Abstract
Dominant missense mutations in leucine-rich repeat kinase 2 (LRRK2) are the most common genetic causes of Parkinson disease (PD) and genome-wide association studies identify LRRK2 sequence variants as risk factors for sporadic PD. Intact kinase function appears critical for the toxicity of LRRK2 PD mutants, yet our understanding of how LRRK2 causes neurodegeneration remains limited. We find that most LRRK2 PD mutants abnormally enhance LRRK2 oligomerization, causing it to form filamentous structures in transfections of cell lines or primary neuronal cultures. Strikingly, ultrastructural analyses, including immuno-electron microscopy and electron microscopic tomography, demonstrate that these filaments consist of LRRK2 recruited onto part of the cellular microtubule network in a well-ordered, periodic fashion. Like LRRK2-related neurodegeneration, microtubule association requires intact kinase function and the WD40 domain, potentially linking microtubule binding and neurodegeneration. Our observations identify a novel effect of LRRK2 PD mutations and highlight a potential role for microtubules in the pathogenesis of LRRK2-related neurodegeneration.
INTRODUCTION
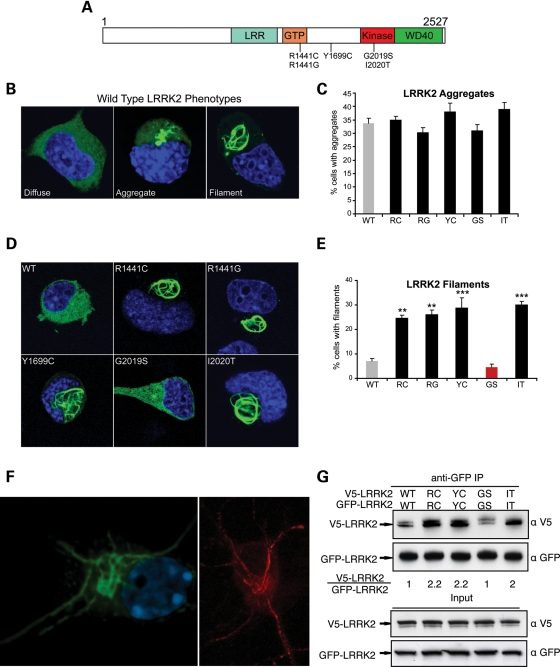
The currently understood biology of leucine-rich repeat kinase 2 (LRRK2) suggests that studies of this protein may provide new insights into neurodegeneration that are broadly relevant to sporadic Parkinson disease (PD) and amenable to therapeutic targeting. Genome-wide association studies demonstrate that common variation around the locus that encodes LRRK2 segregates with increased risk for PD (1,2), and missense mutations in LRRK2 cause a clinical and neuropathological syndrome that is indistinguishable from typical-appearing sporadic PD (3). LRRK2 contains GTPase and kinase domains, as well as leucine-rich repeat (LRR) and WD40 protein–protein interaction domains (Fig. 1A). Many potentially pathogenic sequence alterations have been identified in LRRK2, but only five missense mutations (Fig. 1A) clearly segregate with PD in large family studies (4). Two of these mutations (R1441G, R1441C) are located in the GTPase domain (termed Ras of complex proteins, or ‘Roc’ domain), a third (Y1699C) falls in a region between the GTPase and kinase domains (termed the C-terminal of Roc, or ‘COR’ domain) and two other mutations (G2019S and I2020T) are in the kinase domain.
Figure 1.
Multiple pathogenic mutations enhance LRRK2 oligomerization and filament formation in a kinase-dependent manner. (A) Domain structure and Parkinson's disease mutations of LRRK2. LRR, leucine-rich repeat; GTP, GTPase domain (also called Roc domain, Ras of complex proteins). Five PD-causing missense mutations are shown. (B–E) The formation of LRRK2 filaments is enhanced by multiple PD mutations. Cath.a-differentiated (CAD) cells were transfected with WT (B) or PD mutant forms (D; R1441C, R1441G, Y1699C, G2019S, I2020T) of GFP-tagged LRRK2. WT-LRRK2 adopted either a diffuse, aggregated or filamentous pattern of subcellular localization (B; labeled using anti-GFP antibody). The frequency of cells bearing LRRK2 aggregates or filaments was quantified 48 h after transfection (C and E). Multiple PD mutations increased the percentage of cells with LRRK2 filaments (labeled using anti-GFP antibody). Data are means ± SE of four to five independent experiments (**P< 0.01, ***P< 0.001, n.s., non-significant; ANOVA with Tukey's test). (F) Expression of LRRK2 in neuronal processes. Yellow fluorescent protein (YFP)-LRRK2-Y1699C was transfected into primary cortical neurons and imaged by confocal microscopy (left image, YFP fluorescence shown in green). A similar pattern of filament formation was observed in neurites in primary neurons transfected with untagged LRRK2-I2020T (right image, labeled using an anti-LRRK2 antibody shown in red). (G) WT LRRK2 oligomerizes and multiple LRRK2 PD mutations enhance its oligomerization. V5-LRRK2 was co-expressed in CAD cells with GFP or GFP-LRRK2. Lysates were immunoprecipitated with anti-GFP 48 h after transfection, and the immunoprecipitates were analyzed with anti-V5 and anti-GFP immunoblots. The oligomeric state of LRRK2 is shown as a relative ratio of co-purified V5-LRRK2 to GFP-LRRK2, normalized to the WT-LRRK2 ratio.
