD of at least 1.95 × 104 copies/mL. Application of the generic PCR on 136 patient samples revealed a sensitivity of 95.8% and specificity of 100%. A newly developed multiplex real-time PCR with serotype-specific probes allowed the serotyping of DENV for 80 out of 92 (87%) generic real-time PCR positive patients. Combined these real-time PCRs offer a convenient diagnostic tool for the sensitive and specific quantification of DENV in clinical specimens with the possibility for serotyping.
1. Introduction
Mosquito-borne flavivirus infections such as dengue have rapidly spread and are the most significant and dreaded infectious diseases in the world, in terms of morbidity and mortality [1, 2]. Recent estimates indicate that over 3.5 billion people (∼55%) of the world population live in areas at risk for dengue [3]. Worldwide there are 50–100 million cases of dengue infections per year, which result in 25,000 deaths. Dengue has become a major international public health problem due to the expanding geographic distribution of the vector in tropical and subtropical countries [1]. Increased international travel accompanied with increasing transmission or re-emergence and changing epidemiology of dengue in various (sub) tropical countries contribute to a steady rise in confirmed dengue among ill-returned travelers [4]. Dengue is caused by an RNA virus (DENV). DENV is primarily transmitted through bites of the infected Aedes aegypti mosquito vector. The majority of DENV infections, with any of the four different virus serotypes (DENV-1, DENV-2, DENV-3, and DENV-4), are mildly asymptomatic and often difficult to recognize in the early phase of infection because signs and symptoms are nonspecific and resemble other febrile illnesses. Only a small number of DENV infections (~5%) will result in severe forms of the disease [5, 6]. The most used diagnostic tools for confirmation of DENV infections are based on detection of antibodies (Ab) or, recently, NS1 antigen (Ag) detection [7]. However, both Ab and Ag detection methods do not distinguish the respective DENV serotypes. Tests based on detection and serotyping of dengue virus offer the possibility to look better at the association between complications of the life threatening DHF and DSS, serotype, and sequential infections [8, 9]. Other real-time PCR assays for DENV infections are based upon type-specific primers but all without internal control (IC) [8, 10, 11]. For reliable detection and to rule out false negative results the use of an IC is essential [12]. Serotyping of DENV is important because the lack of cross-reactive immunity for the four different DENV serotypes may lead to a life threatening complication [9]. In this study we developed and validated an internally controlled generic real-time PCR for detection and quantification of all known DENV serotypes and a multiplex PCR for discrimination of serotypes based upon serotype-specific probes.
2. Materials and Methods
2.1. Primers and Probes
All known complete genome sequences of DENV found in GenBank were aligned using Vector NTI Advance version 10 and ClustalW. The generic primer pair used for amplification of DENV and of DENV-1 in vitro RNA for use as positive control (DENV in vitro RNA) is located in the 3′-UTR region. The DENV generic primers, the primers of the noncompetitive internal control RNA (IC) (MS2 RNA; Roche Diagnostics), and the probes sequences for amplification and detection are listed in Table 1. The specific MS2 RNA primers, probe, and characteristics were designed by us. All primers, probes, and linkers were obtained from Biolegio (Biolegio, Nijmegen, The Netherlands).
Table 1.
Primers and fluorescence-labeled probes of DENV and noncompetitive IC (MS2 RNA).
| Primer or probe | Sequence (5′–3′) | 5′ reporter | 3′ quencher |
|---|---|---|---|
| DENV-F (10546)-generic | GGTTAGAGGAGACCCCTCCC | ||
| DENV-R (10711)-generic | GAGACAGCAGGATCTCTGGTCT | ||
| MS2-F | CATAAGTTAGATGGCCGTCTGT | ||
| MS2-R | TAGAGACGACAACCATGCCAAAC | ||
| DENV-generic-MGB probe | AAACAGCATATTGACGCTGGGA | FAM | BHQ1 |
| MS2 probe | TCCAGACAACGTGCAACATATCGCGACGTATCGTGATATGG | HEX | BHQ1 |
| DENV-1-MGB probe | AACACCATGGGAAGCTGTACCCTG | FAM | BHQ1 |
| DENV-2-MGB probe | GAGATGAAGCTGTAGTCTCACTGG | HEX | BHQ1 |
| DENV-3-MGB probe | TGAGGGAAGCTGTACCTCCTTGCA | Texas Red | BHQ2 |
| DENV-4-MGB probe | CCCGAAGCCAGGAGGAAGCTGTACTCC | CY5 | BHQ2 |
Nucleotide numbering for generic primers DENV-F and -R is relative to DENV-1 (reference strain AF226685).
F: forward primer; R: reverse primer; MGB: minor groove binding; BHQ1 or 2: black hole quencher 1 or 2.
