Abstract
OBJECTIVE
Multiple single nucleotide polymorphisms (SNPs) associated with type 2 diabetes (T2D) susceptibility have been identified in predominantly European-derived populations. These SNPs have not been extensively investigated for individual and cumulative effects on T2D risk in African Americans.
RESEARCH DESIGN AND METHODS
Seventeen index T2D risk variants were genotyped in 2,652 African American case subjects with T2D and 1,393 nondiabetic control subjects. Individual SNPs and cumulative risk allele loads were assessed for association with risk for T2D. Cumulative risk was assessed by counting risk alleles and evaluating the difference in cumulative risk scores between case subjects and control subjects. A second analysis weighted risk scores (ln [OR]) based on previously reported European-derived effect sizes.
RESULTS
Frequencies of risk alleles ranged from 8.6 to 99.9%. Eleven SNPs had ORs >1, and 5 from ADAMTS9, WFS1, CDKAL1, JAZF1, and TCF7L2 trended or had nominally significant evidence of T2D association (P < 0.05). Individuals carried between 13 and 29 risk alleles. Association was observed between T2D and increase in risk allele load (unweighted OR 1.04 [95% CI 1.01–1.08], P = 0.010; weighted 1.06 [1.03–1.10], P = 8.10 × 10−5). When TCF7L2 SNP rs7903146 was included as a covariate, the risk score was no longer associated with T2D in either model (unweighted 1.02 [0.98–1.05], P = 0.33; weighted 1.02 [0.98–1.06], P = 0.40).
CONCLUSIONS
The trend of increase in risk for T2D with increasing risk allele load is similar to observations in European-derived populations; however, these analyses indicate that T2D genetic risk is primarily mediated through the effect of TCF7L2 in African Americans.
Type 2 diabetes (T2D) is a complex disease resulting from lifestyle, environmental, and genetic factors. Prevalence rates differ across ethnicities, with the higher rates occurring in African Americans at a prevalence of 18% (1). Although multiple genetic risk variants for T2D have been identified in predominantly European-derived populations (2–6), prior studies have shown little evidence for association of these single nucleotide polymorphisms (SNPs) in African Americans other than TCF7L2 variants (7–9). Several risk assessment studies have been performed to assess the cumulative effect of multiple T2D “risk” variants on diabetes incidence (10–17), although few have included African Americans (11,16) and none have focused solely on this population.
We evaluated the cumulative risk effect of 17 index variants in a well-characterized sample of African American T2D case subjects and nondiabetic control subjects.
RESEARCH DESIGN AND METHODS
A total of 2,652 African Americans with T2D and 1,393 nondiabetic control subjects were evaluated. Only individuals with complete age data and proportions of African ancestry >0.50 were included. Case subjects consisted of DNA samples from 2,652 self-described African American individuals with T2D, including 1,502 ascertained through having end-stage renal disease (T2D-ESRD). Control subjects included DNA samples from 1,393 nondiabetic African Americans. Ascertainment criteria and recruitment methods have been described (18). Recruitment and sample collection procedures were approved by the Wake Forest School of Medicine Institutional Review Board, and written informed consent was obtained from all participants. Subjects were unrelated, self-described African Americans born in North Carolina, South Carolina, Georgia, Virginia, or Tennessee. Subjects with T2D-ESRD were recruited from dialysis facilities.
T2D was diagnosed as diabetes development after the age of 25 years without prior diabetic ketoacidosis. In addition, T2D-ESRD case subjects met at least one of the following criteria for inclusion: 1) T2D diagnosed ≥5 years before initiating renal replacement therapy, 2) background or greater diabetic retinopathy, or 3) ≥100 mg/dL proteinuria on urinalysis in the absence of other causes of nephropathy (set 1 case subjects). Subjects with T2D without evidence of nephropathy were recruited from medical clinics, churches, health fairs, and community resources using the above criteria (set 2 case subjects). African American control subjects without a current diagnosis of diabetes or renal disease were recruited from the community and internal medicine clinics (set 1 and 2 control subjects). DNA extraction was performed using the PureGene system (Gentra Systems, Minneapolis, MN).