LRRK2 appears to exist as a dimer (5–7), and studies of fragments of LRRK2 or its prokaryotic homolog indicate that dimerization occurs in the Roc-COR region (5). Structural analyses of these fragments indicate that the R1441C PD mutation (or mutation of the analogous residue in the prokaryotic protein) can destabilize the dimer formed by these fragments (5,8). Yet, available data do not define the functional significance of LRRK2 self-association and it is unclear whether this property is altered in PD mutant forms of the full-length protein. One possibility is that LRRK2 self-association regulates its kinase activity, but only the G2019S mutation clearly increases kinase activity (by ∼3–5-fold), whereas the other mutations appear to have little or no effect on kinase function (9–13), at least in the in vitro assays used thus far. Intact kinase function does appear necessary for LRRK2 toxicity in vitro and in vivo (9,12,14). LRRK2-induced neurodegeneration of primary neuronal cultures is caspase-dependent, and may involve activation of the fas-associated protein with death domain (FADD)–caspase-8 pathway (15). One site of LRRK2 toxicity may be in neuronal processes, as overexpression of LRRK2 in vitro or in vivo causes neurite shortening, whereas the loss of LRRK2 function leads to increased neuron length and branching (11). The molecular details that underlie LRRK2 effects in neuronal processes are not clear, but may involve interactions with the cytoskeleton or cytoskeletal-related molecules (10,16–18).
Here, we provide evidence that four PD mutations (R1441C, R1441G, Y1699C, I2020T) cause LRRK2 to decorate microtubules in a well-ordered periodic fashion, as evidenced by immunofluorescence, immuno-electron microscopy (EM) and EM tomographic studies. Microtubule-associated LRRK2 appears as filamentous structures in transfected cell lines or primary neuronal cultures, and the frequency of these structures increases with microtubule stabilization (taxol) and decreases with microtubule dissolution (nocodazole). Similar to LRRK2-induced neurodegeneration, filament formation requires intact kinase function and the WD40 domain, potentially linking LRRK2-microtubule association to neurodegeneration. Our observations identify a novel effect of LRRK2 PD mutations, and provide a platform to further dissect LRRK2-related signals by identifying factors that modulate the interaction between LRRK2 and microtubules.
RESULTS
LRRK2 PD mutations enhance LRRK2 filament formation
In multiple cell lines (including Cath.a-differentiated (CAD), human embryonic kidney (HEK) and HeLa) transfected with wild-type (WT) LRRK2, we observed that LRRK2 adopted a primarily diffuse cytosolic distribution, but we also observed LRRK2-positive aggregates and a distinct pattern of filament formation (Fig. 1B). These patterns were observed with different epitope tags (e.g. GFP and V5) as well as with untagged protein (data not shown). To determine whether PD mutations alter the subcellular distribution of LRRK2, we quantified the frequency of these patterns (diffuse, aggregate or filament) for WT and all PD mutant alleles (Fig. 1C and E). This analysis was performed in the neuronal catecholaminergic CAD cell line because of the robust transfection efficiency possible with this system. The percentage of cells with punctate aggregates was not increased by any of the PD mutant alleles (Fig. 1C), so these structures may represent a non-specific effect of LRRK2 overexpression. In contrast, filament formation was significantly enhanced by all LRRK2 PD mutations that do not consistently enhance kinase function (9,10,13), whereas no effect on filament formation was observed for the single mutant, G2019S, that consistently and significantly increases kinase activity (Fig. 1D and E; Supplementary Material, Fig. S1; 9,10,13). Notably, steady-state protein levels of WT and PD mutant LRRK2 are similar (data not shown), so these filament-forming mutations do not appear to act by altering LRRK2 stability. Morphologically, these filaments are reminiscent of death-effector filaments formed by FADD, caspase-8, tumor necrosis factor receptor type 1-associated death domain protein and BCL10 (19–21). This observation intrigued us because all of these filament-forming proteins participate in cell death-related signaling, potentially providing a clue to LRRK2 function relevant to neurodegeneration. We found that LRRK2 filaments also occur in LRRK2-transfected primary neurons, both in the cell body and in neurites, and with both yellow fluorescent protein (YFP)-tagged as well as untagged protein (Fig. 1F).
For previously characterized filament-forming proteins, filament formation reflects a homotypic protein–protein interaction that is required for their normal signaling function (19,20,22). Oligomerization is also a signaling mechanism employed by RIP1 and RIP2 kinases (23), close phylogenetic relatives of LRRK2. These facts led us to hypothesize that the filaments represent enhanced LRRK2 oligomerization. To investigate whether LRRK2 oligomerizes, we co-expressed GFP- and V5-LRRK2 in CAD cells and tested whether they co-immunoprecipitate (co-IP). Lysates were immunoprecipitated with a GFP antibody and probed for associated V5-LRRK2. Differentially tagged WT-LRRK2 molecules did co-purify, indicating that LRRK2 can oligomerize (Fig. 1G). Moreover, LRRK2 oligomerization was clearly enhanced by filament-forming PD mutations (R1441C, Y1699C, I2020T), whereas the non-filament-inducing mutant (G2019S) did not differ from the WT-LRRK2 control (Fig. 1G). The oligomerization status of LRRK2 mutants therefore correlates with the level of filament formation, indicating that the cytosolic filaments are related to this feature of LRRK2 biology.