2.2. Construction of the DENV-1 In Vitro RNA Positive Control (DENV In Vitro RNA)
For the construction of DENV in vitro RNA two linkers were designed, named DENV-hyb-1 and DENV-hyb-2. The sequence of DENV-hyb-1 is (DENV generic primers sequences are in Italic) 5′-GGTTAGAGGAGACCCCTCCCAAGTCACAACGCAGCAGCGGGGCCCAACACCAGGGGAAGCTGTACCCTGGTGGTAAGGACTAGAGGTTAGAGGAGACCCCCCGCGCAACAATAAACAGCATATTGACGCTGGGAGAGACCAGAGATCCTGCTGTCTC-3′ and of DENV-hyb-2 is 5′-GAGACAGCAGGATCTCTGGTCTCTCCCAGCGTCAATATGCTGTTTATTGTTGCGCGGGGGGTCTCCTCTAACCTCTAGTCCTTACCACCAGGGTACAGCTTCCCCTGGTGTTGGGCCCCGCTGCTGCGTTGTGACTTGGGAGGGGTCTCCTCTAACC-3′. DENV in vitro RNA was constructed as described earlier [12, 13].
2.3. RNA Quantification
The DENV in vitro RNA and MS2 RNA were quantified by measuring the optical density at 260 nm. Serial dilutions of the RNAs were stored at −70°C until use.
2.4. Nucleic Acid Extraction, cDNA, and Generic Real-Time PCR
DENV in vitro RNA and the 136 patient samples were extracted in the presence of a fixed amount of IC and cDNA was made. The IC was spiked before extraction into lysis buffer containing the sample. The extraction of nucleic acids (NA) was performed on 200 μL serum or plasma sample and 5 μL IC and eluted in 100 μL TE-buffer [14]. 80 μL of the NA extract was used for cDNA synthesis (RT-reaction) as earlier described [12] using random primers (Roche Diagnostics) and the cDNA products were stored at −20°C. For the preparation of cDNA of DENV in vitro RNA the DENV in vitro RNA and IC were spiked into the lysis buffer containing DENV negative human serum before extraction. The generic PCR mix contained 12.5 μL of 2x Probes Master Mix (Roche Diagnostics), 500 nM of primer (each), 300 nM of DENV-generic-MGB probe and 100 nM of MS2 probe, and 10 μL of cDNA (corresponding to 1/12.5 of extraction) in a 25 μL volume. PCR reactions were performed in Bio-Rad iQ5 real-time machine (Bio-Rad), as followed: 10 minutes at 50°C and 10 minutes at 95°C, followed by 45 cycles of 20 s at 95°C, and 1 minute at 60°C for annealing and extension. Data were analyzed using the Bio-Rad iQ5 software version 2.1 (Bio-Rad). A patient sample was considered positive in the generic PCR if the Cq value was below 40. The negative results were truly negative if the IC was detected in the samples. Otherwise the sample was considered to be unsuitable for analysis and the PCR was repeated for a new extraction.
2.5. Serology
Antibodies against DENV were measured using DENV IgM and IgG Capture ELISA (PANBIO) and by the method earlier described by Tran et al. [15].
2.6. Clinical Samples
Samples (serum or plasma) from 109 patients with an antibody positive dengue serology and 27 with an antibody negative dengue serology were included. The 109 patients with an anti-DENV positive serology stratified in 26 patients with a primary DENV infection and 83 patients with a secondary DENV infection. The 136 patients could be further divided into 3 groups: 46 ill-returned travelers (21 primary infections, 16 secondary infections, and 9 antibodies negative) [16], 72 patients from Vietnam (5 primary infections and 67 secondary infections), [17] and 18 patients from whom 16 patients were antibody and PCR positive for hepatitis C (HCV) and 2 patients were antibody and PCR positive for hepatitis A (HAV). The 118 of 136 patients (46 ill returned travellers and 72 patients from Vietnam) were tested by our generic PCR and previous by the earlier described type-specific primer PCR [11]. The 18 patients with HCV or HAV were tested only in our generic PCR.
2.7. Serotyping by SYBR-Green
The SYBR Green reactions were performed in the Bio-Rad iQ5 real-time machine (Bio-Rad) using the cDNA (10 μL) in a 25 μL volume consisting of 12.5 μL of iQ SYBR Green Supermix (Bio-Rad), 500 nM generic DENV primer pair. For amplification the following PCR program was used: 10 minutes at 95°C, followed by 45 cycles of 20 s at 95°C, and 1 minute at 60°C for annealing and extension. The melting temperature (T m) curve analysis was performed following the amplification and included one cycle of denaturation at 95°C for 1 minute, followed by 60°C for 1 minute and a ramp to 95°C at a rate of 0.1°C with continuous fluorescence measurement, using the iQ5 real-time PCR machine, and the data were analyzed using Bio-Rad software version 2.1 (Bio-Rad).