Genotyping and quality control
Seventeen index SNPs with prior compelling evidence of association with T2D in European-derived populations (2–6) were genotyped in 2,652 African Americans with T2D (1,502 with T2D-ESRD, 1,150 with T2D lacking nephropathy) and in 1,393 nondiabetic control subjects. SNPs were genotyped on the Affymetrix Genome-wide Human SNP array 6.0 (Affy 6.0) at the Center for Inherited Disease Research (CIDR) as part of a Genome Wide Association Study or by using the MassARRAY genotyping system with PCR primers designed using MassARRAY Design 3.4 software (Sequenom, San Diego, CA). For 913 T2D-ESRD case subjects and 826 control subjects, 12 of 17 SNPs were genotyped on the Affy 6.0, methods and quality control for which have been previously described (18). SNPs had <5% missing data, and there was no significant difference in missing data between case and control subjects. Forty-six blind duplicates were included in genotyping and had a concordance rate of 99.6%. Genotyping of remaining SNPs and samples was done on the iPLEX Sequenom MassARRAY platform (Sequenom). Genotyping efficiency exceeded 93.5%, and the 95 blind duplicate samples included in genotyping had a concordance rate of >99.5%. Hardy-Weinberg proportions were >0.0005 in all case and control subjects analyzed.
Admixture estimates
Ancestral allele proportions were estimated from allele frequencies of 70 ancestry informative markers (19) genotyped in 44 Yoruba Nigerians and 39 European Americans. Individual ancestral proportions were calculated for each subject using the frequentist estimation of individual ancestry proportion (FRAPPE) program (20) under a two-population model. Individuals with proportions of African ancestry <0.50 were excluded from analyses. Admixture estimates were used as covariates in all statistical analyses.
Statistical analysis
Individual SNP association analyses.
The 17 SNPs were individually tested for association with T2D in the overall cohort using a logistic regression model and then evaluated individually for association with T2D by disease type in sets 1 and 2 using the SNPGWA program (www.phs.wfubmc.edu). The additive genetic model is reported here. Set 1 and 2 results were subsequently combined in a meta-analysis. The inverse variance weighted method was implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal/) to determine the overall association of each index variant with T2D. Because of the statistical significance and potential confounding effects, all analyses were adjusted for age (P = 5.6 × 10−119), sex (P = 1.3 × 10−4), and percentage of African ancestry (P = 1.6 × 10−4). In evaluating individual variants for association with T2D, a supplemental analysis was performed that also adjusted for BMI. Multiple comparison correction was not performed owing to the a priori hypothesis of association between the variants examined and T2D and the primary hypothesis of a single cumulative risk for these loci.
Cumulative risk scores.
Risk allele loads were initially determined in an unweighted approach where the number of risk variants carried by an individual at each SNP was summed to create a cumulative risk score based on the reported risk allele. A second risk score was calculated by a weighted method in which published effect sizes (natural logarithm of odds ratio [OR]) for each risk variant (identified in predominantly European studies) were used to adjust for the relative contribution of each SNP in the cumulative risk score calculation. In both methods, missing values were replaced with the average-risk allele load at each SNP (<4.5% missing data for all SNPs), and cumulative scores were rounded to the nearest value; the maximum possible score in each analysis was 34. Risk score differences between case and control subjects were compared using a Wilcoxon rank sum test.
Cumulative risk assessment.
Cumulative risk scores in the unweighted and weighted analyses were grouped into nine bins: ≤17, 18, 19, 20, 21, 22, 23, 24, and ≥25. Binned risk scores of case and control subjects were compared using a Wilcoxon rank sum test. Bins from the unweighted and weighted analyses were both tested for association with T2D using logistic regression analysis, adjusting for the covariates of African ancestry, age, and sex. Binned risk scores were modeled as ordinal categoric variables and, in a separate analysis, as nine nominal categories; the latter is less parsimonious but allows for greater flexibility (i.e., nonlinearity) in the estimation of the individual bin risk score effects. A model was computed that tested for evidence of a departure from linearity in the bin risk score effect on T2D affection status. Additional analyses included the number of TCF7L2 rs7903146 risk variants as covariates in the logistic regression model. This model estimates and tests the effect of the other loci after accounting for the strong TCF7L2 influence on risk of T2D. Theoretically, this latter model will have greater statistical power to detect the effects of the other loci.
