Abstract

Fluorescent microscopy and flow cytometry are widely used tools in biomedical sciences. Cost-effective translation of these technologies to remote and resource-limited environments could create new opportunities especially for telemedicine applications. Toward this direction, here we demonstrate the integration of imaging cytometry and fluorescent microscopy on a cell phone using a compact, lightweight, and cost-effective optofluidic attachment. In this cell-phone-based optofluidic imaging cytometry platform, fluorescently labeled particles or cells of interest are continuously delivered to our imaging volume through a disposable microfluidic channel that is positioned above the existing camera unit of the cell phone. The same microfluidic device also acts as a multilayered optofluidic waveguide and efficiently guides our excitation light, which is butt-coupled from the side facets of our microfluidic channel using inexpensive light-emitting diodes. Since the excitation of the sample volume occurs through guided waves that propagate perpendicular to the detection path, our cell-phone camera can record fluorescent movies of the specimens as they are flowing through the microchannel. The digital frames of these fluorescent movies are then rapidly processed to quantify the count and the density of the labeled particles/cells within the target solution of interest. We tested the performance of our cell-phone-based imaging cytometer by measuring the density of white blood cells in human blood samples, which provided a decent match to a commercially available hematology analyzer. We further characterized the imaging quality of the same platform to demonstrate a spatial resolution of ~2 μm. This cell-phone-enabled optofluidic imaging flow cytometer could especially be useful for rapid and sensitive imaging of bodily fluids for conducting various cell counts (e.g., toward monitoring of HIV+ patients) or rare cell analysis as well as for screening of water quality in remote and resource-poor settings.
Cell phones are now widely used all over the world. According to the International Telecommunication Union (ITU), by the end of 2010 the number of cell-phone subscribers reached almost 5 billion worldwide, and the majority of these users are actually located in developing countries.1 Because of this massive volume, cell phones are now having a central role in telemedicine applications, especially for developing countries and resource-limited locations. Among several examples, the use of cell phones for remote monitoring of various medical data/images has already been well-established in, e.g., dermatology, wound care, or even ophthalmology.2–4
In addition to these, together with the recent advances in hardware and software capabilities of cell phones, there is also a growing interest in converting cell phones into microscopic analysis tools for imaging of, e.g., bodily fluids or water samples.5–8 These previous efforts, however, focused on microscopic imaging of static objects (with significantly limited sample volumes), using a cell-phone-enabled optical microscope. On the other hand, there is also an important need today for translating flow-based fluorescent cytometry concepts9–12 into cell phones to enable specific and sensitive analysis of large sample volumes through, e.g., integrated microfluidic devices. Such an imaging flow cytometry device running on a cell phone could extend our microanalysis capabilities in resource-limited settings for, e.g., conducting whole blood analysis or for screening of water-borne parasites in drinking water.
Toward this goal, here we demonstrate the integration of an optofluidic fluorescent cytometry platform onto a cell phone using compact optical attachments. This cell-phone attachment, weighing only ~18 g, includes a simple lens (cost, <3 USD/ piece), a plastic color filter (cost, <0.1 USD/piece), two light-emitting diodes (LEDs) (cost, ~0.3 USD/piece), and batteries (cost, ~0.5 USD/piece), which make it rather cost-effective. In our design, the sample solution of interest is continuously delivered/pumped to our imaging volume through a disposable microfluidic channel using a syringe pump. This microfluidic device assembly is placed above a separate inexpensive lens that is put in contact with the existing camera unit of the cell phone (see Figure 1) such that the entire cross section of our microfluidic device can be mapped onto the CMOS sensor chip of the cell phone. Once inserted into our cell-phone-based imaging flow cytometer, each microfluidic device is illuminated from the side by inexpensive LEDs using simple butt-coupling, i.e., without the use of any bulky light-coupling optics or lenses as illustrated in Figure 1. This excitation light is then guided within the cross section of the microfluidic device (which acts as a multilayered optofluidic waveguide composed of poly(dimethylsiloxane)–liquid–glass interfaces surrounded by air) uniformly exciting the labeled specimens within the imaging volume (see, e.g., Figure 1C). Since the guided excitation light propagates perpendicular to the detection path, this optofluidic pumping scheme permits the use of an inexpensive plastic absorption filter to create the dark-field background that is required for fluorescent imaging. With this optofluidic design, we continuously deliver the sample solution of interest to our imaging flow cytometer using, e.g., a simple syringe pump, such that a fluorescent microscopic movie of the flowing particles or cells can be acquired using the cell-phone camera unit. The digital frames of these fluorescent movies are then rapidly processed to determine the count of labeled particles/cells, which can be used to estimate the density of the target fluorescent objects within the sample solution. We have tested the performance of this cell-phone-based fluorescent imaging cytometer platform by measuring white blood cell density of human blood samples, the results of which provided a decent agreement with a commercially available hematology analyzer (Sysmex KX-21N). We have also quantified the imaging performance of our cell-phone-based fluorescent microscopy platform to demonstrate that ~2 μm spatial resolution can be reached with the same optofluidic design.
