Abstract
X-linked myotubular myopathy (MTM) is a severe neuromuscular disease of infancy caused by mutations of MTM1, which encodes the phosphoinositide lipid phosphatase, myotubularin. The Mtm1 knockout (KO) mouse has a severe phenotype and its short lifespan (8 weeks) makes it a challenge to use as a model in the testing of certain preclinical therapeutics. Many MTM patients succumb early in life, but some have a more favorable prognosis. We used human genotype–phenotype correlation data to develop a myotubularin-deficient mouse model with a less severe phenotype than is seen in Mtm1 KO mice. We modeled the human c.205C>T point mutation in Mtm1 exon 4, which is predicted to introduce the p.R69C missense change in myotubularin. Hemizygous male Mtm1 p.R69C mice develop early muscle atrophy prior to the onset of weakness at 2 months. The median survival period is 66 weeks. Histopathology shows small myofibers with centrally placed nuclei. Myotubularin protein is undetectably low because the introduced c.205C>T base change induced exon 4 skipping in most mRNAs, leading to premature termination of myotubularin translation. Some full-length Mtm1 mRNA bearing the mutation is present, which provides enough myotubularin activity to account for the relatively mild phenotype, as Mtm1 KO and Mtm1 p.R69C mice have similar muscle phosphatidylinositol 3-phosphate levels. These data explain the basis for phenotypic variability among human patients with MTM1 p.R69C mutations and establish the Mtm1 p.R69C mouse as a valuable model for the disease, as its less severe phenotype will expand the scope of testable preclinical therapies.
INTRODUCTION
Myotubular myopathy (MTM) is an X-linked centronuclear myopathy that presents at birth with profound weakness, hypotonia, external ophthalmoplegia and compromised respiratory function (1–3). Muscle biopsy shows numerous small myofibers with an increased proportion of centrally nucleated myofibers. MTM is due to mutations in MTM1, which encodes a phosphoinositide lipid phosphatase called myotubularin (4). The Mtm1 knockout (KO) mouse nicely models the clinical and pathologic features of severe MTM associated with null mutations of MTM1, but its lifespan of ∼8 weeks complicates the testing of certain experimental therapeutics (5). A murine model of MTM with a longer lifespan would expand the variety of preclinical therapeutic approaches that could be explored. Such an animal would be an ideal complement to the Mtm1 KO mouse in efforts to understand the pathogenesis of MTM and test treatment strategies.
After a review of the human genotype–phenotype correlation data, we ultimately decided that the best approach would be to model a human MTM1 point mutation known to cause a mild MTM phenotype (6,7). Mutations in MTM1 are likely to cause similar effects in mice and humans due to the high degree of homology between mice and humans at both the gene and protein levels. An added benefit of this approach would be that all of the tissues in the mouse would bear the Mtm1 mutation, which may afford an opportunity to study the medical complications of MTM experienced by some long-term survivors, such as pyloric stenosis, gallstones, kidney stones and fatal liver hemorrhages (8,9).
MTM is caused by a number of different MTM1 mutations, and genotype–phenotype correlations are challenging due to the genetic and clinical heterogeneity seen in this disease (1). Nonetheless, one generalization that can be made is that MTM1 missense mutations involving the phosphatase domain, or mutations that truncate the protein, are associated with a severe phenotype (7) and would be of limited benefit to model since the complete KO is already available (5). Analysis of human genotype–phenotype correlation studies revealed the recurrent c.205C>T base pair change, which is predicted to introduce the p.R69C missense alteration in myotubularin, as a possible candidate to model in mice.
The c.205C>T mutation is consistently associated with a less severe phenotype, as many reported patients have consistently lived beyond infancy and some well into childhood (6,10–14). This mutation occurs in exon 4, which encodes part of the pleckstrin homology, glucosyltransferases, Rab-like GTPase activators and myotubularin (PH-GRAM) domain, which is not a catalytically active domain. The PH-GRAM domain binds the two substrates phosphatidylinositol 3-phosphate (PI3P) and phosphatidylinositol 3, 5-bisphosphate (PI3,5P2) with high affinity, and this interaction is important for proper late endosomal trafficking (15). Myotubularin-bearing p.R69C binds PI3,5P2 with less affinity than wild-type myotubularin, so it is predicted that this myotubularin mutant is hypofunctional (15). Other PH-GRAM domain missense alterations, such as p.L70P and p.L87P, are also associated with a mild phenotype (6,13) and PI3,5P2 binding of p.R69C is intermediate to that of p.L70P and p.L87P (15). Not all missense mutations involving the PH-GRAM domain lead to a mild MTM phenotype, however. Mutations introducing the p.R69S and p.V49F amino acid changes are associated with a more severe clinical course (6,10,13) and dramatically decreased PI3,5P2-binding capacity (15). Given the consistent findings of a relatively mild phenotype in MTM patients with the p.R69C mutation and the experimental data supporting hypofunctionality of this mutant protein, we predicted that modeling this missense change would result in a mouse with a less severe MTM phenotype.