2.8. Serotyping in a Multiplex Real-Time PCR Assay
Serotypes were determined using the generic DENV primer pair and 4 different serotype-specific DENV-probes in one multiplex PCR reaction. Multiplex PCR reactions were performed as described for the generic PCR with 500 nM of DENV primer (each) and 300 nM of each serotype-specific DENV-probe 1–4 (Table 1).
2.9. Statistical Analysis
Statistical analyses were performed using SPSS software for windows (version 16.0). Frequencies, means, or medians were calculated to describe background variables. Agreement was expressed by Cohen's kappa value. The type-specific primer PCR by Laue et al. was used as gold standard for calculation of sensitivity and specificity of our assays [11].
3. Results
3.1. Determination of the Dynamic Range of the Internally Controlled, Generic Real-Time PCR for DENV
For reliable detection and to rule out false negative results an IC was used throughout the whole process of extraction, cDNA synthesis, and PCR reaction. To determine the dynamic range of the internal control the extraction, cDNA synthesis, and PCR procedure were performed on serial dilutions of IC spiked into the lysis buffer containing negative serum. Limiting dilution results for the IC revealed a detection limit of 10 copies/PCR with a 100% hit rate (Table 2). However, for reliable detection of the IC in a duplex assay an input corresponding to 500 copies IC/PCR was chosen which had no influence on the detection limit of DENV in vitro RNA (results not shown). In subsequent experiments a fixed amount of 6.25 × 103 IC copies was spiked in the lysis buffer prior to extraction and cDNA synthesis. The IC showed good stable Cq-values (mean Cq-value of 33.34) during the validation.
Table 2.
Dynamic range of ICa.
| IC copies in extraction | IC copies in RT | IC copies in PCRb | Number positives IC (%; mean Cq-value & SD) |
|---|---|---|---|
| 1.25 × 107 | 107 | 106 | 4/4 (100%; 20.74; 0.23) |
| 1.25 × 106 | 106 | 105 | 4/4 (100%; 24.80; 0.43) |
| 1.25 × 105 | 105 | 104 | 4/4 (100%; 27.92; 0.48) |
| 1.25 × 104 | 104 | 103 | 4/4 (100%; 32.08; 0.71) |
| 1.25 × 103 | 103 | 102 | 4/4 (100%; 34.53; 0.56) |
| 1.25 × 102 | 102 | 101 | 4/4 (100%; 36.93; 0.72) |
| 0 | 0 | 0 | 0/4 |
aThe dynamic range was determined on 10-fold serial dilution of IC spiked into lysis buffer containing DENV negative human serum as background with 4 replicates for each dilution.
bNumber of copies cDNA/PCR has been calculated as 1/12.5 of the extracted RNA, assuming 100% efficiency in extraction, reverse transcription (RT), and PCR.
SD: standard deviation.
To determine the dynamic range of the generic PCR, we spiked 10-fold serial dilutions of DENV in vitro RNA together with a fixed amount of IC into lysis buffer containing DENV negative serum prior to extraction and cDNA synthesis. Subsequently 10 μL of the cDNA was used in PCR. Limiting dilution of DENV in vitro RNA resulted in a quantitative detection limit of 103 copies/PCR with a 66.7% hit-rate (Table 3) and a regression coefficient of 0.9967 with a dynamic range of 6.25 × 103–6.25 × 107 copies/mL (Figure 1).
Table 3.
Dynamic range of generic real-time PCRa.
| DENV in vitro RNA copies in extraction | DENV in vitro RNA copies in RT | DENV in vitro RNA copies in PCRb | IC copies in PCRb | Number positives DENV in vitro RNA (%; mean Cq-value: SD) | Number positives IC (%; mean Cq-value: S.D) |
|---|---|---|---|---|---|
| 1.25 × 107 | 107 | 106 | 500 | 12/12 (100%; 26.59; 0.54) | 12/12 (100%; 35.82; 1.08) |
| 1.25 × 106 | 106 | 105 | 500 | 12/12 (100%; 30.59; 0.46) | 12/12 (100%; 32.67; 0.79) |
| 1.25 × 105 | 105 | 104 | 500 | 12/12 (100%; 34.35; 1.09) | 12/12 (100%; 32.97; 0.74) |
| 1.25 × 104 | 104 | 103 | 500 | 8/12 (66.7%; 37.22; 0.73) | 12/12 (100%; 33.73; 0.63) |
| 1.25 × 103 | 103 | 102 | 500 | 1/12 (8%; 40.71) | 12/12 (100%; 32.24; 0.60) |
| 0 | 0 | 0 | 0 | 0/12 | 0/12 |
aThe dynamic range of the generic PCR was determined on 10-fold dilution series of DENV in vitro RNA in the presence of a fixed amount of IC spiked into lysis buffer containing DENV negative human serum as background and for 12 replicates per dilution.
bNumber of copies cDNA/PCR has been calculated as 1/12.5 of the extracted RNA. assuming 100% efficiency in extraction, reverse transcription (RT), and PCR.