Because the LRRK2 filaments appear similar to cytoskeletal structures, we explored whether LRRK2 was templating onto existing cytoskeletal elements or forming novel ‘LRRK2 only’ structures. To determine which, if any, cytoskeletal structures LRRK2 may interact with, we examined cells transfected with FLAG-LRRK2-I2020T with both anti-FLAG and a panel of antibodies against the major cytoskeletal elements, as well as with mito-tracker, since mitochondria exist in filamentous strands. However, LRRK2 filament staining did not colocalize with any of these markers (data not shown).
LRRK2 decorates microtubules in an organized manner
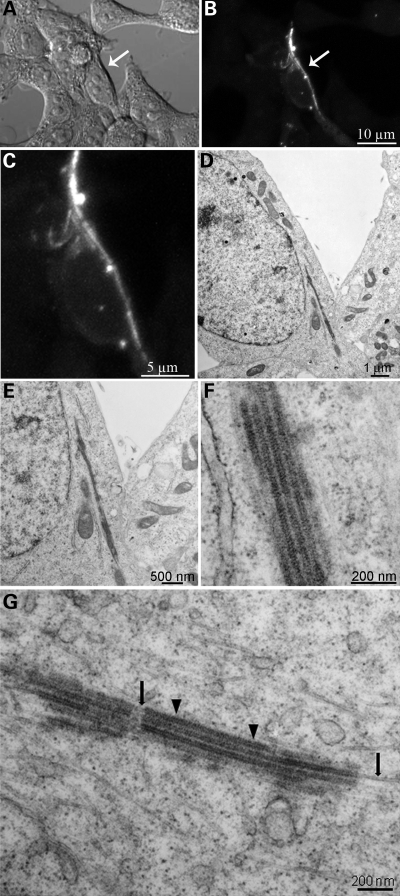
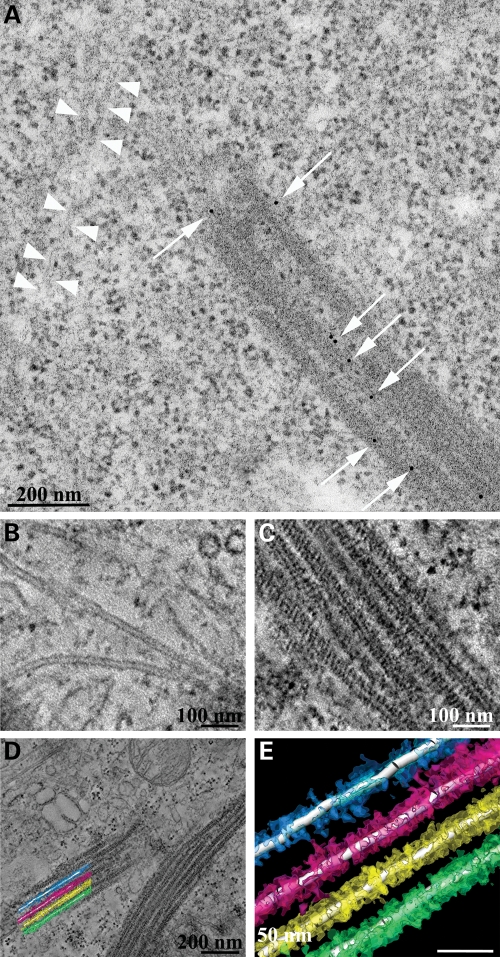
To further explore the nature of LRRK2 filaments, we employed EM to define the ultrastructure of these structures. We performed correlated light and EM by transfecting HEK293T cells with YFP-LRRK2-I2020T, using YFP fluorescence to identify filament-bearing cells, and then processed the same samples for EM to obtain high-resolution information on LRRK2 distribution in specific subcellular domains. Figure 2 shows filament formation identified by YFP fluorescence (Fig. 2A–C) and the corresponding low-magnification electron micrograph of a thin section from the same area (Fig. 2D). Higher magnifications (Fig. 2E and F) reveal filamentous structures that are organized in parallel arrays. The filaments consist of microtubules, identified by their characteristic structure in high-resolution EM as well as the correlated immunofluorescence, and these consist of LRRK2 apparently recruited onto parts of these unusually arrayed microtubules (Fig. 2F and G). We observed areas of naked microtubules (indicated by black arrows in Fig. 2G) interrupted by bundles of microtubules decorated by electron densities around them (indicated by black arrowheads in Fig. 2G). Immuno-EM using a specific antibody to LRRK2 confirmed the specificity of LRRK2 densities around the microtubules (Fig. 3A). To further examine the 3D electron microscopic organization of the filaments, we performed electron tomography on 250 nm thick sections. The tomograms show that LRRK2 appears to interact closely with the microtubules in a well-ordered, periodic fashion (Fig. 3B–E).
