Abstract
Enzyme-responsive protein nanocapsules are synthesized to release their protein cargoes in response to specific enzymes secreted in certain cellular events not only with specificity but also with controlled rate by composition tuning. The unique nanocapsule structures protect the encapsulated proteins with robustness against reacting reaction system, providing a new direction towards responsive protein delivery according to specific cellular events or local environment.
Keywords: nanocapsule, responsive delivery, protein delivery
Biological systems rely on synergic actions of proteins and other biomolecules to maintain their normal functions. Delivering proteins to replace the malfunctioned ones or to direct or regulate normal biological functions holds great promises for a broad spectrum of applications ranging from therapeutics, tissue engineering to other areas[1]. To date, although various delivery approaches have been established, developing the capability to deliver proteins according to local environmental changes (e.g. pH, [2][3] glucose[4] and enzyme concentrations[5] [6] ) has been of great demand but remains highly challenging.
It is known that biological system often secret specific enzymes in response to certain cellular events such as injury or disease. [7][8] If such enzymes can be used to trigger a delivery system to release its protein cargoes, proteins may be delivered effectively based on specific cellular events or environmental changes. Moreover, enzymes are generally highly specific and secreted with precisely spatial and temporal control; such an enzyme-responsive delivery may allow protein delivery with spatial and temporal control, opening huge opportunities for tissue engineering and other applications. However, despite of such advantages, the current state of the art for protein delivery from hydrogel scaffolds mainly relies on attaching protein-containing polymer microspheres to hydrogel scaffolds or directly immobilizing protein covalently or electrostatically to hydrogel scaffolds. [9][10]) The use of enzymatic action for protein delivery is limited to bulk hydrogels containing enzyme-responsive linkers, which can be cleaved off by specific enzymes and release their protein cargoes. [10] [11] [12] Note that nano-hydrogels have been synthesized using microemulsion polymerization for general purpose of drug delivery. [13] [14] Generally, enzyme-responsive linkers may also be incorporated within the nano-hydrogels to provide enzyme-responsive delivery capability. However, the synthesis of such nano-hydrogels was generally achieved in reaction media containing significant amount of organic solvent and surfactant, forbidding effective incorporation of active proteins within such nano-hydrogels.
Herein, we report a novel enzyme-responsive delivery platform with controlled-releasing capability and specificity based on protein nanocapsules illustrated in Scheme 1. In this work, bovine serum albumin (BSA) and vascular endothelial growth factor (VEGF) were used as the model proteins; VEGF has been proven to be specific and critical for angiogenesis. [15] It has also been proven that matrix metalloproteinases (MMP) [16] and serine proteases, such as plasmin, [17][18] are generally up-regulated in diseased or injured tissues. Using such proteases as the trigger, we demonstrated here controlled release of VEGF from the nanocapsules to induce and guide blood vessel formation.
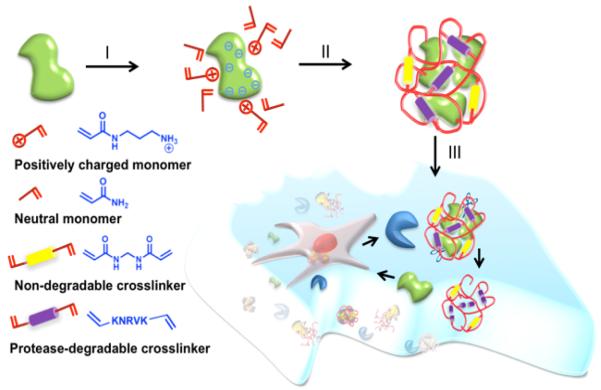
Scheme 1.
Schematics of synthesis of the nanocapsules and their controlled release process: (I) enriching the monomers and crosslinkers around the protein surface, (II) forming a polymer shell around the protein molecules by in-situ polymerization leading to the formation of protein nanocapsules, and (III) degrading of the polymer shell by protease releasing their protein cargo.