SD: standard deviation.
Figure 1.
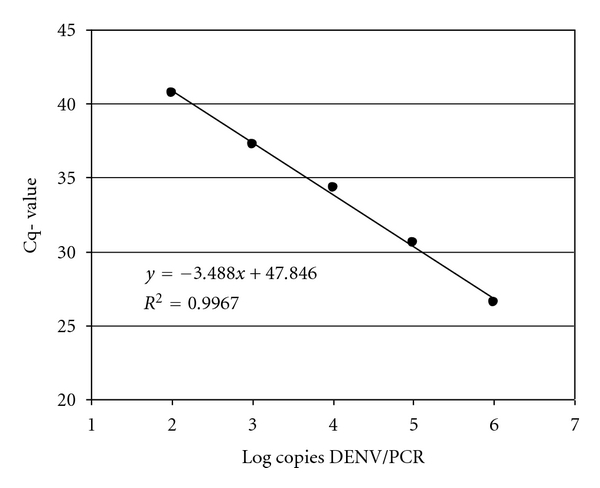
The dynamic range of generic of PCR. Mean Cq-values of twelve replicates per each 10-fold dilutions of DENV in vitro RNA extracted with a fixed amount of 6.25 × 103 IC copies in extraction in the back ground of DENV negative serum and tested in generic real-time PCR resulting in a dynamic range of 6.25 × 103–6.25 × 107 copies/mL of DENV in vitro RNA with a regression coefficient of 0.9967.
3.2. Determination of the Lower Limit of Detection (LOD)
To determine the LOD of the generic PCR assay 2-fold serial dilutions of DENV in vitro RNA were performed with 20 replicates per dilution with fixed amount of IC in the background of DENV negative human serum each separately spiked in the lysis buffer prior to extraction. The LOD for DENV in vitro RNA was at least 312 copies/PCR corresponding to 1.95 × 104 copies/mL (Table 4).
Table 4.
LOD of the generic real-time PCRa.
| DENV in vitro RNA copies in PCRa | IC copies in PCRb | Number positive DENV in vitro RNA (%; mean Cq-value: SD) | Number positive IC (%; mean Cq-value: SD) |
|---|---|---|---|
| 2500 | 500 | 20/20 (100%; 35.98; 0.99) | 20/20 (100%; 33.43; 0.65) |
| 1250 | 500 | 12/20 (60%; 36.96; 1.13) | 20/20 (100%; 32.84; 0.53) |
| 1000 | 500 | 10/20 (50%; 37.33; 0.94) | 20/20 (100%; 33.66; 0.53) |
| 625 | 500 | 8/20 (40%; 38.72; 1.82) | 20/20 (100%; 32.61; 0.59) |
| 312 | 500 | 5/20 (25%; 39.17; 2.67) | 20/20 (100%; 32.69; 0.34) |
| 0 | 0 | 0/20 | 0/20 |
aLOD of the generic PCR was determined by 2-fold dilution series with 20 replicates per dilution of DENV in vitro RNA in the presence of a fixed amount of IC spiked into lysis buffer containing DENV negative human serum as background.
bnumber of copies cDNA/PCR has been calculated as 1/12.5 of the extracted RNA, assuming 100% efficiency in extraction, reverse transcription (RT) and PCR.
LOD: lower limit of detection; SD: standard deviation.
3.3. Intra- and Inter-Assay Variation
The intra- and inter-assay variation was measured in four replicates per dilution of 10-fold dilution series with fixed amount of IC in a background of DENV negative serum: For intra-assay variation determined in three runs performed at one day (results not shown) and for inter-assay variation performed in three 3 runs over 3 consecutive days. The assay variation showed stable Cq-values for the IC and each serial dilution of DENV in vitro RNA (Table 5).
Table 5.
Inter-assay variation of generic real-time PCRa.
