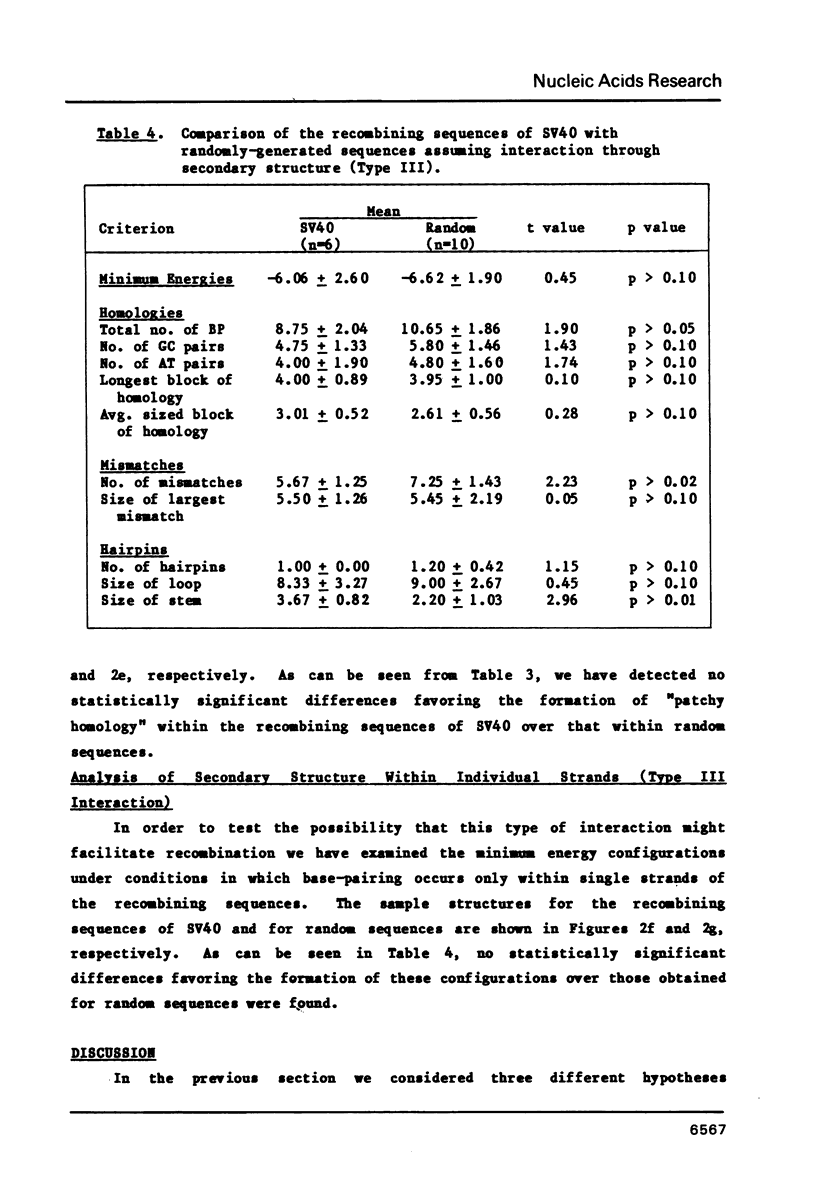
Abstract
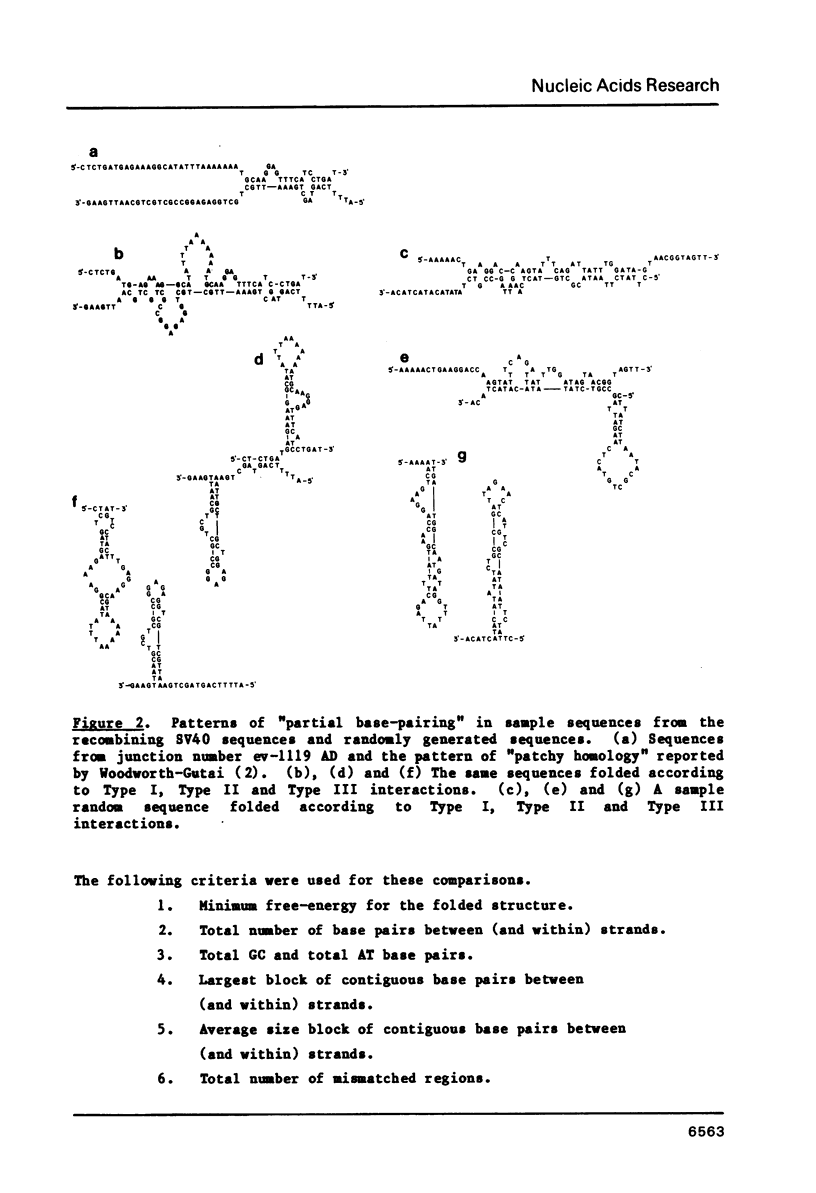
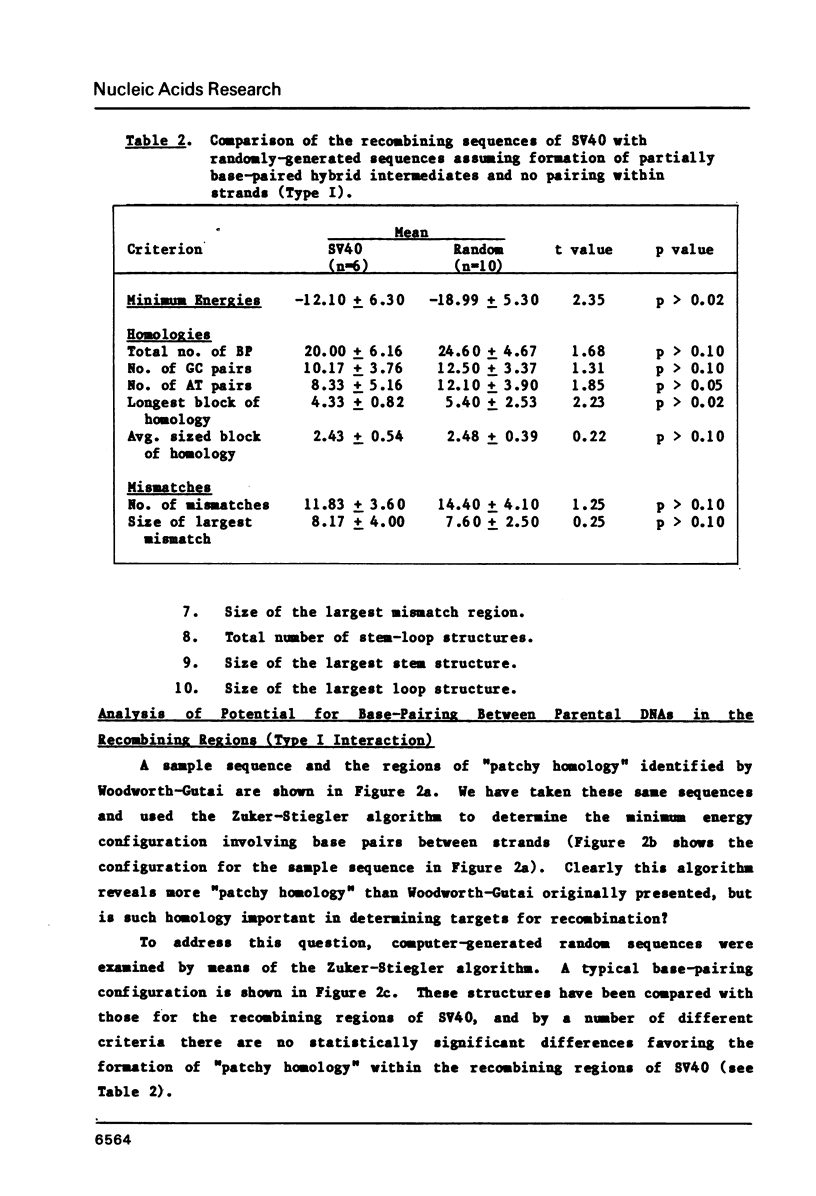
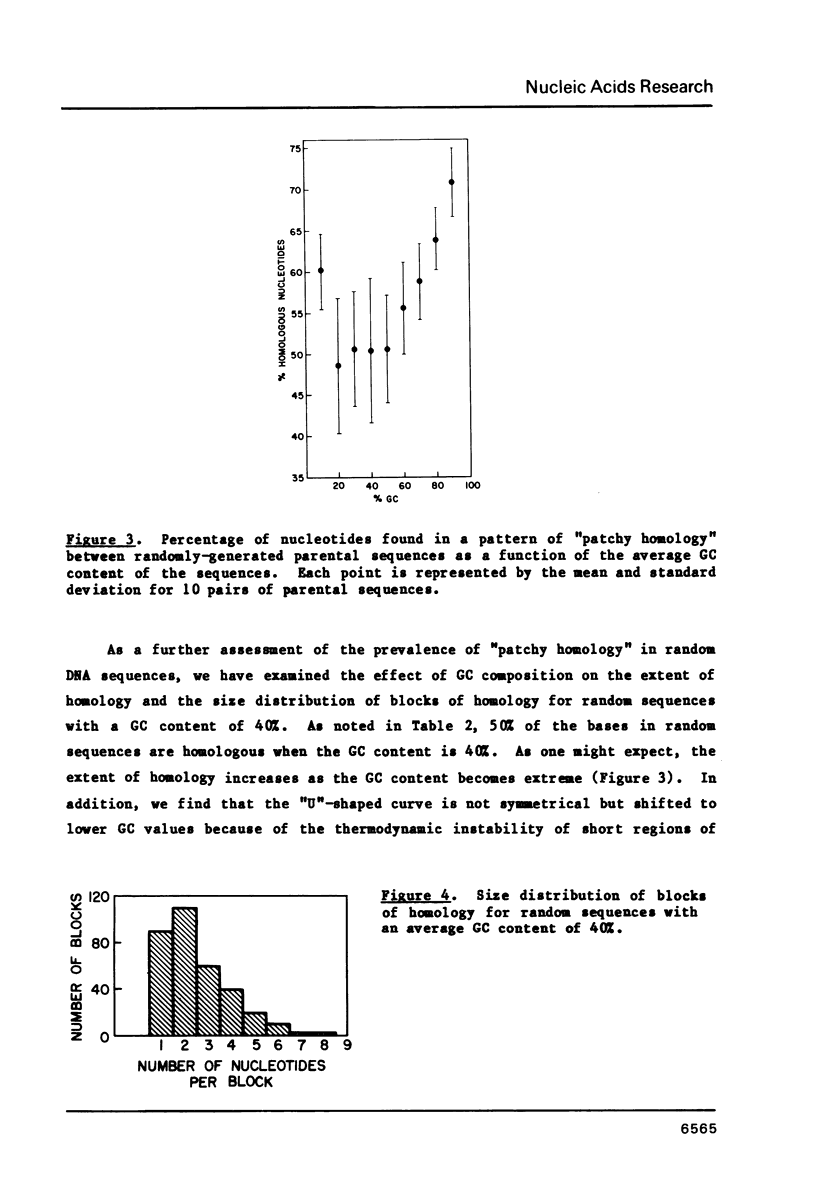
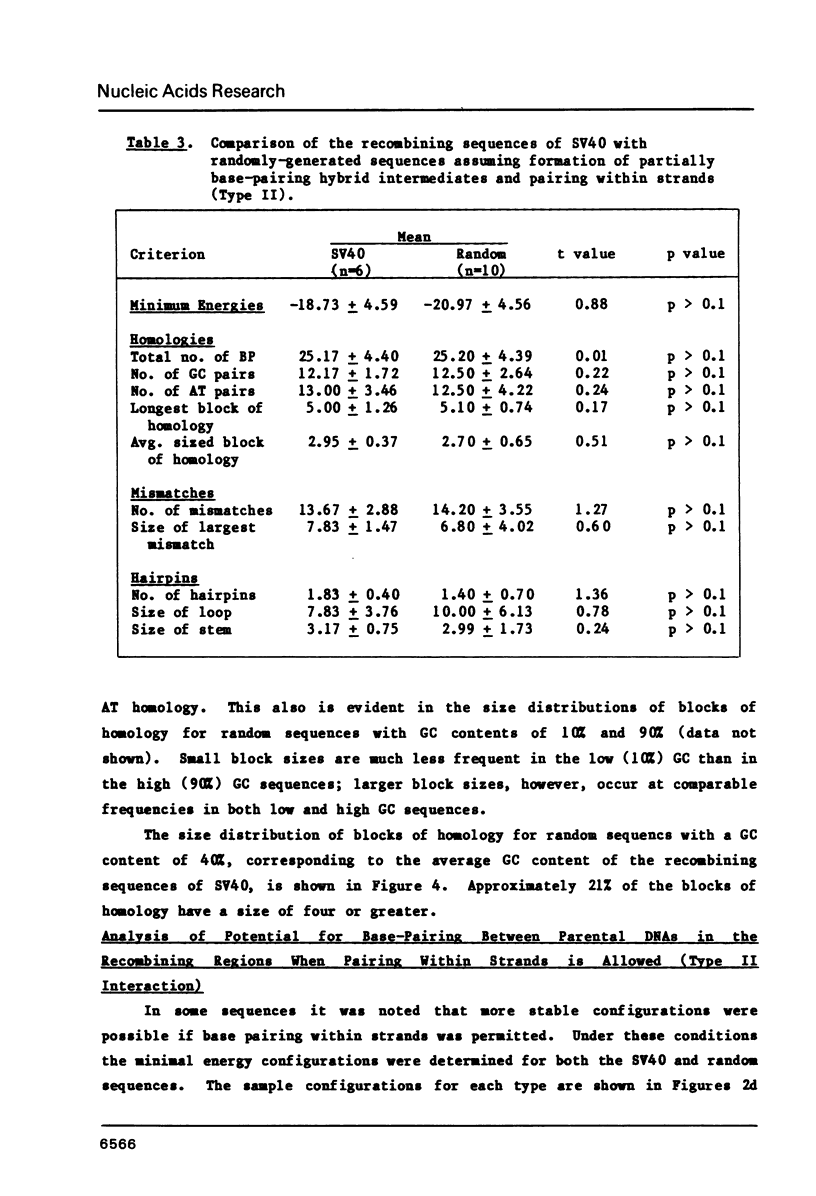
The frequent detection of "patchy homology" between recombining DNAs in eukaryotic systems suggests that partial sequence homology might facilitate formation of partially base-paired hybrid structures and thus define a specific target for recombination. Indeed, the extent of such "patchy homology" initially appears impressive. The question of whether such homology is statistically significant, however, has not been addressed. In this paper we compare the extent of "patchy homology" within the sequence of SV40 recombination sites and within randomly-generated sequences with the same average GC content. We have found no statistically significant differences favoring the existence