Figure 1.
(A–C) Various schematic diagrams of the designed optical attachment for optofluidic fluorescent imaging cytometry on a cell phone are illustrated. This lightweight attachment has dimensions of ~35 × 55 × 27.9 mm3. The optofluidic unit can be repeatedly attached or detached to the cell-phone body without the need for fine alignment. (D) The picture of our optofluidic fluorescent imaging cytometer on a cell phone. The entire attachment to the cell phone weighs ~18 g.
Our results demonstrate the feasibility of a timely opportunity to translate some basic functions of imaging flow cytometers into field use through integration of an optofluidic microimaging unit onto a cell phone. Therefore, we believe that this compact and cost-effective optofluidic imaging cytometry platform attached to a cell phone could provide a useful toolset for, e.g., telemedicine applications in resource-limited settings, such as for the measurement of CD4+ T cell density of HIV+ patients or for screening of water-borne parasites in drinking water.
EXPERIMENTAL METHODS
Materials
Poly(dimethylsiloxane) (PDMS) was purchased from Dow Corning (U.S.A.). SYTO16 green fluorescent nucleic acid stain (product no. S7578, excitation/emission 488 nm/ 518 nm(+DNA) and 494 nm/525 nm (+RNA)) and fluorescent beads (yellow-green fluorescence: excitation/emission 505 nm/ 515 nm) with 4 μm diameter (product no. F8859), 2 μm diameter (product no. F8827), and 1 μm diameter (product no. F13081) were purchased from Invitrogen (Carlsbad, U.S.A.). The 1× phosphate-buffered saline (PBS) buffer (product no. BP 2438-4) and microscope slides (product no. 12-5500) were purchased from Fisher Scientific (Pittsburgh, PA, U.S.A.). The Harris Uni-Core hole puncher was purchased from Ted Pella (Redding, CA, U.S.A.). The high-frequency plasma generator (model BD-10 AS) was purchased from Electro-Technic Products Inc. (Chicago, IL, U.S.A.). Tygon tubing (i.d., 0.01 in./o.d., 0.03 in.) (product no. 06418-01) was purchased from Cole-Parmer (Vernon Hills, IL, U.S.A.). The plano-convex lens (f = 0.6 mm) (product no. NT45-588) and yellow Kodak Wratten color filter (product no. NT54-467) were purchased from EdmundOptics (Barrington, NJ, U.S.A.). The aspherical lens (f = 4.5 mm) (product no. C230TME-A) was purchased from Thorlab (Newton, NJ, U.S.A.).