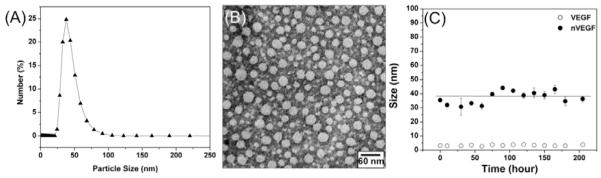
The synthesis of the nanocapsules was achieved using a simple encapsulating technique as illustrated in Scheme 1. We started from buffer solutions containing positively charged monomer (N-(3-aminopropyl) methacrylamide (APM), neutral monomer (acrylaminde (AAM)), non-degradable and protease-degradable crosslinkers. Electrostatic and hydrogen-bonding interactions enrich the monomers and crosslinkers around the protein molecules (Step I). In-situ free-radical polymerization forms a thin polymer layer around the proteins, forming the protein nanocapsules (denoted as nVEGF and nBSA, respectively) with controlled composition (Step II). [19] Tuning the ratio of the positive-charged and the neutral monomers allows the control of nanocapsule surface charge; while turning the ratios of the degradable peptide and non-degradable crosslinkers allows the synthesis of nanocapsules with tunable degradability. Upon exposed to proteases, the peptide crosslinkers are cleaved off and release their protein cargo with controlled release rate (Step III). Figure 1B shows a representative transmission electron microscopic (TEM) image of the nanocapsules prepared with a VEGF/monomer ratio of 1:6000 and an APM: AAM: degradable crosslinker molar ratio of 5/5/1. These nanocapsules exhibit a zeta potential of +1.4 mV and diameters ranging from 20 to 45 nm, which is consistent with the dynamic light scattering (DLS) measurement (25~60 nm) (Figure 1A). The nanocapsule is expected to have multiple proteins per nanocapsule; however, it is very difficult to measure the exact number of protein molecules within each capsules. Note that surface charge of the nanocapsules can be readily tuned from −7 to +5 mV by adjusting the ratios of the positively charged monomer to other building molecules used (Table S1). The stability of these nanocapsules was examined by monitoring their size variation in PBS buffer at 4 °C (Figure 1C). Similar to the native protein, these nanocapsules with an average diameter of 35 nm exhibit no obvious aggregation in 8 days, indicating an excellent stability in solution.
Figure 1. Characterization of nVEGF.
A, Size of the VEGF nanocapsules (nVEGF) synthesized at VEGF/monomer (V/M) molar ratio of 1:6000. B, Transmission electron microscopic (TEM) image of nVEGF synthesized at V/M molar ratio of 1:6000. C, Stability of a 35-nm-size nVEGF monitored by DLS in pH 7.4 PBS buffer at 4°C.
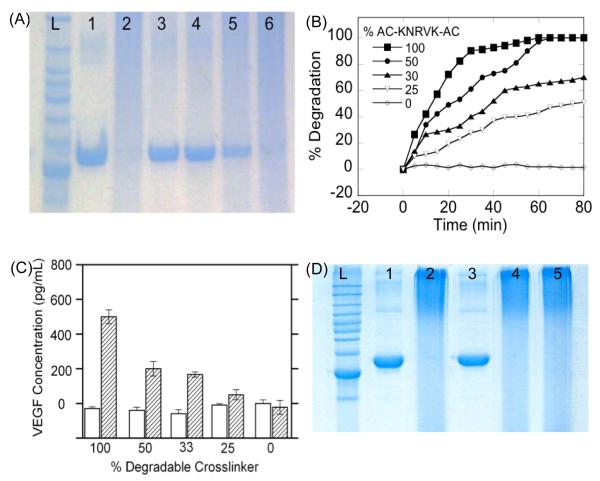
The release of the protein from the nanocapsules can be readily controlled by the ratios of degradable crosslinkers to non-degradable crosslinker. In this work, plasmin-cleavable crosslinker was synthesized by bisacryloylating a plasmin-specific peptide Lys-Asn-Arg-Val-Lys (KNRVK); [20] while N, N’-methylene bisacrylamide was chosen as the non-degradable crosslinker. Figure 2A shows the SDS-PAGE image of (1) native BSA, (2) nBSA prepared with 100% degradable crosslinker without plasmin treatment, and (3-6) nBSA prepared with 100, 50, 33, 25, and 0% degradable crosslinkerafter treating with 0.2 mg/mL plasmin for 30 min. Compared with the native BSA exhibiting a well-defined band (line 1), the nBSA is broadly dispersed within the gel, possibly because of their dispersive size and charge distribution (line 2). Exposing the nanocapsules to plasmin degrades the polymer shell, leading to a subsequent release of the BSA that exhibits bands (line 3-5) similar as that of native BSA (line 1). As expected, nBSA prepared using the non-degradable crosslinker (line 6) exhibits similar pattern as that of the degradable one prior to exposing to plasmin (line 2). This work demonstrates the feasibility to encapsulate proteins within the nanocapsules and release them responsively by enzymatic degradation of the nanocapsules.