of "patchy homology" within the recombining regions of SV40. On the average 50% of the bases in randomly-generated sequences with the same GC content can be paired in a pattern of "patchy homology". We have also assessed the ability of sequences near recombination junctions in SV40 to form partially base-paired intra-strand secondary structures. Again, the ability of these sequences to form such configurations was unremarkable when compared with random sequences. Thus, the notion that partial base-pairing provides specificity for sites of recombination must be considered with caution.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender M. A., Christensen J., Brockman W. W. Characterization of a T-antigen-negative revertant isolated from a mouse cell line which undergoes rearrangement of integrated simian virus 40 DNA. J Virol. 1983 Jul;47(1):115–124. doi: 10.1128/jvi.47.1.115-124.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):709–719. doi: 10.1101/sqb.1979.043.01.079. [DOI] [PubMed] [Google Scholar]
- Brockman W. W. Evolutionary variants of simian virus 40. Prog Med Virol. 1977;23:69–95. [PubMed] [Google Scholar]
- Graessmann A., Graessmann M., Topp W. C., Botchan M. Retransformation of a simian virus 40 revertant cell line, which is resistant to viral and DNA infections, by microinjection of viral DNA. J Virol. 1979 Dec;32(3):989–994. doi: 10.1128/jvi.32.3.989-994.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Jahn C. L., Hutchison C. A., 3rd, Phillips S. J., Weaver S., Haigwood N. L., Voliva C. F., Edgell M. H. DNA sequence organization of the beta-globin complex in the BALB/c mouse. Cell. 1980 Aug;21(1):159–168. doi: 10.1016/0092-8674(80)90123-3. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- McCutchan T., Singer M., Rosenberg M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. II. The structure of variant 1103 and its comparison to variant CVPS/1P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3592–3597. [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamiya T., McCutchan T., Rosenberg M., Singer M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. I. The structure of variant CVPS/1/P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3584–3591. [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: nucleotide sequences at viral-viral recombinant joints in naturally arising variants. Virology. 1981 Mar;109(2):344–352. doi: 10.1016/0042-6822(81)90505-5. [DOI] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: viral DNA sequences at sites of circularization of transfecting linear DNA. Virology. 1981 Mar;109(2):353–365. doi: 10.1016/0042-6822(81)90506-7. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