Microfluidic Device Fabrication and Preparation
PDMS-based microfluidic devices were fabricated using standard soft lithographic techniques.13 Briefly, the PDMS prepolymer mixture (PDMS prepolymer; curing agent (v/v) = 10:1) was poured onto the mold and degassed at room temperature for half an hour. Then it was baked at 70 °C for an hour. After curing, the PDMS was peeled off from the mold and holes were punched to form the inlet and outlet. The PDMS and glass slide (1 mm thick) were simply cleaned with soap water first, and then they were treated with the high-frequency plasma generator for 15 s. Immediately after plasma treatment, the PDMS was bonded to the glass substrate to be baked at 70 °C for another hour to strengthen the bonding. Finally, the Tygon tubing (i.d., 0.01 in.) was inserted into the chip inlet/outlet and sealed with epoxy. The dimensions of the microfluidics chamber were 44 μm × 3mm × 15 mm (height × width × length). Due to the final step of high-temperature baking, the PDMS–glass chip interior surface becomes hydrophobic. In order to reduce the nonspecific cell adsorption to the surface and bubble generation in the chamber, the microfluidic chamber was treated with the plasma generator for 1 min before each experiment.
Fluorescent Bead Sample Preparation
To prepare the fluorescent bead samples, 10 μL of beads (4, 2, or 1 μm diameter) was mixed with 40 μL of deionized (DI) water. Then 10 μL of this mixture was sandwiched between two glass slides (12.5 mm × 17 mm) using a micropipet. This sample was inserted into the sample tray and slid into the cell-phone attachment for imaging.
Fluorescent Labeling of White Blood Cells in Whole Blood
To start with, SYTO16 was warmed to room temperature and then briefly centrifuged in a microcentrifuge tube to bring the dimethyl sulfoxide (DMSO) solution to the bottom of the vial. Whole blood sample was gently rotated and mixed well. Following this, 20 μL of SYTO16 was pipetted from the supernatant and added to 200 μL ofwhole blood. This mixture was then incubated in dark for ~30 min. To avoid cell sedimentation, the sample was gently rotated during incubation. No red blood cell lysing step was performed. In addition, note that, since the intrinsic fluorescence quantum yield of SYTO16 is extremely low (<0.01), the unbound SYTO16 was not separated from the whole blood after incubation. This labeled whole blood sample was directly diluted 10-fold with PBS buffer and was continuously delivered/pumped into our microfluidic chip through a syringe pump with a typical flow rate of ~1 μL/min. During the experiment, the blood sample vial was slowly agitated to avoid sedimentation of cells.
Design of the Optofluidic Fluorescent Microscopy and Cytometry Platform on a Cell Phone
We used a Sony-Erickson U10i Aino as the base cell-phone device for our optofluidic fluorescent microscopy and cytometry unit. This cell phone has an 8 Mpixel color RGB sensor installed on it, which is used to capture fluorescent images/movies of the specimens. In addition, the camera has a built-in lens in front of the CMOS sensor of the cell phone, which has a focal length of f ~ 4.65 mm.
To build an optofluidic fluorescent microscopy unit on this cell phone, a single inexpensive lens with a focal length of f2 (typically varying between 0.6 and 10 mm) was directly placed in front of the existing camera lens of the cell phone as shown in Figure 1. This imaging geometry gives a magnification of f/f2 between the sample plane (located at the focal plane of f2) and the CMOS sensor. Depending on the application and its requirements on resolution and field of view (FOV), different magnification factors can be achieved by varying the f2 value. Note also that this magnification factor is theoretically independent of the distance between the two lenses, which makes alignment of our attachment to the cell phone rather easy.
In order to increase the imaging throughput in our fluorescent cytometry platform we used unit magnification such that an aspherical lens with f2 = 4.5mmwas placed in front of the existing cell-phone camera lens as shown in Figure 1. For achieving higher spatial resolution in our optofluidic design, however, we used a plano-convex lens with f2 =~0.6mmsuch that a magnification of ~7.8 × was used, which enabled us to improve the resolution down to ~2 μm in fluorescent imaging mode.