Figure 2. VEGF nanocapsules (nVEGF) control and release.
A, SDS-PAGE of the degradation of BSA nanocapsules prepared with different ratios of degradable and non-degradable crosslinkers with and without 30 min plasmin treatment (left to right) : ladder; (1) native BSA; (2) nBSA before plasmin treatment; (3) 100% nBSA; (4) 50% nBSA; (5) 33% nBSA and (6) 0% nBSA. B, Degradation rates of nVEGF prepared with different percentages of degradable crosslinker after 30 min plasmin treatment. C, Concentrations of VEGF released from the nVEGF prepared with different percentages of degradable crosslinkers with (shadow) and without (blank) the presence of plasmin. D SDS-Page of the specific degradation of 100% nBSA (left to right): ladder; (1) BSA; (2) nBSANRV, (3) nBSANRV+ 0.2 mg/mL plasmin, (4) nBSANRV+ 50 unit/mL collagenase; (5) nBSALGPA+ 0.2 mg/mL plasmin.
The degradation kinetics of the nanocapsules can be quantified using the DLS technique. Using nVEGF as an example, Figure 2B shows the degradation profiles of nVEGF prepared with different ratios of degradable crosslinkers in the presence of plasmin. The initial shell thickness of the nVEGF can be estimated from R0-RVEGF, where R0 is the initial diameter of nVEGF and RVEGF is the diameter of a VEGF molecule (~ 4nm). The degree of degradation can be described as [R0-Rt]/[R0-RVEGF]*100%, where Rt is the time-depended nVEGF diameters. nVEGF prepared with 100% degradable crosslinkers was degraded completely after 40 min, while those prepared with 0% degradable crosslinker, no degradation was observed. Note that the slopes of the degradation profiles consistently increase with the % of degradable crosslinkers, confirming increasing degradation rates with increasing percentage of the degradable crosslinkers.
Consistent with the degradation kinetics, as-encapsulated VEGF can be released with controlled rates, which were determined using enzyme-linked immunosorbent assay (ELISA). It was found that more that 90% of VEGF was encapsulated within the capsules (Figure S1-2). Figure 2C shows the concentrations of VEGF released from the nVEGF prepared with different percentages of degradable crosslinkers with and without the presence of plasmin. In the absence of plasmin, no free VEGF was detected in all nVEGF samples, suggesting that no VEGF was released and VEGF antibody could not bind to nVEGF. In the presence of plasmin, nVEGF degradation leads to release of their encapsulated VEGF. As expected, the amount of VEGF released systematically increases with increasing percentage of degradable crosslinker used during the nanocapsule synthesis, indicating that the rate of VEGF release can be modulated effectively. It is worth pointing out that nVEGF formed with 100% degradable crosslinker released 95% of the VEGF used to form the nanocapsules, indicating that VEGF can be effectively encapsulated and released using this technology.