Figure 2.
Pattern of expression of LRRK2 using correlated light and EM. (A–C) The distribution of LRRK2 filaments was revealed in HEK293T cells expressing YFP-LRRK2-I2020T by the YFP fluorescence, (D) correlated image at low-power EM and (E–G) intermediate magnifications. Intermediate magnification of filaments (G) shows areas of naked microtubules (arrows) as well as electron densities (arrowheads) around them.
Figure 3.
Immuno-EM and electron tomography of LRRK2 filaments. (A) Immuno-EM was performed with a LRRK2-specific antibody. Immunogold signals appear as black dots (indicated by white arrows) on bundles of microtubules. For comparison, white arrowheads indicate a microtubule with the absence of LRRK2 staining. (B–E) Electron tomography of LRRK2 filaments. Higher magnification images of non-decorated tubules (B) and decorated microtubules (C) from electron tomograms are shown as well as a low power field from the area reconstructed by electron tomography (D) from which a graphical reconstruction was produced using Amira (E). These data are available for viewing and download from the Cell Centered Database either as images or 3D movies.
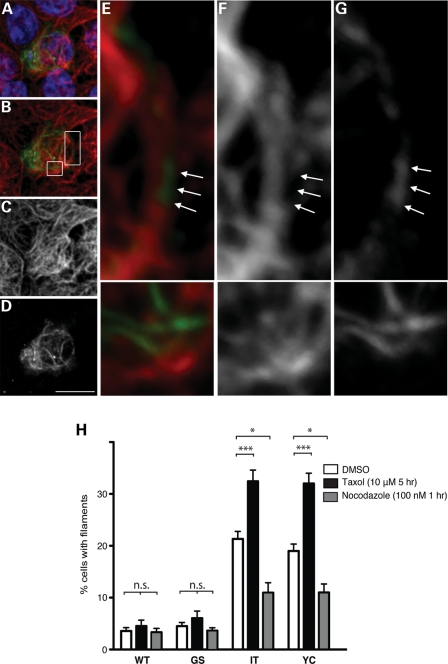
The tomographic data showing LRRK2 ‘coating’ microtubules suggested to us that masking of the tubulin epitope by LRRK2 may explain the lack of co-localization between tubulin and LRRK2 immunofluorescence (noted above). Indeed, a re-examination of individual Z-images from the immunofluorescent staining (rather than the maximal intensity projection) showed distinct regions in which LRRK2 filaments appear as direct continuations of microtubules (Fig. 4A–G). Moreover, we further explored the LRRK2-microtubule association by testing whether the frequency of filament formation was altered by microtubule stability. To accomplish this, we treated LRRK2-transfected CAD cells with the microtubule-stabilizing drug taxol (10 μm for 5 h), the microtubule-destabilizing drug nocodazole (100 nm for 1 h) or vehicle [dimethyl sulfoxide (DMSO)] control. An examiner blinded to genotype and drug treatment then counted the proportion of cells with filaments. We found a significant effect of genotype, drug treatment and the interaction between the two [two-way analysis of variance (ANOVA): F = 14.36, P < 0.0001]. In cells transfected with either GFP-LRRK2-I2020T or Y1699C, taxol significantly increased the percentage of filament-bearing cells, whereas this percentage was significantly decreased by nocodozole (Fig. 4H). In contrast, taxol or nocodazole did not significantly change the percentage of filament-bearing cells transfected with GFP-LRRK2-WT or G2019S. This distinction between the behavior of the G2019S and the other PD mutations is similar to that observed for their kinase activity (Supplementary Material, Fig. S1; 9–13). Taken together with the observations of LRRK2-coating microtubules by EM tomography, these data further indicate that multiple LRRK2 PD mutations (but not G2019S) enhance its ability to associate with microtubules. As cellular stressors, it is possible that taxol and nocodazole treatment alter LRRK2 behavior through indirect mechanisms, although the fact that they modulate filament formation in opposite directions makes this unlikely.
Figure 4.
LRRK2 filaments associate with microtubules and are modified by microtubule-altering drugs. (A–G) LRRK2 filaments localize to microtubules. HEK293T cells transfected with FLAG-LRRK2-I2020T were double labeled with anti-FLAG (D and G) and anti-alpha tubulin (C and F) antibodies. LRRK2 filaments appear as continuations of microtubules (A, B and E, merged images showing tubulin in red, LRRK2-FLAG in green and nuclei in blue; insets in B are shown in E, F and G). Arrows denote portions of microtubules positive for LRRK2 staining, but lacking tubulin staining. Scale bar is 10 μm. (H) Treatment with microtubule-altering drugs changed the filament proportion. CAD cells were transfected with WT or PD mutant forms (G2019S, I2020T or Y1699C) of GFP-LRRK2. Forty-eight hours post-transfection, cells were treated with the microtubule-stabilizing drug taxol (10 μm for 5 h), the microtubule-destabilizing drug nocodazole (100 nm for 1 h) or dimethyl sulfoxide (DMSO) control. Following drug treatment, cells were fixed, stained for GFP and the frequency of cells with filaments was quantified. Data are means ± SE of three independent experiments. (*P< 0.05, ***P< 0.001, n.s., non-significant; two-way analysis of variance (ANOVA) with post-hoc Tukey's test).