On the basis of the principles of optofluidic wave-guiding,14,15 our fluorescent imaging cytometry unit utilizes blue LEDs that are directly butt-coupled to a microfluidic chip as illustrated in Figure 1. Apart from continuously delivering the labeled specimens to our imaging volume, this microfluidic chip also acts as an optofluidic multimode slab waveguide, which has a three-layered refractive index structure (PDMS–liquid–glass) surrounded by air on both sides as illustrated in Figure 1C. Due to the stronger refractive index contrast of air–glass and air–PDMS interfaces compared to the glass–liquid or PDMS–liquid interfaces, this butt-coupled LED excitation light is tightly confined inside this multimode optofluidic waveguide structure (despite optical losses) which results in uniform and efficient pumping of our imaging volume within the microchannel. In addition, since the excitation light propagates perpendicular to the fluorescence detection path, a simple plastic absorption filter is sufficient to reject the scattered pump photons, creating, as desired, a strong dark-field background as shown in Figure 4. We should also emphasize that, in our optofluidic design, the illumination light source (LEDs) and the plastic absorption filter can be easily changed to different colors, and therefore the system is compatible with fluorophores that have different excitation/emission wavelengths.
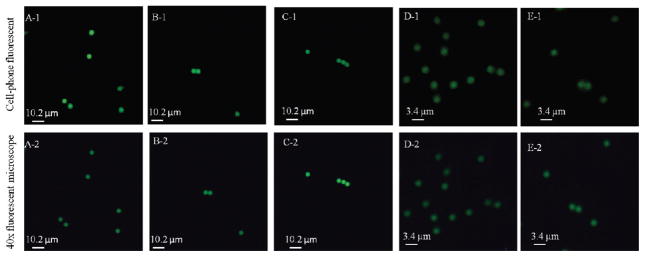
Figure 4.
Spatial resolution of the optofluidic cell-phone fluorescent microscope shown in Figure 1 is quantified using green fluorescent beads with diameters of 4 and 2 μm. The top row shows the raw cell-phone images of these fluorescent beads, which demonstrate ~2 μm resolution; see, e.g., the touching 2 μm particles in panel E-1 that can be resolved. For comparison purposes, the bottom row shows the images of the same particles obtained using a 40× microscope objective lens (NA = 0.65) on a benchtop fluorescent microscope.
During our flow cytometry measurements, we used near high-definition (nHD) mode of the cell phone to record fluorescent videos of the flowing specimens, which provided a resolution of 640 × 352 pixels per frame at a frame rate of ~7 frames/s (fps). In these flow cytometry experiments, the microfluidic chip was connected to a syringe pump through Tygon tubings and fluorescently labeled samples were pumped into the microfluidic chamber continuously at a typical flow rate of ~1 μL/min. Digital processing of these fluorescent video frames enabled us to automatically count the labeled cells/particles and then calculate their density for a given sample.
For imaging experiments that we quantified the spatial resolution of our optofluidic cell-phone attachment, the objects were kept stationary (i.e., without any fluidic flow) such that conventional fluorescent microscope images of the same samples can be obtained for comparison purposes. In this static microscopy mode, the fluorescent images captured by our optofluidic platform were stored at the cell-phone memory in jpg format and could be viewed through the cell-phone screen directly. These jpg files (typically ~3–4 MB for an ~8 Mpixel image) can also be transferred to a computer (e.g., through memory cards or using wireless communication) for further digital processing or analysis.
Digital Analysis of Fluorescent Flow Videos
We used video postprocessing in order to count the labeled cells/particles passing through our microfluidic chip. This involved a repeating sequence of particle detection and tracking. Starting with the first frame, every visible fluorescent microparticle is detected using the contour detection algorithm outlined in ref 16. For every detected contour, the image center of mass is computed and is thereon assumed to be the coordinates of the corresponding particle. Each particle is then assigned a unique identification number which is preserved throughout the analysis process for a given video stream. For every subsequent frame in the video, the particle detection process is repeated. The coordinates of the newly detected particles (in the second frame and onward) are then compared to those of the particles in the previous frame. Assuming that the instantaneous flow rate (between two frames) is reasonably constant for each particle, the location of the particle from one frame to the next can be predicted to be within a reasonable spatial range in the image. On the basis of their proximity to the particles in the previous frame, the newly detected particles are assigned new identification numbers, or in other words, the coordinates of the particles are updated in the new frame, thus allowing particles to be tracked from the moment they first appear in the video.