| Run and DENV in vitro RNA copies in PCRa | IC copies in PCRb | Number positive DENV in vitro RNA (%; mean Cq-value: SD) | Number positive IC (%; mean Cq-value: SD) | SD between the 3 runs |
|---|---|---|---|---|
| Run 1 | ||||
| 106 | 500 | 4/4 (100%; 26.28; 0.43) | 4/4 (100%; 36.21; 1.21) | 0.27 |
| 105 | 500 | 4/4 (100%; 30.40; 0.39) | 4/4 (100%; 33.10; 1.37) | 0.35 |
| 104 | 500 | 4/4 (100%; 33.78; 0.38) | 4/4 (100%; 32.17; 0.30) | 0.22 |
| 103 | 500 | 2/4 (50%; 36.99; 0.37) | 4/4 (100%; 33.64; 0.29) | 0.65 |
| 102 | 500 | 0/4 | 4/4 (100%; 32.47; 0.20) | — |
| 0 | 0 | 0/4 | 0/4 | — |
|
| ||||
| Run 2 | ||||
| 106 | 500 | 4/4 (100%; 26.71; 0.56) | 4/4 (100%; 36.11; 0.84) | 0.27 |
| 105 | 500 | 4/4 (100%; 30.35; 0.19) | 4/4 (100%; 32.66; 0.65) | 0.35 |
| 104 | 500 | 4/4 (100%; 34.31; 0.64) | 4/4 (100%; 33.32; 0.39) | 0.22 |
| 103 | 500 | 3/4 (75%; 37.08; 0.86) | 4/4 (100%; 33.64; 0.92) | 0.65 |
| 102 | 500 | 1/4 (25%; 37.83) | 4/4 (100%; 32.73; 1.09) | — |
| 0 | 0 | 0/4 | 0/4 | — |
|
| ||||
| Run 3 | ||||
| 106 | 500 | 4/4 (100%; 26.79; 0.60) | 4/4 (100%; 35.13; 1.06) | 0.27 |
| 105 | 500 | 4/4 (100%; 30.98; 0.50) | 4/4 (100%; 32.47; 0.10) | 0.35 |
| 104 | 500 | 4/4 (100%; 34.27; 0.78) | 4/4 (100%; 33.24; 0.54) | 0.22 |
| 103 | 500 | 1/4 (25%; 38.17) | 4/4 (100%; 33.91; 0.67) | 0.65 |
| 102 | 500 | 0/4 | 4/4 (100%; 32.59; 0.23) | — |
| 0 | 0 | 0/4 | 0/4 | — |
aThe inter-assay variation was determined with 10-fold dilution series of DENV in vitro RNA in the presence of a fixed amount of IC spiked into lysis buffer containing DENV negative human serum as background and for 4 replicates per each dilution. The variation was determined in three separate runs performed at three different days.
bNumber of copies cDNA/PCR has been calculated as 1/12.5 of the extracted RNA, assuming 100% efficiency in extraction, reverse transcription (RT), and PCR.
SD: standard deviation.
3.4. Testing of Clinical Samples
A fixed amount of IC (5 μL) and 200 μL of the patient sample were added to the lysis buffer prior to extraction and cDNA synthesis. Overall, the generic PCR showed a sensitivity of 95.8% and a specificity of 100% (Table 6). All patient samples with detected IC and DENV negative were truly negative. Just four patient samples that tested positive in the type-specific primer PCR tested negative in the generic PCR. The patient samples had been tested earlier by the type-specific primer PCR [16, 17] and the extra freeze-thaw steps may have affected the quality of the RNA in these four samples that showed already a high Cq-value close to 37 or higher in the type-specific primer PCR. A lower percentage (62.2%) of the anti-DENV positive ill-returned travelers had a positive test result in the generic PCR compared with the anti-DENV positive Vietnamese patients (95.8%) which correlates with the longer duration of illness in the ill-returned travelers before seeing a clinician (Table 6).
Table 6.
Summary of patient characteristics and sensitivity and specificity of PCR assays.
| Patient characteristics and test parameters | Vietnamese patients with DENV | Ill-returned travelers with DENV | Other causes of fever in travelers | Patients positive for | |||
|---|---|---|---|---|---|---|---|
| Primary infection | Secondary infection | Primary infection | Secondary infection | OFI | HCV/HAV | Total | |
| Patients (n) | 5 | 67 | 21 | 16 | 9 | (18)d | 136 |
| Age; median (range) | 10.9 (6.6–12.8) | 18.9 (5.6–53.1) | 38.7 (17.4–62.6) | 33.2 (24.6–73.6) | 39.5 (26.3–64.0) | unknown | 26.3 (6.6–73.6) |
| Sex (M/F) | 3/2 | 48/19 | 9/12 | 6/10 | 6/3 | unknown | 72/46 |
| Days of illness at presentation; median (range) | 1 (1-1) | 1 (1–4) | 6 (1–12) | 8.5 (4–22) | — | — | 2 (1–22) |
| Days of DENV viremia; median (range) | 1 (1-1) | 1 (1–4) | 4 (1–11) | 7 (4–9) | — | — | 1 (1–11) |
| Patients (n) with a positive result in | |||||||
| Type-specific PCRa | 5 | 66 | 16 | 9 | 0 | ND | 96 |
| Generic PCRb | 3 | 66 | 15 | 8 | 0 | 0 | 92 |
| Multiplex PCRc | 3 | 63 | 11 | 3 | 0 | ND | 80 |
| Assay characteristics of Generic PCR | |||||||
| Sensitivity; % (95% CI) | 60.0 (23.1–88.2) | 100.0 (94.5–100.0) | 93.8 (71.7–99.0) | 88.9 (56.5–98.0) | — | — | 95.8 (89.8–98.4) |
| κ-value | 0.736 | 1.000 | 0.944 | 0.924 | 0.846 | ||
| Specificity; % | 100.0 | 100.0 | 100.0 | ||||
aThe results were earlier generated by the type-specific primer PCR according to according Laue et al. [11, 16, 17].
bInternally controlled generic real-time PCR for DENV.
cMultiplex PCR with type-specific probes was only done on samples with a positive generic PCRbresult.
d16 patients were antibody and PCR positive for HCV and 2 patients were antibody and PCR positive for HAV.