This method can be generalized for protein delivery not only with rate control but also with specificity to the enzyme present. To demonstrate this concept, nBSA containing plasmin-cleavable crosslinker KNRVK (denoted as nBSANRV) or MMP-cleavable crosslinker KLGPAK (Lys-Leu-Gly-Pro-Ala-Lys) (denoted as nBSALGPA) were used as the model system. Plasmin, a serine protease in blood, is commonly secreted by tissue cells during the formation of vessels, while MMP plays an important role in tissue remodeling to degrade extracellular matrix proteins during the angiogenesis. [21] [16] Figure 2D shows SDS-page image of native BSA, nBSANRV and nBSALGPA without and with exposing to plasmin or collagenase [22] (a bacterial equivalent of MMP with capability to degrade MMP sensitive peptides) for 30 min. Compared to the characteristic band of native BSA (line 1), nBSANRV without exposing to plasmin (line 2) or after exposing to collagenase (line 4) does not show any well-defined band, but does exhibit a well-defined band (line 3) similar to that of native BSA upon exposing to plasmin. This observation clearly suggests the release of BSA is highly specific to the enzyme exposed and confirms a feasibility to deliver protein with enzyme specificity. Similarly, exposing nBSALGPA to plasmin (line 5) results in broad dispersion in the gel, confirming that plasmin is incapable to trigger release of BSA from MMP-specific nBSALGPA; as expected, exposing nBSALGPA to collagenase results in specific degradation of the polymer shells, which is confirmed by DLS study (Figure S2). This study indicates that the specific enzyme-degradation of nanocapsules can be achieved by choosing judicious choice of enzymes and particular peptide crosslinkers with specific sequences.
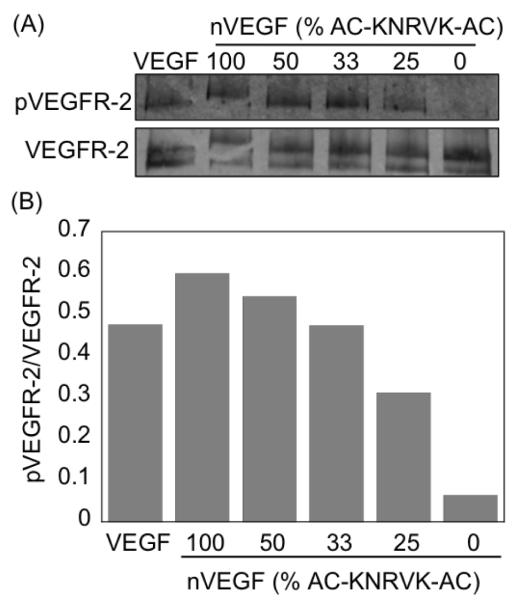
To further confirm the bioactivity of the encapsulated VEGF, the ability of the released VEGF to phosphorylate its receptor was assessed and compared with native VEGF. VEGF nanocapsules with different percentages of the plasmin degradable crosslinkers (100% to 0%) were exposed to plasmin for 15 min and added to confluent HUVECs cells for another 5 min. A western blot of the exposed cells was run and probed for phosphorylated VEGFR-2 as well as the total VEGFR-2 (Figure 3A). Clearly, all nVEGF that contained the degradable crosslinker showed receptor phosphorylation, indicating that the VEGF released from the nanocapsules was active. Further, these data show that the nanocapsules were degraded to a sufficient extent by plasmin to release their VEGF cargoes. Care was taken to expose the cells to the same amount of VEGF for all the conditions. Figure 3B shows a histogram of normalized pVEGFR-2-band intensities vs. that of total VEGFR-2 for each sample (IpVEGFR-2/IVEGFR-2). Native VEGF and the 100% degradable nVEGF show a similar level of receptor phosphorylation, indicating the nanocapsules well preserve the VEGF activity. Further, nVEGF prepared without the plasmin degradable crosslinker (0%) shows a ratio less than 0.1, confirming the degradation of the nanocapsules is required for receptor phosphorylation. This finding further confirms the possibility to controllably release growth factors from the nanocapsules.