LRRK2-microtubule association requires intact kinase function and the WD40 domain
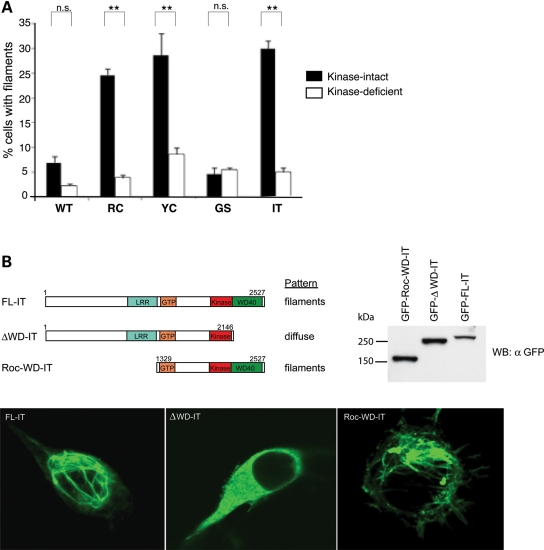
Because intact kinase activity is required for the enhanced neurotoxicity of LRRK2 PD mutants (9,12; Supplementary Material, Fig. S1), we tested whether this activity was also required for the enhanced microtubule association caused by LRRK2 PD mutations. Consistent with the neurotoxicity data, blocking kinase activity by introducing a second mutation in the kinase domain, K1906R, completely abolished the effect of LRRK2 PD mutations on microtubule association (Fig. 5A). To further elucidate the mechanism whereby LRRK2 PD mutations provoke enhanced microtubule association, we attempted to define a microtubule association domain by performing a structure–function study, testing which regions of LRRK2 are required (or are dispensable) for filament formation. We used the I2020T mutant as a representative mutation for these studies since the behavior of this mutant does not differ from R1441C or Y1699C (Figs 1 and 4). This analysis demonstrated that the WD40 domain was necessary for filament formation (Fig. 5B), whereas the entire N-terminus (prior to the LRR domain) is dispensable for microtubule binding. We reported previously (7) that ΔWD40-LRRK2 is unable to autophosphorylate yet retains the ability to trans-phosphorylate the model substrate MBP, suggesting a potential relationship between autophosphorylation and microtubule association. Such a relationship could contribute to an explanation of why blocking kinase function virtually abolishes microtubule association (Fig. 5A).
Figure 5.
Kinase activity and WD40 domain are required for LRRK2-microtubule association. (A) The microtubule-association effect of LRRK2 PD mutations is kinase-dependent. Blocking LRRK2 kinase function blocks the effect of filament formation caused by PD mutations. CAD cells were transfected with WT LRRK2-GFP or ‘double mutant’ constructs containing a LRRK2 PD mutation and a kinase-deficient mutation (K1906R) and stained for filaments with an anti-GFP antibody. Quantification of filament formation was done as in Figure 1E. Data are means ± SE of three independent experiments (**P< 0.01, n.s.: non-significant; ANOVA with Tukey's test). (B) Filament formation requires the WD40 domain of LRRK2. CAD cells were transfected with full-length GFP-LRRK2-I2020T (FL-IT), GFP-LRRK2-I2020T lacking the WD40 domain (ΔWD40) or the Roc and WD40 domains of LRRK2-I2020T (Roc-WD-IT). Forty-eight hours after transfection, the cells were assessed for filament formation. Anti-GFP immunoblot of transfected cell lysate demonstrates that truncated proteins are produced at the same or greater level of expression as full-length LRRK2.
DISCUSSION
Our study is the first to provide evidence that multiple PD mutations enhance the association of LRRK2 with microtubules. Several characteristics of this interaction indicate that it may contribute to LRRK2-dependent neurodegeneration. First, similar to LRRK2 neurotoxicity, this effect requires intact LRRK2 kinase function and the presence of the WD40 domain (7,9,12,14). Secondly, our colocalization and ultrastructural analyses show that LRRK2 interacts closely with microtubules in a well-ordered, periodic fashion, suggesting the existence of a LRRK2-binding site on microtubules or microtubule-bound proteins. Conversely, the specific requirement of the WD40 domain (but not the N-terminus) for microtubule-binding indicates that specific regions of LRRK2 mediate microtubule association. Our studies also show that LRRK2 filaments appear similar to linear arrays of electron dense structures observed in human post-mortem substantia nigra tissue from a patient carrying the filament-forming Y1699C mutation (24). With the development of better LRRK2 antibodies for immunohistochemical studies, it will be interesting to see if similar filaments are observed in the brains of PD patients with LRRK2 mutations.