OFI: other febrile illness; HCV: hepatitis C virus; HAV: hepatitis A virus; M: male; F: female; 95% CI: 95% confidence interval, ND: not done.
3.5. Serotyping by SYBR-Green
Since the SYBR-green real-time PCR requires no type-specific primers or probes, results can be obtained with one real-time PCR program for the detection of the specific melting curve (T m). We used the cDNA from a panel of patients which were known to be infected with DENV serotype 1 to 4 and who tested previously positive in our generic PCR. The T m curves however for DENV-1, DENV-2, DENV-3, and DENV 4 were too close to each other (84.90°C, 83.60°C, 85.80°C, and 84.70°C) to enable discrimination between all four serotypes. Also, differences in the viral loads influenced melting temperatures make serotyping of DENV by SYBR-green PCR unreliable (results not shown).
3.6. Serotyping by Multiplex Real-Time PCR with Serotype-Specific Probes
The multiplex PCR with serotype-specific probes was only applied on the cDNAs from the 92 patient samples which had a positive test result in the generic PCR. The DENV serotype from these samples was previously determined using the type-specific primer PCR (Table 7) and was tested blindly in our multiplex PCR. The generic PCR detected all 4 DENV serotypes. The multiplex PCR with the type-specific probes correctly identified the DENV serotype in 80 out of the 92 (87%) patient samples. These 12 patient samples which could not be serotyped in our multiplex assay had Cq-values close to 38 or higher in the generic PCR. In three patient samples an infection by two serotypes was detected with the type-specific primer PCR, whereas by the multiplex PCR with serotype-specific probes an infection by two serotypes was detected in seven out of the 80 patient samples (Table 7). The viremia levels in first serotype were higher than in the second serotype.
Table 7.
Serotyping by multiplex real-time PCR with serotype-specific probes.
| Serotype | No. of patients positive in the following assays | No. of infections by two serotypes detected by the following assays (DENV serotypes in infections)c | |||
|---|---|---|---|---|---|
| Type-specific PCRa | Generic PCR | Multiplex PCRb | Type-specific PCRa | Multiplex PCRb | |
| 1 | 39 | Pos (39) | 30 | 3 (1 + 2) | 4 (1 + 4) |
| 2 | 19 | Pos (19) | 17 | 0 | 1 (2 + 1) |
| 3 | 3 | Pos (3) | 2 | 0 | 1 (3 + 4) |
| 4 | 31 | Pos (31) | 31 | 0 | 1 (4 + 2) |
| total | 92 | 92 | 80 | 3 | 7 |
Pos: generic PCR can make no distinction between the serotypes.
aThe results were earlier generated by the type-specific primer PCR according to Laue et al. [11, 16, 17].
bMultiplex real-time PCR with serotype-specific probes was only done on positive generic PCR results.
cA DENV infection by two serotypes has been assigned to the serotype which signal came up first and showed the highest viral load of the two.
4. Discussion
DENV infections have become a major international public health problem due to the increasing geographic distribution of the vector [1], the increased international travel accompanied with increasing confirmed DENV infection among ill-returned travelers [4], and the lack of cross-reactive immunity for the four different DENV serotypes and hyperendemic circulation of the four serotypes in the same regions. These all are factors that play a significant role in the threat posed by DENV.
An inherent problem in real-time PCR is the presence of amplification inhibitors which may cause false-negative results. MS2 RNA is an ideal “noncompetitive” internal control for use as a process control which have already been proven by other studies. MS2 RNA is commercially available for any user, and the RNA is a stable, noninfectious and absent from human clinical samples, and is a noncompetitive control which does not react with the selected primers and probe for DENV. Our generic real-time PCR assay was capable to amplify the four known serotypes in one PCR reaction and showed a good linearity (R 2 = 0.9967) with a dynamic range of 6.25 × 103–6.25 × 107 copies/mL and a LOD of at least 1.95 × 104 copies/mL. The intra- and inter-assay variation showed stable Cq-values (SD < 0.70) for each 10-fold serial dilution for DENV in vitro RNA. A viral load of 6.25 × 103 copies/mL can still be detected with certainty at a Cq-value of 40 in our generic PCR in 8% of the cases (Table 3). Therefore, a patient sample was considered positive in the generic PCR if the Cq-value was below 40. The DENV generic primers and the various probes are based on a highly conserved region in the 3′-UTR of DENV (Figure 2). DENV genetic diversity and quasi-species could contribute to PCR accuracy as has been shown for HCV, another member of the family of Flaviviruses. However, the variation and the proportion of biological variants are very low in DENV (unpublished data). The target of the described DENV PCR is very small (157 bp) and we cannot describe quasi-species—like in, for example, HCV, and moreover there is no external selection pressure like DENV-specific antivirals or a vaccine affecting the sequence of the amplicon we used for detection of DENV.