RESULTS
Study samples
Clinical characteristics of the study samples are reported in Table 1. The case group in set 1 had more women than the control group. The mean age at enrollment for the T2D-ESRD group was higher (P < 0.05), but the age of onset of T2D in the case subjects was >7 years earlier than the age of enrollment for control subjects. Mean BMI was not significantly different between T2D-ESRD case and nondiabetic control subjects (P = 0.19). Set 2 case subjects also included a higher percentage of women. Mean age at enrollment for the T2D group was higher (P < 0.05); the mean BMI for T2D case subjects was higher than for control subjects (P < 0.05). Overall, the samples have characteristics that reflect the general population of African Americans in the southeast. The mean (SD) African ancestry proportions in the 1,502 T2D-ESRD case subjects, 1,150 T2D case subjects, and 1,393 combined control subjects were 0.79 ± 0.10, 0.78 ± 0.10, and 0.78 ± 0.09, respectively, as estimated by the frequentist estimation of individual ancestry proportion program.
Table 1.
Clinical characteristics of study samples
Analysis of individual index variants
Risk allele frequencies and individual SNP association results are reported in Table 2. When the 2,652 T2D case subjects and 1,393 control subjects were combined, frequencies of risk alleles ranged from 8.64 to 99.97%. Notably, we observed an association of rs7903146 in TCF7L2 (OR 1.38 [95% CI 1.23–1.54], P = 1.25 × 10−8). In addition, rs10010131 in WFS1 (OR 1.13, P = 0.029), rs10946398 in CDKAL1 (OR 1.14, P = 0.014), and rs864745 in JAZF1 (OR 1.20, P = 2.30 × 10−3) were associated with directions of association consistent with prior reports in European-derived samples. SNP rs4607103 in ADAMTS9 was also associated (P = 2.40 × 10−3); however, the direction of association was opposite that reported in a European sample (5) (OR 0.84 in this analysis). Results of an additional analysis adjusting for BMI were comparable (Supplementary Table 1).
Table 2.
Individual SNP associations by logistic regression analysis in entire cohort
SNPs were evaluated in case groups (sets 1 and 2) and subsequently combined in a meta-analysis to determine if associations differed between the overall and individual analyses (Supplementary Table 2). The TCF7L2 variant rs7903146 was associated in sets 1 and 2. In set 1, rs4607103 in ADAMTS9 was associated; however, the effect was in the opposite direction of that reported in a European sample (5). In set 2, rs10946398 in CDKAL1 was associated, as was rs864745 in JAZF1. In the meta-analysis, results were consistent with the combined analysis (described above) (Table 2); rs10010131 in WFS1, rs10946398 in CDKAL1, rs864745 in JAZF1, and rs7903146 in TCF7L2 replicated association with risk, whereas rs4607103 in ADAMTS9 replicated association in the opposite direction of effect of that previously reported in a European sample (5).
Cumulative risk score
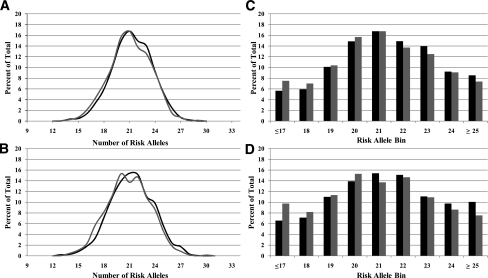
Unweighted and weighted cumulative risk scores were evaluated in the overall cohort. Figure 1A and B show the percentage of T2D case and nondiabetic control subjects plotted by the cumulative risk score of each individual; values were significantly different when scores were determined by both unweighted and weighted methods (P < 0.005 for both analyses). Figure 1C and D show binned distributions of cumulative risk scores determined by unweighted and weighted methods in T2D case and nondiabetic control subjects. Figure 1C shows that the nondiabetic control group had higher percentages of individuals carrying lower-risk allele loads (<21); whereas the T2D groups generally had greater percentages of individuals carrying higher-risk allele loads. Figure 1D highlights the more pronounced distribution differences in risk allele bins between the two groups. Differences in binned distributions for both unweighted and weighted analyses were significant (P < 0.005).
Figure 1.