The link between visible microtubule association (filaments) and toxicity mirrors those observed for protein aggregates seen with synuclein and other neurodegenerative-related proteins: neurotoxicity occurs in the population of LRRK2-transfected neurons, whereas visible filaments are seen in a minority of cells. In all of these cases, the visible protein lesions (filaments or aggregates) appear to reflect toxic events that also occur in soluble (and therefore not visualizable) protein. Our co-IP results (Fig. 1G) support such a scenario since these studies, which assess the entire cell population (including untransfected cells), show a markedly increased association for filament-forming LRRK2 PD mutations.
Similar filaments have been reported by Alegre-Abarrategui et al. (25) for R1441C-LRRK2. That report suggested that LRRK2 filaments were a form of multivesicular bodies, yet used a LRRK2 antibody for immuno-EM that has not been validated using LRRK2 knockout tissue, and no multivesicular body markers were shown to colocalize with LRRK2 filaments. In contrast, we used a validated LRRK2 antibody for detection (as well as an epitope tag), and employed correlated light and EM to ensure that we were observing the ultrastructural features of the same structures we visualized using immunofluorescence. We never observed membrane in association with LRRK2 filaments, either by EM or using membrane dyes on LRRK2-transfected cells (data not shown).
Dzamko et al. (26) reported that the treatment of LRRK2-transfected HEK293 cells with the kinase inhibitor H-1152 (which inhibits the LRRK2 kinase) caused G2019S-LRRK2 to colocalize with microtubules, a finding that would appear to be inconsistent with our finding that intact kinase function is required for microtubule association. However, key differences preclude comparison of these studies. H-1152 is not a LRRK2-specific inhibitor, and is therefore likely to inhibit a variety of kinases in living cells. Thus, the localization effect that Dzamko et al. report may be the indirect effect of inhibiting a different kinase, and may not reflect inhibition of the LRRK2 kinase per se. Moreover, Dzamko et al. do not quantify the apparent effect of H-1152 treatment on subcellular localization, which is necessary to confirm suspected effects, since a variable low rate of filament formation is seen with both wild-type and G2019S-LRRK2 (Fig. 1E). Notably, we do not observe an effect of H-1152 on the subcellular localization of wild-type or PD mutant forms of LRRK2 (data not shown).
The interaction of LRRK2 with microtubules could affect microtubule dynamics or microtubule-related transport, or microtubules could serve as a scaffold to concentrate LRRK2 signaling events such as the recruitment of the FADD/caspase-8 complex. In vitro, LRRK2 can bind alpha/beta-tubulin heterodimers (16) and can phosphorylate beta-tubulin (17), and active (but not kinase dead) LRRK2 enhances the polymerization of tubulin (isolated from bovine brain) in the presence of microtubule-associated proteins (17). Further support for LRRK2 interaction with the microtubule cytoskeleton includes the findings of deficient microtubule associated protein Tau phosphorylation in brain lysates from LRRK2 null mice (17), and multiple reports of alterations of neurite length and branching in situations of LRRK2 deficiency or overexpression (11,27–30). LRRK2 is reported to signal via the FADD/caspase-8 pathway (15), and in olfactory receptor neurons, degeneration of the axon and soma require synaptically activated caspase-8 that is retrogradely transported along microtubules (31). LRRK2 might therefore modulate the axonal transport of caspase-8 or other cargo, a notion consistent with the potentially central role of axonal degeneration in PD (32).
One potential mechanism for the enhanced microtubule association of mutant LRRK2 is that PD mutations may directly enhance the affinity of LRRK2 for microtubules or microtubule-bound proteins. Alternatively, PD mutations may enhance LRRK2 homo-oligomerization or complex formation with other cytosolic proteins, and these homo-oligomers or protein complexes may themselves have enhanced affinity for microtubules. Our data provide evidence that multiple pathogenic mutations enhance LRRK2–LRRK2 interactions, an effect that requires intact kinase activity and that is correlated with filament formation. Studies from Wittinghofer and colleagues (8) suggest that the COR domain is important for LRRK2 dimerization, and data from Cookson and colleagues (5) suggest that PD mutations that replace the arginine at position 1441 with either cysteine or glycine (which cause filaments) weaken the interaction in the Roc-COR region. The structural alteration produced by the weakening of this interaction may expose a protein–protein interaction domain, providing a potential mechanism for how PD mutations enhance LRRK2-microtubule association. The fact that multiple mutations provoke LRRK2 filaments indicates that other insults, such as those that occur in idiopathic PD, might also promote abnormal LRRK2 association with microtubules.
MATERIALS AND METHODS
Plasmids
LRRK2 cDNA from HEK 293 cells was cloned in pCMS-EGFP (Clontech), pcDNA-DEST53 and -DEST40 (Invitrogen). All subsequent mutants were generated using site-directed mutagenesis and all mutant clones were re-sequenced to confirm their accuracy.
Cell lines and primary cortical neuron cultures
CAD cells were grown in Dulbecco's modified eagle medium (DMEM)/F12 (GIBCO) supplemented with 8% fetal bovine serum. HEK293T cells were grown in DMEM (GIBCO) with 10% serum. CAD cells were transfected with Lipofectamine/PLUS (Invitrogen), whereas 293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cultures of cortical neurons from E16 mice were maintained in the Neurobasal medium containing B-27 supplements (GIBCO), and transfected with Lipofectamine 2000 four days after being plated. Primary neurons were transfected with LRRK2 expression constructs and pCMS-EGFP at 10:1 ratio. Each experiment was performed on cover slips in triplicate three times, with >100 cells on each cover slip being quantified.