Figure 2.
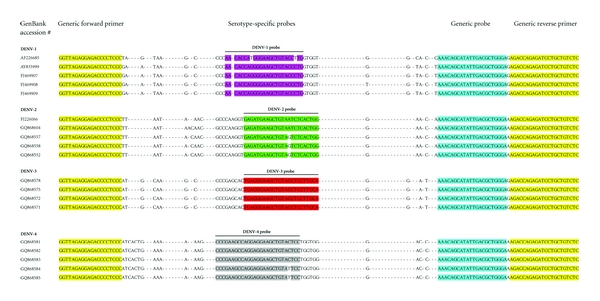
Multiple sequence alignments of DENV 3′UTR region deduced from complete genome sequences of dengue serotypes 1–4 (DENV 1–4) obtained from GenBank showing primers and probes sequences used in generic and/or multiplex real-time PCR with serotype-specific probes for detection of DENV in vitro RNA and DENV in patient samples.
The comparison between the described generic real-time PCR and the earlier described type-specific primer PCR using 118 patient samples showed an excellent concordance between the two assays with a sensitivity of 95.8% and specificity of 100% [11]. The patient samples that were not detected in our generic PCR were quantified with a very low viral load (about 4.16 × 104 copies/mL) in the type-specific primer PCR. The extra freeze-thaw steps caused by using the same sample tube for serological tests, for the type-specific primer PCR assay, and for our generic PCR assay may lead to degradation of DENV RNA by RNases in the sample. Patient samples may be divided into aliquots to minimize the effect of freeze-thawing.
We tried the SYBR-green real-time PCR for serotyping because it is less expensive and easier to perform compared to type-specific primers or probes assays. The SYBR-green assay results showed that both the viral load and the close similarity between the four serotypes influenced the melting temperatures of each serotype (data not shown), making it difficult to obtain reliable results, which were also seen in previous studies [10, 18]. Therefore, we developed a multiplex real-time PCR assay using four serotype-specific probes in one PCR reaction which showed reliable results to discriminate between the 4 DENV serotypes in patient samples. We were able to differentiate between the serotypes in 80 out of 92 patient samples, which had positive generic PCR result, with the multiplex assay (sensitivity of 87%), and 7 coinfections with two serotypes were found. The 12 patient samples which could not be serotyped had Cq-values close to 38 or higher in the generic PCR which was probably due to the differences in sensitivity between the generic and multiplex PCR and competition within the multiplex PCR. A total of 109 patients had positive antibodies against DENV and 92 patients were positive in our generic PCR. This difference may be explained by the relatively high median days of ill-returned travelers after onset of symptoms before visiting a clinician and multiple freeze-thaw steps. The ill-returned travelers which had a negative PCR result but positive antibodies against DENV visited a clinician after a median of 6 days (primary infection; range 1 to 12 days) and 8.5 days (secondary infection; range 4 to 22 days) of illness and possibly the antibodies had emerged after DENV had already disappeared from the blood. It has been postulated that DENV can be detected approximately until 5 days after onset of symptoms by PCR, regardless of the serotype [11].
Our study describes a specific, sensitive, and internally controlled generic real-time PCR assay for early detection of DENV and the possibility for serotyping by multiplexing. Screening patients with the internally controlled, generic PCR assay for DENV and applying the multiplex PCR assay for typing only on positive sample will be more cost-effective than performing a typing PCR on all samples. Many of the published multiplex assays use mixtures of serotype type-specific primers and probes which makes the assay less sensitive and more expensive [10]. Other multiplex PCRs are not genuine multiplex PCR assays because they consist of two separate duplex PCR assays, each with a probe pair for DENV 1–3 or DENV 2–4 [18].
The described assays are an important complementary tool for diagnosis of DENV in patients in the early phase of disease.
Acknowledgments
The authors would like to thank Hakima Belkasim for technical assistance, Dr. Edou Heddema for providing them with DENV negative but HAV and HCV positive samples, and Nicholas Griffin for review. K. T. D. Thai is supported by a Mosaic fellowship from the Netherlands Organization for Scientific Research (NWO), Grant no. 017.004.074.