To summarize, the tracking algorithm is initialized in the first frame by detecting all the visible particles in the chamber. After initialization, a process of detection and matching is repeated to find the coordinates of the particles as they move through the chamber. In order to reduce errors in detection and tracking, we averaged the number of detected particles and their velocities over multiple frames (i.e., longer time periods), and over predefined distances in the chamber. To do this, our custom-designed software allows for setting up a “cascade” of virtual counters at specific cross sections of the chamber, such that each virtual counter monitors the particles that go through it independent of the others. Setting up multiple cascade lines to track the particles allows us to more accurately count the number of cells passing through the microfluidics chamber (see, e.g., the supplementary movie in the Supporting Information).
It must be noted that the tracking algorithm used in this work is kept simple since themost important parameter for our work is the count of cells per unit time of flow. In more complicated settings different components of our algorithms need to be adapted to cope with the added complexities. For instance, in cases where the physical patterns of the particle flow also need to be measured and tracked as a function of time/flow, we need a more complicated tracking algorithm which could be achieved by, e.g., addition of Kalman filtering to our current implementation. It is also worth mentioning that the algorithm used in this work was implemented in OpenCV, which is available both for the iOS platform and for the Android OS. As such, it lends itself for embedded implementation on modern smart phones and tablets, thus potentially eliminating the need for a PC for video postprocessing.
RESULTS AND DISCUSSION
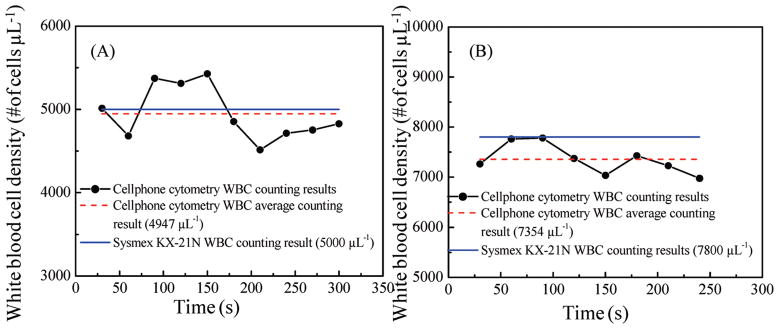
In order to demonstrate the proof of concept of our cell-phone-based optofluidic cytometry platform, white blood cells (WBCs) in human whole blood samples were chosen as our model system. White blood cells density in whole blood is routinely tested for clinical diagnosis of various diseases, including infections, leukemia, HIV, and bone marrow deficiencies.17 Various point-of-care-based hematology analyzers have also been developed for WBC counting.18–29 To test our flow cytometry platform on the cell phone, white blood cells in fresh whole blood samples were labeled with SYTO16 fluorescent dyes and diluted as described in the Experimental Methods section without lysing red blood cells. These 10× diluted and labeled whole blood samples were then flushed through our microfluidic chamber using a syringe pump at a flow rate of ~1 μL/min, and a video of the fluorescent emission arising from the labeled WBCs was recorded continuously as shown in the supplementary movie in the Supporting Information. As illustrated in this supplementary movie, our custom-designed video analysis software defines a cascade of five counters with a separation distance of ~270 μm from each other; and these digital counters dynamically monitor the number of WBCs that go through the microchannel. In order to improve our cell-counting accuracy, the program counted the number of WBCs that passed through each counter line over a period of 210 frames (i.e., ~30 s) and we averaged the number of the counted WBCs from these five independent counters (see the supplementary movie in the Supporting Information) to get the number of WBCs within this 30 s time frame. On the basis of the volume flow rate and the average number of the