Distribution of cumulative risk allele loads in T2D case subjects (black bars) and nondiabetic control subjects (gray bars) by risk score (A and B) and by bin (C and D) in unweighted analysis (A and C) and in weighted analysis by published effect size (B and D).
Cumulative risk assessment
The OR and 95% CI for each of the nine bins assessed by the full logistic regression model and the model with adjustment for rs7903146 are represented in Fig. 2A (unweighted) and Fig. 2B (weighted); results are summarized in Supplementary Table 3. There was an increase in the OR with an increase in risk allele category. There was no evidence of departure from a linear trend in the association of the number of risk alleles and disease (P = 0.71). In a model in which data were reanalyzed, treating the risk allele categories as a continuous variable, there was evidence of an association between cumulative risk allele load and disease status in the unweighted and weighted analyses (unweighted OR 1.04 [95% CI 1.01–1.08], P = 0.011; weighted 1.06 [1.03–1.10], P = 8.10 × 10−5). In both types of analysis (i.e., nominal and ordinal categories for the count of risk alleles), association was no longer significant after adjusting for the number of TCF7L2 risk variant rs7903146: nominal analysis results (Supplementary Table 3 and Fig. 2A and B); ordinal analysis results (unweighted 1.02 [0.98–1.05], P = 0.33; weighted 1.02 [0.98–1.06], P = 0.40).
Figure 2.
The OR and 95% CI (range bars) for each of the nine bins assessed in the cumulative risk assessment using the full logistic regression model (black diamonds) and the model with adjustment for rs7903146 (gray squares) are represented from unweighted (A) and weighted (B) analyses. All models were adjusted for age, sex, and proportion of African ancestry.
We found no evidence for interaction with age (P = 0.20), sex (P = 0.59), BMI (P = 0.47), or degree of African Ancestry (P = 0.74). Recall that only individuals with >0.50 proportions of African ancestry are included in these analyses.
CONCLUSIONS
T2D risk variant associations have been identified in European-derived populations, although the functional variants are not all known. Also, there have been few reports in the high-risk African American population. This study evaluated 17 SNPs that have shown association with risk for T2D in mostly European-derived populations (2–6). Five risk variants are nominally associated with T2D in this African American sample, including rs4607103 (ADAMTS9), rs10010131 (WFS1), rs10946398 (CDKAL1), rs864745 (JAZF1), and rs7903146 (TCF7L2); all risk alleles are consistent with those reported in European-derived samples except for ADAMTS9. SNPs rs4607103 (ADAMTS9), rs10010131 (WFS1), rs10946398 (CDKAL1), and rs864745 (JAZF1) remained associated when accounting for the presence of TCF7L2 SNP rs7903146 risk variants, suggesting these are independent loci (data not shown). Inferences were not changed when adjustment for BMI was performed. In addition, this study demonstrated that a genetic risk score based on 17 European-derived T2D-associated risk variants was not predictive of T2D status in African Americans after removing the effects of the important TCF7L2 risk variant rs7903146.
There are a number of potential explanations for the observed association in the ADAMTS9 SNP rs4607103 that is in the opposite direction of what was previously described (5). The actual functional variant in the ADAMTS9 region is not known, and the rs4607103 SNP may not be the functional variant in this region. It is presumed that rs4607103 is in linkage disequilibrium (LD) with the functional variant in individuals of European descent. An independent mutation may have arisen in African Americans in the ADAMTS9 region on a different haplotype, resulting in a change in direction of association. In addition, with the exception of rs7903146 in TCF7L2, the evidence for association of other SNPs is modest, and given the number of SNPs, rs4607103 barely meets evidence for association (Bonferroni corrected P = 0.0029 vs. P = 0.0024 for rs4607103), and the resulting inverse association could be a chance false-positive. In addition, the LD pattern in Africans is sufficiently different from Europeans so that LD relationships in African Americans may be substantially different with a trait-defining variant, thus obscuring the true relationship.