Abbreviations
- DENV:
Dengue virus
- IC:
Internal control
- HCV:
Hepatitis C
- HAV:
Hepatitis A
- PCR:
Polymerase chain reaction
- RT:
Reverse transcription
- DHF:
Dengue hemorrhagic fever
- DSS:
Dengue shock syndrome
- NS1:
Nonstructural 1 protein
- Ab:
Antibody
- Ag:
Antigen
- LOD:
Lower limit of detection
- Cq-value:
Cycle for quantification.
References
- 1.Kroeger A, Nathan MB. Dengue: setting the global research agenda. The Lancet. 2006;368(9554):2193–2195. doi: 10.1016/S0140-6736(06)69873-5. [DOI] [PubMed] [Google Scholar]
- 2. WHO guidelines Treatment, prevention and control, 2009, http://whqlibdoc.who.int/publications/2009/9789241547871_eng.pdf.
- 3.Beatty M, Letson W, Edgil D, Margolis H. Estimating the total population at risk for locally acquired dengue infection. American Society of Tropical Medicine and Hygiene. 2007;77(supplement 5):170–257. [Google Scholar]
- 4.Schwartz E, Weld LH, Wilder-Smith A, et al. Seasonality, annual trends, and characteristics of dengue among Ill returned travelers, 1997–2006. Emerging Infectious Diseases. 2008;14(7):1081–1088. doi: 10.3201/eid1407.071412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke DS, Nisalak A, Johnson DE, Scott McN. R. R. A prospective study of dengue infections in Bangkok. American Journal of Tropical Medicine and Hygiene. 1988;38(1):172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 6.Endy TP, Chunsuttiwat S, Nisalak A, et al. Epidemiology of inapparent and symptomatic acute dengue virus infection: a prospective study of primary school children in Kamphaeng Phet, Thailand. American Journal of Epidemiology. 2002;156(1):40–51. doi: 10.1093/aje/kwf005. [DOI] [PubMed] [Google Scholar]
- 7.Phuong HL, Thai KTD, Nga TTT, et al. Detection of dengue nonstructural 1 (NS1) protein in Vietnamese patients with fever. Diagnostic Microbiology and Infectious Disease. 2009;63(4):372–378. doi: 10.1016/j.diagmicrobio.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Leparc-Goffart I, Baragatti M, Temmam S, et al. Development and validation of real-time one-step reverse transcription-PCR for the detection and typing of dengue viruses. Journal of Clinical Virology. 2009;45(1):61–66. doi: 10.1016/j.jcv.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Fink J, Gu F, Vasudevan SG. Role of T cells, cytokines and antibody in dengue fever and dengue haemorrhagic fever. Reviews in Medical Virology. 2006;16(4):263–275. doi: 10.1002/rmv.507. [DOI] [PubMed] [Google Scholar]
- 10.Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJJ. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. Journal of Clinical Microbiology. 2006;44(4):1295–1304. doi: 10.1128/JCM.44.4.1295-1304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laue T, Emmerich P, Schmitz H. Detection of dengue virus RNA in patients after primary or secondary dengue infection by using the TaqMan automated amplification system. Journal of Clinical Microbiology. 1999;37(8):2543–2547. doi: 10.1128/jcm.37.8.2543-2547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beld M, Minnaar R, Weel J, et al. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. Journal of Clinical Microbiology. 2004;42(7):3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M. Rapid detection of human parechoviruses in clinical samples by real-time PCR. Journal of Clinical Virology. 2008;41(2):69–74. doi: 10.1016/j.jcv.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim-Van Dillen PME, Van Der Noordaa J. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran TNT, de Vries PJ, Hoang LP, et al. Enzyme-linked immunoassay for dengue virus IgM and IgG antibodies in serum and filter paper blood. BMC Infectious Diseases. 2006;6, article 13 doi: 10.1186/1471-2334-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thai KTD, Wolthers KC, van Vugt M, de Vries PJ. Dengue fever among ill returned travelers and concurrent infection by two dengue virus serotypes. Dengue Bulletin. 2009;33:60–69. [Google Scholar]
- 17.Thai KTD, Phuong HL, Thanh Nga TT, et al. Clinical, epidemiological and virological features of dengue virus infections in vietnamese patients presenting to primary care facilities with acute undifferentiated fever. Journal of Infection. 2010;60(3):229–237. doi: 10.1016/j.jinf.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naze F, Le Roux K, Schuffenecker I, et al. Simultaneous detection and quantitation of Chikungunya, Dengue and West Nile viruses by multiplex RT-PCR assays and Dengue virus typing using High Resolution Melting. Journal of Virological Methods. 2009;162(1-2):1–7. doi: 10.1016/j.jviromet.2009.03.006. [DOI] [PubMed] [Google Scholar]