counted WBCs, we can calculate the WBC density in the blood sample within this time frame (i.e., 30 s). The same process was repeated for 5–6 min with continuous blood flow, and the WBCs density for each sample was plotted as a function of time as shown in Figure 2, parts A and B. The average WBC density for each sample was finally calculated by further averaging each one of these dynamic curves over 5–6 min, and our results were compared to the standard test results obtained from a commercially available hematology analyzer (Sysmex KX-21N). As shown in Figure 2, parts A and B, our WBC density results matched well with the standard test results with <5% error. To further evaluate our cell-phone-based imaging cytometry platform and its counting accuracy, we imaged 12 different patients’ blood sample. Each sample was imaged for 5–6 min, and average WBC density of the whole blood was estimated by processing its corresponding fluorescent video captured by our cell-phone cytometer. The cell-phone-based imaging cytometry results were compared to the parallel results obtained from the Sysmex KX- 21N hematology analyzer. Figure 3A compares the WBC densities obtained using our cell-phone imaging cytometer to the standard results obtained with the Sysmex KX-21N hematology analyzer, which showed a good correlation to our measurements. For these 12 patients’ blood samples (with WBC densities ranging from ~4000 to 8000 μL−1), we obtained a correlation coefficient of ~0.93 between the two methods. In addition to this, we also performed Bland–Altman analysis30 on our results (see Figure 3B), which shows a bias of −339 cells μL−1 with 95% limits of agreement of −1026 and 347 cells μL−1 for a wide range of WBC concentrations. This negative bias indicates that our cell-phone cytometry platform is undercounting the WBCs, which may be caused, e.g., by the partial loss of cells within the microfluidic channel and tubing.31
Figure 2.
Automated WBC-counting results obtained using our cell-phone-based imaging flow cytometer are demonstrated for two different patients’ whole blood: (A) for a lower WBC density sample (5000 cells μL−1) and (B) for a higher WBC density sample (7800 cells μL−1).
Figure 3.
(A) Comparison of WBC density measurement results obtained with our cell-phone-based imaging flow cytometer is provided against the results of a commercially available hematology analyzer (Sysmex KX-21N) for 12 different patients. A linear regression of the experimental data (n = 12, red line) with WBC concentrations ranging from ~4000 to 8000 μL−1 demonstrates a good agreement between the two modalities with a correlation coefficient of ~0.93. (B) The Bland –Altman analysis results, evaluating the accuracy of the cell-phone-based flow cytometry for WBC density measurements vs the standard hematology analyzer. The blue line shows the bias of −339 cells μL−1, and the red lines show the 95% limits of agreement of −1026 (lower limit) and 347 cells μL−1 (upper limit).
Besides counting accuracy, throughput is also an important parameter for an imaging cytometer. The throughput in our flow cytometry system is mainly determined by the cell phone’s camera frame rate. In our current implementation, the camera has a relatively slow frame rate of ~7 fps. To further increase the throughput, we can use a cell-phone camera with a higher frame rate, e.g., LG Dare VX9700, which can achieve a frame rate of ~120 fps. This could potentially further improve our flow rate and thus the counting throughput by, e.g., >15-fold, which would reduce the imaging time for, e.g., a whole blood sample to <20 s per test.