Figure 3. Activity of released VEGF from nanocapsules.
A, Activity of released VEGF tested by adding to HUVEC cells and running western blotting. B, Analysis of the western blotting based on the band intensities showing phosphorylated percentage of pVEGF-2.
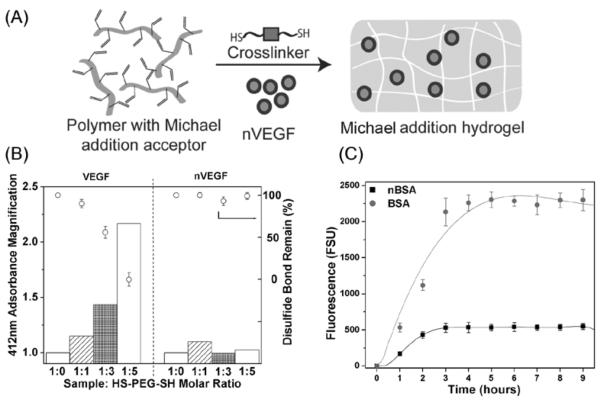
Such protein nanocapsules with highly retained activity, controlled released capability and specificity are of great interest for a board range of applications ranging from therapeutics to tissue engineering. For example, as illustrated in Figure 4A, hydrogel scaffolds are commonly synthesized by reacting a polymer containing vinyl groups with crosslinkers containing dithiol groups, during which Michael addition reaction between the vinyl and thio groups effectively crosslinks the polymer into a hydrogel. [23] Proteins including growth factors can be directly added to the mixture of the polymer and crosslinker and incorporated within the hydrogels[10] [12][24].Although such hydrogels are extensively used for tissue regeneration application, the crosslinkers may react with disulfide bridges of the proteins, resulting in structural miss-folding and loss of activity. Since the nanocapsules contain the protective shells that prevent the encapsulated protein from reacting with the crosslinkers, the nanocapsules can be effectively incorporated within the hydrogel with better-retained activity.
Figure 4. Protection of nanocapsules.
A, Scheme of hydrogel formation by Michael addition reaction between bis-cysteine containing crosslinkers and vinyl-group-containing polymers in the presence of nVEGF. B, Percentages of disulfide bonded remained and normalized adsorption intensities of VEGF and nVEGF after expose to HS-PEG-SH and DTNB vs those exposed to DTNB alone at different VEGF/HS-PEG-SH and nVEGF/HS-PEG-SH molar ratios. C, Fluorescence intensities of PBS buffer after placing HA hydrogels that incorporated 3.5 pmol FITC-labeled BSA or its nanocapsules (nBSA) in 400μL buffer solution at different times.
To demonstrate the enhanced stability of the encapsulated VEGF, native VEGF and nVEGF were incubated with crosslinker HS-(CH2CH2O)75-SH (HS-PEG-SH) for 30 min, allowing exchange reaction between the disulfide bonds within the VEGF with the thiol groups of the crosslinker molecules. Such an exchange reaction would result in increasing number of thiol groups within the VEGF, which can be quantified using the Ellman’s test. In a typical assay, as generated thiol groups were reacted with 5,5′-Dithio-bis(2-nitrobenzoic acid) (DTNB) and stoichiometric released 2-nitro-5-thiobenzoate ions with a characteristic absorbance at 412 nm. Figure 4B compares the relative absorbance intensity after incubating native VEGF and nVEGF (made with 100% degradable crosslinker) with different amounts of HS-PEG-SH for 30 min followed by addition of DTNB. For the native VEGF, the absorbance increases systematically with increasing the amount of HS-PEG-SH used, suggesting increasing degree reaction between the VEGF disulfide bonds with the crosslinker molecules. The disulfide bonds survived from the reaction rapidly decrease; at VEGF/HS-PEG-SH molar ratio of 1/5, all the disulfide bonds were breakdown. In contrast, the absorbance for nVEGF remains constant and the disulfide bonds were retained at all nVEGF/HS-PEG-SH ratios, suggesting the encapsulated VEGF is immune from the crosslinker attack. This study unambiguously proves that encapsulating proteins within the nanocapsules can effectively prevent such inactivating reactions, providing a novel approach for the design and fabrication of regenerative bio-scaffolds.