Previous studies assessing European-identified T2D variants have shown little evidence that these SNPs are associated with T2D in African Americans, with the exception of TCF7L2 variant rs7903146 (7,8,21,22). This issue is difficult to convincingly address given differences in allele frequency, sample sizes, and attendant power in African American studies (7). The current analyses tested variants that were also analyzed by Waters et al. (16) in the Population Architecture Using Genomics and Epidemiology study. Many variants were associated in a pooled analysis of multiple ethnic groups, but results from the African American subset (1,077 case subjects, and 1,469 control subjects) were significant for four variants (rs1801282 in PPARG, rs4402960 in IGF2BP2, r864745 in JAZF1, rs7903146 in TCF7L2). The larger sample size evaluated here provides a nominal increase in power compared with prior reports and found an association with SNPs in ADAMTS9, WFS1, CDKAL1, and JAZF1, in addition to the well-replicated rs7903146 in TCF7L2. An important feature of the analysis reported here is a detailed evaluation of admixture proportions to exclude DNA samples with <0.50 African ancestry. Admixture was a major influence on our inferences (P = 1.6 × 10−4) in analysis of these data and was therefore included as a covariate to eliminate potential stratification.
We observed a significant increase in T2D risk with increased risk allele load in analyses using unweighted and weighted methods to generate a risk score. Similar risk score analyses have been performed in other studies, most often in individuals of European descent. We observed similar results (i.e., increased risk with increasing risk allele load) (10–17). Two such studies included African Americans and detected an increased risk with increase in risk allele load in their overall samples (11,16). When analyses were limited to African American individuals, however, results were less significant (if at all) in contrast to European-only and multiethnic samples. Waters et al. (16) evaluated increased progression to T2D with risk allele load and observed association in a multiethnic cohort and African American subset. Hivert et al. (11) also observed association between risk allele load and progression to T2D in the Diabetes Prevention Program (DPP) cohort; however, results were not significant when limited to the smaller African American subset. Neither Waters et al. nor the DPP studies accounted for admixture proportions in African American participants.
The major observation here is that removing the most significantly associated variant (rs7903146) from the risk prediction model substantially changed the inferences. Significance was no longer observed in the risk score model after adjustment for the effect of the TCF7L2 variant. Although risk for T2D increases with increasing risk allele load, similar to Europeans, this was primarily driven by the rs7903146 variant of TCF7L2. The other risk variants do not appear to jointly significantly contribute to disease risk because of the lower or insignificant effect of these SNPs.
Previous T2D genetic risk models have failed to be more predictive of disease risk than conventional factors that contribute to risk (10,13,15,23). In addition, it has been reported that the variants studied here, along with other European T2D “risk” variants, do not explain a significant portion of the racial/ethnic disparities in T2D prevalence, although African Americans on average carry more risk alleles compared with European Americans (16). Genetic risk models for T2D, such as the one in this study, may not include causal variants and therefore must be regarded carefully (16). Causal variants or variants in high LD with causal variants for T2D may differ across ethnicities, further complicating the search for genetic contributors to this polygenic disease. Differences in LD architecture encompassing the variants among different populations may help to fine-map the causal variant if shared across ethnicities, as shown in Palmer et al. (8).
In addition, only 4 of the remaining 16 variants were associated, highlighting that SNPs associated with T2D risk in individuals of European descent may not consistently represent critical risk variants in non-European populations, African Americans in particular. Alternatively, the SNPs we studied may not represent variants in LD with causal variants in this population, supporting the variability of disease-risk architecture across different ethnicities. This effect may be a consequence of ancestral exposure to different selective pressures and/or environments, resulting in the conservation of disease-causing alleles in LD with other genetic variants necessary for survival, such as the case of APOL-1 risk variants that are protective against trypanosomes that cause African sleeping sickness (24).
This study represents a comprehensive evaluation of cumulative genetic risk for T2D in a large African American cohort and demonstrates compelling evidence that the TCF7L2 SNP rs7903146 is the most significant genetic contributor to T2D risk in African Americans. Differences in LD structure and genetic architecture across different ethnic groups, in addition to diverse evolutionary pressures and environmental modifiers, are important factors to consider as the search for genetic variants contributing to T2D in African Americans progresses.
Supplementary Material
Acknowledgments
Genotyping services were provided by the Center for Inherited Disease Research (CIDR). The CIDR is fully funded through a federal contract from the National Institutes of Health (NIH) to Johns Hopkins University, contract number HHSC268200782096C.