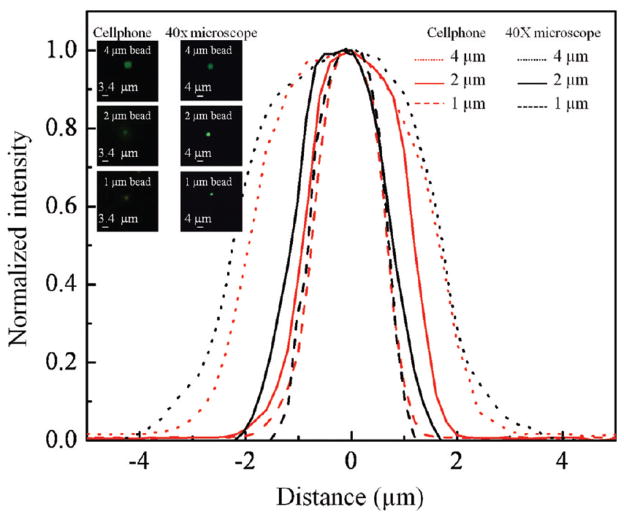
We also tested the spatial resolution of our optofluidic design by imaging static fluorescent objects. By changing the external lens of our attachment to the cell phone to a focal length of ~0.6 mm, a geometrical magnification of ~7.8× was achieved, such that the effective pixel size at the sample plane was <0.3 μm. The system spatial resolution was characterized using green fluorescent beads with various sizes including 4, 2, and 1 μm. Figure 4, top row, illustrates the imaging performance of our cell-phone-based optofluidic fluorescent microscope for several sets of beads. For comparison purposes, the same beads were also imaged by a conventional benchtop fluorescent microscope using a 40× (NA = 0.65) microscope objective as shown in Figure 4, bottom row. On the basis of Figure 4, parts B-1 and C-1, we can easily resolve two fluorescent beads that are separated by 4 μm (center to center). In Figure 4, part E-1, two beads with a center-to-center distance of 2 μm are also successfully resolved by our cell-phone fluorescent microscope. This spatial resolution level is also validated through cross-sectional profiles of isolated 1 μm fluorescent particles as illustrated in Figure 5, which illustrates a full width at half-maximum (fwhm) of ~1.8 μm.
Figure 5.
Cross-sectional profiles of 4, 2, and 1 μm fluorescent beads obtained using our optofluidic cell-phone fluorescent microscope (red lines) and a benchtop fluorescent microscope using a 40× objective lens (black line) are provided. These cross-sectional profiles are obtained from the inset bead images.
And finally, the captured fluorescent flow data can be digitally analyzed on modern smart phones and tablets, on a local computer (e.g., an inexpensive laptop), or at a remote PC located in, e.g., a hospital or a clinic. Depending on the setting and the local resources, one approach can be preferred over the other.
CONCLUSION
In conclusion, we demonstrated, for the first time, the integration of optofluidic fluorescent microscopy and flow cytometry on a cell phone using a compact, lightweight, and cost-effective attachment to the existing camera unit of the cell phone. In this cell-phone-based optofluidic imaging cytometer, fluorescently labeled particles/cells are continuously delivered to our imaging volume through a disposable microfluidic chip that is placed above the existing camera unit of the cell phone. The same microfluidic device also acts as a multilayered optofluidic wave-guide and efficiently guides our excitation light, which is butt-coupled from the side facets of our microfluidic channel using inexpensive LEDs. We tested the performance of our cell-phone-based imaging cytometer by measuring the density of WBCs in whole blood samples, providing a good match to a commercially available hematology analyzer. We further characterized the imaging performance of our platform to demonstrate a fluorescent resolution of ~2 μm. This cell-phone-enabled optofluidic imaging flow cytometer could especially be useful for rapid and sensitive imaging of bodily fluids for, e.g., conducting various cell counts or for screening of water quality in resource-limited locations.
Supplementary Material
Footnotes
ASSOCIATED CONTENT
Supporting Information. Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1. [Accessed on August, 20, 2010];International Telecommunication Union, Market Information and Statistics. 2010 http://www.itu.int/ITU-D/ict/statistics/index.html.