Furthermore, rapid leaching of the incorporated proteins from hydrogels has been a challenge that is required to be addressed. [25] One may consider decreasing the pore size of the hydrogels to decrease or avoid leaching of incorporated proteins. Nevertheless, to ensure cellular growth and nutrient diffusion into the hydrogels from surrounding media, hydrogels with large enough pore size are often required, which inevitably causes leaching of incorporated protein. Fortunately, the protein nanocapsules described here generally contain residual vinyl groups on their surface, which can be immobilized to the hydrogel network through the Michael addition reaction with the thio-containing crosslinkers. Such an immobilization process effectively prevents leaching of the nanocapsules. To prove this concept, fluorescence-labeled BSA and BSA nanocapsules were incorporated within the HA hydrogels, which were then placed in PBS buffer for different time. Figure 4C compares fluorescent intensities of the BSA or nBSA leached into the buffer solution. Clearly, fluorescent intensity of the leached BSA rapidly increases with time, which is much higher than those of the leached nBSA. This study suggests that using protein nanocapsules rather than native ones can effectively reduce leaching of the protein from the hydrogels.
In conclusion, we have demonstrated a novel class of protein nanocapsules that can release the encapsulated proteins with controllable rate and specificity to specific enzyme. This work provides a new direction towards responsive protein delivery according to specific cellular events or local environment, which are of great importance for therapeutic and other applications. Furthermore, the unique nanocapsule structures protect the encapsulated proteins with robustness against reacting reaction system, enabling the synthesis of a series of hydrogel scaffolds for broad regenerative tissue applications.
Experimental
Synthesis of nanocapsules
250 μg of protein bovine serum albumin (BSA) was dissolved into 500 μL of 10 mM pH 8.5 sodium bicarbonate buffer. Firstly, the positively charged monomer N-(3-Aminopropyl) methacrylamide (APmTAAm), prepared in a 10 mg/mL aqueous solution, was added into protein solution with stirring for 10 min at 4°C. Then the acrylamide (AAm) monomer and crosslinkers (combination of bisacryloylated Lys-Asn-Arg-Val-Lys (KNRVK) peptide and N,N’-methylene bisacrylamide (BIS)) were added to protein solution with stirring sequentially. The molar ratio of AAm/ APM/ crosslinker was adjusted to 5/5/1. Radical polymerization was initiated by adding both ammonium persulfate (1/10 molar ratio of total monomers) dissolved in deionized water and the same volume of 10% N,N,N’,N’-tetramethylethylenediamine into reaction solution. The size and zeta potential of nanocapsules were tuned by adjusting the ratios of protein to monomers used. The polymerization was allowed to proceed for 90~120 min in a nitrogen atmosphere at 4°C. Finally, unreacted monomers, crosslinker and initiators were removed by dialysis in 10 mM pH 7.4 phosphate buffer. Synthesis of growth factor vascular endothelial growth factor (VEGF) nanocapsules was similar to that of BSA nanocapsules. Ten times higher amount of monomers was required when the reaction volume was smaller than 50 μL and final protein concentration was lower than 0.05 μg/ μL. Detailed information regarding nanocapsule synthesis and characterization are provided as Supplementary Information.
Supplementary Material
Acknowledgements
This work was partially supported by DTRA program (YL), the support of the Josson Comprehensive Cancer center (TS), and 1R21EB009516-01A1 (TS).