This work was supported by NIH grants K99-DK-081350 (N.D.P.), R01-HL-56266 (B.I.F.), R01-DK-070941 (B.I.F.), R01-DK-084149 (B.I.F.), R01-DK-066358 (D.W.B.), and R01-DK-053591 (D.W.B.), and in part by the General Clinical Research Center of the Wake Forest School of Medicine grant M01 RR07122. Computing was provided by the Wake Forest School of Medicine Center for Public Health Genomics.
No potential conflicts of interest relevant to this article were reported.
As the corresponding author and guarantor of this article, D.W.B. takes full responsibility for the work as a whole, including the study design, access to data, and the decision to submit and publish the article. J.N.C. researched data, contributed to discussion, and wrote the manuscript. M.C.Y.N. and N.D.P. researched data, contributed to discussion, and reviewed and edited the manuscript. S.S.A. and J.M.H. researched data. B.I.F. contributed to recruitment and phenotyping of study samples, contributed to manuscript discussion, and reviewed and edited the manuscript. C.D.L. researched data, contributed to discussion, and reviewed and edited the manuscript. D.W.B. contributed to discussion and reviewed and edited the manuscript.
The authors acknowledge the analytic assistance of Jiang Li, MS, and Jianzhao Xu, BS, both of the Center for Diabetes Research, and Lingyi Lu, MS, of the Department of Public Health Sciences at Wake Forest School of Medicine, Winston-Salem, North Carolina.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc11-0957/-/DC1.
See accompanying commentary, p. 193.
References
- 1.Centers for Disease Control and Prevention. National Diabetes Fact Sheet: National Estimates and General Information on Diabetes and Prediabetes in the United States, 2011 Atlanta, GA, U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2011
- 2.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research; Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 10.1126/science.1142358 [DOI] [PubMed] [Google Scholar]
- 3.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 10.1126/science.1142382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 10.1038/nature05616 [DOI] [PubMed] [Google Scholar]
- 5.Zeggini E, Scott LJ, Saxena R, et al. ; Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 10.1038/ng.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeggini E, Weedon MN, Lindgren CM, et al. ; Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 10.1126/science.1142364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis JP, Palmer ND, Hicks PJ, et al. Association analysis in African Americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes 2008;57:2220–2225 10.2337/db07-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer ND, Hester JM, An SS, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 2011;60:662–668 10.2337/db10-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sale MM, Smith SG, Mychaleckyj JC, et al. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes 2007;56:2638–2642 10.2337/db07-0012 [DOI] [PubMed] [Google Scholar]
- 10.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med 2009;150:541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hivert MF, Jablonski KA, Perreault L, et al. An updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the Diabetes Prevention Program. Diabetes 2011;60:1340–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lango H, Palmer CN, Morris AD, et al. ; UK Type 2 Diabetes Genetics Consortium Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes 2008;57:3129–3135 10.2337/db08-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 2008;359:2208–2219 10.1056/NEJMoa0804742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rong R, Hanson RL, Ortiz D, et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes 2009;58:478–488 10.2337/db08-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes 2008;57:3122–3128 10.2337/db08-0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet 2010;6:e1001078. 10.1371/journal.pgen.1001078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weedon MN, McCarthy MI, Hitman G, et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med 2006;3:e374. 10.1371/journal.pmed.0030374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonough CW, Palmer ND, Hicks PJ, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int 2011;79:563–572 10.1038/ki.2010.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene KL, Mychaleckyj JC, Leak TS, et al. Exploration of the utility of ancestry informative markers for genetic association studies of African Americans with type 2 diabetes and end stage renal disease. Hum Genet 2008;124:147–154 10.1007/s00439-008-0532-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 2005;28:289–301 10.1002/gepi.20064 [DOI] [PubMed] [Google Scholar]
- 21.Moore AF, Jablonski KA, McAteer JB, et al. ; Diabetes Prevention Program Research Group Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes 2008;57:2503–2510 10.2337/db08-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helgason A, Pálsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007;39:218–225 10.1038/ng1960 [DOI] [PubMed] [Google Scholar]
- 23.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 10.1056/NEJMoa0801869 [DOI] [PubMed] [Google Scholar]
- 24.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010;329:841–845 10.1126/science.1193032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.