- 2.Wurm EMT, Hofmann-Wellenhof R, Wurm R, Soyer HP. J Dtsch Dermatol Ges. 2008;6:106–112. doi: 10.1111/j.1610-0387.2007.06440.x. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann-Wellenhof R, Salmhofer W, Binder B, Okcu A, Kerl H, Soyer HP. J Telemed Telecare. 2006;12(Suppl 1):15–17. doi: 10.1258/135763306777978407. [DOI] [PubMed] [Google Scholar]
- 4.Pamplona VF, Mohan A, Oliveira MM, Raskar R. Dual of Shack-Hartmann Optometry Using Mobile Phones. Proceedings of Frontiers in Optics, Adaptive Optics for the Eye (FTuB); Oct. 24, 2010; Rochester, New York. [Google Scholar]
- 5.Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthews D, Wachsmann-Hogiu S. PLoS One. 2011;6:e17150. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. PLoS One. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng D, Mudanvali O, Oztoprak C, Iskiman SO, Sencan I, Yaglidere O, Ozcan A. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Yaglidere O, Su TW, Tseng D, Ozcan A. Lab Chip. 2010;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro HM. Practical Flow Cytometry. 4. Wiley-Liss; Hoboken, NJ: 2003. [Google Scholar]
- 10.Nunez R. Flow Cytometry for Research Scientists: Principles and Applications. Horizon Press; Wymondham, U.K: 2001. [Google Scholar]
- 11.Owens MA, Loken MR. Flow Cytometry: Principles for Clinical Laboratory Practice. Wiley-Liss; New York: 1995. [Google Scholar]
- 12.Givan A. Flow Cytometry: First Principles. Wiley-Liss; New York: 2001. [Google Scholar]
- 13.Mcdonald JC, Whitesides GM. Acc Chem Res. 2001;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt O, Bassler M, Kiesel P, Knollenberg C, Johnson N. Lab Chip. 2007;7:626–629. doi: 10.1039/b618879f. [DOI] [PubMed] [Google Scholar]
- 15.Kiesel P, Beck M, Johnson N. Cytometry, Part A. 2011;79A:317–324. doi: 10.1002/cyto.a.21042. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki S, Abe K. Comput Vision Graphics Image Process. 1985;30:32–46. [Google Scholar]
- 17.Heikali D, Di Carlo D. JALA. 2010;15:319–328. [Google Scholar]
- 18.Osei-Bimpong A, Jury C, Mclean R, Lewis SM. Int J Lab Hematol. 2009;31:657–664. doi: 10.1111/j.1751-553X.2008.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao LV, Ekberg BA, Connor D, Jakubiak F, Vallaro GM, Snyder M. Clin Chim Acta. 2008;389:120–125. doi: 10.1016/j.cca.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. PLoS Med. 2005;2:e182. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Lab Chip. 2008;8:2079–2090. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng X, Gupta A, Chen C, Tompkins RG, Rodriguez W, Toner M. Lab Chip. 2009;9:1357–1364. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng X, Irimia D, Dixon M, Ziperstein JC, Demirci U, Zamir L, Tompkins RG, Toner M, Rodriguez W. JAIDS, J Acquired Immune Defic Syndr. 2007;45:257–261. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Chin SY, Chin CD, Sarik J, Harper M, Justman J, Sia SK. Anal Chem. 2010;82:36–40. doi: 10.1021/ac902144w. [DOI] [PubMed] [Google Scholar]
- 26.Holems D, Pettigrew D, Reccius CH, Gwyer JD, van Berkel C, Holloway J, Davies DE, Morgan H. Lab Chip. 2009;9:2881–2889. doi: 10.1039/b910053a. [DOI] [PubMed] [Google Scholar]
- 27.Holmes D, Morgan H. Anal Chem. 2010;82:1455–1461. doi: 10.1021/ac902568p. [DOI] [PubMed] [Google Scholar]
- 28.Watkins NN, Sridhar S, Cheng X, Chen GD, Toner M, Rodriguez W, Bashir R. Lab Chip. 2011;11:1437–1447. doi: 10.1039/c0lc00556h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang JH, Wang CH, Lin CC, Lei HY, Lee GB. Microfluid Nanofluid. 2011;10:531–541. [Google Scholar]
- 30.Bland JM, Altman DG. Lancet. 1986b;1:307–310. [PubMed] [Google Scholar]
- 31.Li P, Tian Y, Pappas D. Anal Chem. 2011;83:774–781. doi: 10.1021/ac102975g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.