References
- [1].Golan DE, Leader B, Baca QJ. Nat Rev Drug Discov. 2008;7:21. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- [2].Irvine DJ, Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N. Nano Lett. 2007;7:3056. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- [3].Irvine DJ, Hu YH, Atukorale PU, Lu JJ, Moon JJ, Um SH, Cho EC, Wang Y, Chen JZ. Biomacromolecules. 2009;10:756. doi: 10.1021/bm801199z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kataoka K, Miyazaki H, Bunya M, Okano T, Sakurai Y. J Am Chem Soc. 1998;120:12694. [Google Scholar]
- [5].Anseth KS, Aimetti AA, Machen AJ. Biomaterials. 2009;30:6048. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Franssen O, Stenekes RJH, Hennink WE. J Control Release. 1999;59:219. doi: 10.1016/s0168-3659(98)00193-x. [DOI] [PubMed] [Google Scholar]
- [7].Powell JS, Clozel JP, Muller RKM, Kuhn H, Hefti F, Hosang M, Baumgartner HR. Science. 1989;245:186. doi: 10.1126/science.2526370. [DOI] [PubMed] [Google Scholar]
- [8].Ji LL. Med Sci Sport Exer. 1993;25:225. [PubMed] [Google Scholar]
- [9].Gopferich AM, Tessmar JK. Adv Drug Deliver Rev. 2007;59:274. doi: 10.1016/j.addr.2007.03.020. [DOI] [PubMed] [Google Scholar]
- [10].Ladewig K. Expert Opinion on Drug Delivery. 2011 doi: 10.1517/17425247.2011.588698. [DOI] [PubMed] [Google Scholar]
- [11].West JL, Moon JJ, Saik JE, Poche RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME. Biomaterials. 2010;31:3840. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Garcia AJ, Phelps EA, Landazuri N, Thule PM, Taylor WR. P Natl Acad Sci USA. 2010;107:3323. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kopelman R, Gao D, Xu H, Philbert MA. Angew Chem Int Edit. 2007;46:2224. doi: 10.1002/anie.200603927. [DOI] [PubMed] [Google Scholar]
- [14].Kopelman R, Gao D, Xu H, Philbert MA. Nano Lett. 2008;8:3320. doi: 10.1021/nl8017274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. J Cell Biol. 2003;161:1163. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Okada Y, Shiomi T, Lemaitre V, D’Armiento J. Pathol Int. 2010;60:477. doi: 10.1111/j.1440-1827.2010.02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Syrovets T, Simmet T. Cell Mol Life Sci. 2004;61:873. doi: 10.1007/s00018-003-3348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roth D, Piekarek M, Paulsson M, Christ H, Krieg T, Bloch W, Davidson JM, Eming SA. Am J Pathol. 2006;168:670. doi: 10.2353/ajpath.2006.050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yan M, Du JJ, Gu Z, Liang M, Hu YF, Zhang WJ, Priceman S, Wu LL, Zhou ZH, Liu Z, Segura T, Tang Y, Lu YF. Nat Nanotechnol. 2010;5:48. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- [20].Hubbell JA, West JL. Macromolecules. 1999;32:241. [Google Scholar]
- [21].Pepper MS. Arterioscl Throm Vas. 2001;21:1104. doi: 10.1161/hq0701.093685. [DOI] [PubMed] [Google Scholar]
- [22].Harrington DJ. Infect Immun. 1996;64:1885. doi: 10.1128/iai.64.6.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Segura T, Lei YG, Gojgini S, Lam J. Biomaterials. 2011;32:39. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lutolf MR, Weber FE, Schmoekel HG, Schense JC, Kohler T, Muller R, Hubbell JA. Nat Biotechnol. 2003;21:513. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- [25].Shireman PK, Hampton B, Burgess WH, Greisler HP. J Vasc Surg. 1999;29:852. doi: 10.1016/s0741-5214(99)70213-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.