Antibodies
We used the following antibodies: rabbit anti-GFP (Abcam, Cat. No. ab6656); mouse anti-GFP (Roche, Cat. No. 11814460001); mouse anti-V5 (Invitrogen, Cat. No. R960-25); rabbit monoclonal anti-LRRK2 (Epitomics, Cat. No. 3514-1); mouse monoclonal anti-alpha tubulin (Sigma, Cat. No. T6074); and rabbit polyclonal anti-FLAG (Abnova, Cat. No. PAB0900). The rabbit anti-LRRK2 used to immunostain untagged LRRK2 was a kind gift from Dr Benoit Giasson (University of Pennsylvania, Philadelphia, PA, USA).
Cell preparation for EM
HEK293T cells transfected with YFP-LRRK2-IT were fixed for 30 min in 2% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.4) on ice. After washes in 0.1 m cacodylate buffer, cells were post-fixed with 1% osmium tetraoxide for 30 min, stained with 2% uranyl acetate and then dehydrated and embedded in Durcupan epoxy resin. Sections were cut using a diamond knife at a thickness of 70–90 nm for thin sections and 250 nm for thick sections. Post-staining of grids was done with 2% uranyl acetate for 5 min and lead citrate for 1 min. Thin sections were examined using a JEOL 1200 FX1 operated at 120 kV.
Electron tomography
Colloidal gold particles (10 nm diameter) were deposited on opposite sides of the thick epoxy sections to serve as fiducial markers. For stability in the beam, the section was coated with carbon on both sides. For reconstruction, a triple series of images at regular tilt (angular increments of 2° from −60° to +60° increments) was collected with a JEOL 4000EX intermediate-voltage electron microscope operated at 400 kV. The specimens were irradiated before initiating a tilt series to limit anisotropic specimen thinning during image collection. Tilt series were recorded using a slow-scan CCD camera. The pixel dimensions of the CCD camera were 4000 × 4000 and the pixel resolution was 0.754 nm. Fine alignment and reconstruction was performed using the TxBR software package (33).
Post-embedding immuno-EM
For ultrastructural analysis of the filaments, we used an anti-LRRK2 antibody using 10 nm gold particles to identify the LRRK2 protein around the microtubules. After labeling, grids were washed and post-stained with 2% uranyl acetate for 5 min and lead citrate for 1 min.
Immunofluorescent labeling
Forty-eight hours after transfection, methanol- or formaldehyde-fixed HEK293T or CAD cells on poly-d-lysine-coated glass cover slips were blocked in phosphate buffered saline (PBS) containing 0.3% Triton X-100, 0.5% bovine serum albumin, 50 mm glycine and 2% normal donkey serum for 30 min. Cover slips were then incubated overnight at 4°C in primary antibodies diluted in block solution. The next day, cover slips were washed, incubated with Alexa488- or Alexa647-conjugated secondary antibodies and washed in PBS before mounting using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Data acquisition was done with an Olympus FluoView1000 laser-scanning confocal microscope (Olympus, Center Valley, PA, USA), using a 60X1.42 NA lens or with a ×100 oil-immersion objective with a Zeiss LSM510 2-photon confocal microscope. Image analysis of z-scan was done using the Imaris Software (Bitplane AG, St Paul, MN, USA) and the Image J software (rsb.info.nih.gov/ij).
Drug treatment
Forty-eight hours after transfection, cells on cover slips were treated with taxol (10 µm in DMSO for 5 h), nocodazole (100 nm in DMSO for 1 h) or DMSO. Cells were then stained as described above.
co-IP analysis
CAD cells or 293T cells transfected with various expression constructs were Dounce homogenized in lysis buffer (20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mm NaCl, 0.1–0.5% NP-40, 2 mm ethylene glycol tetraacetic acid, 2 mm MgCl2, 10% glycerol, 1 mm sodium orthovanadate, 10 mm NaF, 25 mm β-glycerophosphate, pH 7.2 and protease inhibitors). After centrifugation at 20 000g for 15 min, the supernatants were pre-cleared with protein-A agarose (Roche) for 30 min. Lysates containing 2 mg total protein were immunoprecipitated with rabbit anti-GFP for 1 h–overnight followed by incubation with protein-A beads for 2 h at 4°C.
SUPPLEMENTARY MATERIAL
Supplementary Material is availabe at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institute of Neurological Diseases and Stroke grant R01 NS061098-02 (W.D.), National Institutes of Health P41 RR004050 (M.H.E.) and the Branfman Family Foundation (M.H.E.).
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Kana Tsukamoto, Dr Guido Gaietta and Ying Jones for outstanding technical assistance. This work is also linked to activities of a consortium of researchers working with the Institute for Systems Biology in Seattle on Parkinson's disease.
REFERENCES
- 1.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 2.Simon-Sanchez J., Schulte C., Bras J.M., Sharma M., Gibbs J.R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S.W., Hernandez D.G., et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Caine D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Bonifati V. LRRK2 low-penetrance mutations (Gly2019Ser) and risk alleles (Gly2385Arg)-linking familial and sporadic Parkinson's disease. Neurochem. Res. 2007;32:1700–1708. doi: 10.1007/s11064-007-9324-y. [DOI] [PubMed] [Google Scholar]
- 5.Deng J., Lewis P.A., Greggio E., Sluch E., Beilina A., Cookson M.R. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl Acad. Sci. USA. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J.M., Daniels V., Lewis P., Jain S., Ding J., Syed A., et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jorgensen N.D., Peng Y., Ho C.C., Rideout H.J., Petrey D., Liu P., Dauer W.T. The WD40 domain is required for LRRK2 neurotoxicity. PLoS ONE. 2009;4:e8463. doi: 10.1371/journal.pone.0008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotthardt K., Weyand M., Kortholt A., Van Haastert P.J., Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 2008;27:2352. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blankinton J., Thomas K.J., et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., Alessi D.R. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Smith W.W., Pei Z., Jiang H., Dawson V.L., Dawson T.M., Ross C.A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 13.West A.B., Moore D.J., Choi C., Andrabi S.A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K.L., Dawson V.L., et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 14.Lee B.D., Shin J.H., VanKampen J., Petrucelli L., West A.B., Ko H.S., Lee Y.I., Maguire-Zeiss K.A., Bowers W.J., Federoff H.J., et al. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat. Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho C.C., Rideout H.J., Ribe E., Troy C.M., Dauer W.T. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J. Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi P.N., Wang X., Zhu X., Chen S.G., Wilson-Delfosse A.L. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J. Neurosci. Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability—a point of convergence in parkinsonian neurodegeneration? J. Neurochem. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- 18.Parisiadou L., Xie C., Cho H.J., Lin X., Gu X.L., Long C.X., Lobbestael E., Baekelandt V., Taymans J.M., Sun L., et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J. Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fotin-Mleczek M., Henkler F., Samel D., Reichwein M., Hausser A., Parmryd I., Scheurich P., Schmid J.A., Wajant H. Apoptotic crosstalk of TNF receptors: TNF-R2-induces depletion of TRAF2 and IAP proteins and accelerates TNF-R1-dependent activation of caspase-8. J. Cell Sci. 2002;115:2757–2770. doi: 10.1242/jcs.115.13.2757. [DOI] [PubMed] [Google Scholar]
- 20.Guiet C., Vito P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-kappaB activation. J. Cell Biol. 2000;148:1131–1140. doi: 10.1083/jcb.148.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel R.M., Martin D.A., Zheng L., Ng S.Y., Bertin J., Cohen J., Lenardo M.J. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J. Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muppidi J.R., Lobito A.A., Ramaswamy M., Yang J.K., Wang L., Wu H., Siegel R.M. Homotypic FADD interactions through a conserved RXDLL motif are required for death receptor-induced apoptosis. Cell Death Differ. 2006;13:1641–1650. doi: 10.1038/sj.cdd.4401855. [DOI] [PubMed] [Google Scholar]
- 23.Inohara N., Koseki T., Lin J., del Peso L., Lucas P.C., Chen FF., Ogura Y., Nunez G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 24.Wszolek Z.K., Vieregge P., Uitti R.J., Gasser T., Yasuhara O., McGeer P., Berry K., Caine D.B., Vingerhoets F.J., Klein C., et al. German-Canadian family (Family A) with parkinsonism, amyotrophy, and dementia—longitudinal observations. Parkinson. Relat. Disord. 1997;3:125–139. doi: 10.1016/s1353-8020(97)00013-8. [DOI] [PubMed] [Google Scholar]
- 25.Alegre-Abarrategui J., Christian H., Lufino M.M., Mutihac R., Venda L.L., Ansorge O., Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzamko N., Deak M., Hentati F., Reith A.D., Prescott A.R., Alessi D.R., Nichols R.J. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14–3–3 binding and altered cytoplasmic localization. Biochem. J. 2010;430:405–413. doi: 10.1042/BJ20100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cookson M.R. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat. Rev. Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dachsel J.C., Behrouz B., Yue M., Beevers J.E., Melrose H.L., Farrer M.J. A comparative study of Lrrk2 function in primary neuronal cultures. Parkinson. Relat. Disord. 2010;16:650–655. doi: 10.1016/j.parkreldis.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C.H., Tsai P.I., Wu R.M., Chien C.T. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3β. J. Neurosci. 2010;30:13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plowey E.D., Cherra S.J., 3rd, Liu Y.J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carson C., Saleh M., Fung F.W., Nicholson D.W., Roskams A.J. Axonal dynactin p150Glued transports caspase-8 to drive retrograde olfactory receptor neuron apoptosis. J. Neurosci. 2005;25:6092–6104. doi: 10.1523/JNEUROSCI.0707-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng H.C., Ulane C.M., Burke R.E. Clinical progression in Parkinson disease and the neurobiology of axons. Ann. Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawrence A., Bouwer J.C., Perkins G., Ellisman M.H. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J. Struct. Biol. 2006;154:144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.