RESULTS
Generation of the Mtm1 p.R69C mouse
The Mtm1 p.R69C mouse was generated using polymerase chain reaction (PCR) mutagenesis to introduce the c.205C>T base change in Mtm1 exon 4, which was then subcloned into a targeting vector (Supplementary Material, Fig. S1A and B). The wild-type sequence was replaced in embryonic stem (ES) cells by homologous recombination with a targeting vector containing Mtm1 exons 2, 3 and 4. ES cells were selected for antibiotic resistance and surviving clones were expanded for PCR analysis to identify recombinants (Supplementary Material, Fig. S1C). Clones were further confirmed by DNA sequencing (data not shown) to ensure that the c.205C>T change was present prior to microinjection of blastocyst stage embryos (3.5 dpc), which yielded viable F1 mice. PCR analysis of genomic DNA from F1 mice confirmed the presence of the Neo cassette (Supplementary Material, Fig. S1D) and sequencing confirmed the presence of the c.205C>T mutation in Mtm1 exon 4 (data not shown).
Mtm1 p.R69C mice have a less severe MTM phenotype, but still develop muscle atrophy and significant weakness
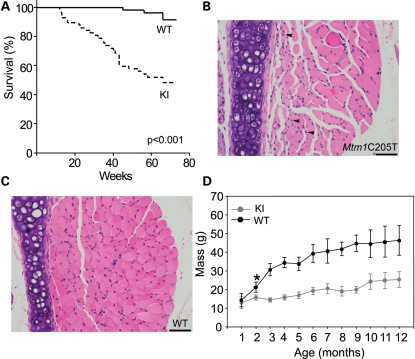
Mtm1 p.R69C mice are asymptomatic at birth and skeletal muscle develops normally. Affected males comprise 22% of all progeny, which is near the Mendelian prediction of 25%, and suggests that, in contrast to the Mtm1 KO mouse (5), there is minimal pre- or perinatal lethality. The mean survival time is 53.8 ± 2.9 weeks (median: 66 weeks) (log-rank: P < 0.001) (Fig. 1A). Unlike Mtm1 KO mice, which show progressive weakness until death, Mtm1 p.R69C mice died suddenly and unexpectedly, consistent with a cardiorespiratory insufficiency as a cause of death. Although death was rare in the perinatal period, when it does occur it is likely due to cardiorespiratory insufficiency as breathing can become labored as it does in the Mtm1 KO model (5). Lung and cardiac tissues are unremarkable histologically (data not shown). Affected animals have a shuffling gait with widely spaced feet and make a chirp-like noise (Supplementary Material, Videos). We believe this noise is apparently related to respiratory activity, as its onset and frequency directly correlates with overall activity level and respiratory rate. Histological examination of the pharyngeal and laryngeal muscles in 3-month-old Mtm1 p.R69C mice showed numerous small fibers with central nuclei, but not in wild-type littermates (Fig. 1B and C). The pulmonary airways, lungs and diaphragm revealed no significant pathological alterations in 3-month-old Mtm1 p.R69C mice (data not shown). The livers were also normal with no evidence of hepatic peliosis or recent liver bleeds that sometimes lead to mortality in older human patients (9).
Figure 1.
(A) The Kaplan–Meier survival analysis of Mtm1 p.R69C (n = 57) and wild-type littermate males (n = 54). Mtm1 p.R69C animals have a median survival of 66 weeks (53.8 ± 2.9 weeks), log-rank P < 0.001. Laryngeal muscles in 3-month-old Mtm1 p.R69C (B) and wild-type littermate (C) showing small fibers with central nuclei in the former (B, arrowheads). Bar = 50 μm. (D) Body mass of Mtm1 p.R69C animals and wild-type littermates over 12 months (n = 12 for each group). At 1 month, there is no significant difference in body mass between Mtm1 p.R69C (13.1 ± 3.2 g) and wild-type (14.3 ± 1.2 g) animals; however, the difference at 2 months (Mtm1 p.R69C: 15.8 ± 1.3 versus wild-type: 21.2 ± 2.7 g) is significant (*P < 0.001), and this coincides with the onset of weakness. Mtm1 p.R69C body mass is ∼50% that of wild-type animals up to 12 months. Abbreviations: KI, Mtm1 p.R69C; WT, wild-type.
Myotubularin is highly expressed in the testes and myotubularin phosphatases are critical regulators of adherens junction dynamics in the seminiferous epithelium governing cell adhesion (16). We tested the effect of the c.205C>T mutation on the reproductive capacity of affected males. Affected males were paired with wild-type females for a 6-month interval and three of the eight breeder pairs produced a total of five viable litters, consistent with diminished, but not abolished fertility. The testes of mature affected males are unremarkable histologically, with mature sperm present in numerous seminiferous tubules (data not shown), suggesting that the reproductive capacity of affected Mtm1 p.R69C males is diminished due to skeletal muscle weakness and not a failure of spermatogenesis.
In our experience, high-fat chow helps maintain the overall health of Mtm1 KO animals so Mtm1 p.R69C and wild-type littermates were also fed this diet starting at weaning. On this diet, Mtm1 p.R69C animal body mass is not significantly different from that of wild-type littermates at 1 month, but at 2 months Mtm1 p.R69C body mass dropped to 74% that of wild-type littermates (Fig. 1D). The body mass of Mtm1 p.R69C animals was 44–55% that of wild-type littermates from 3 to 12 months. Wild-type animals developed generalized obesity at ∼5 months, which did not occur in the Mtm1 p.R69C animals (Fig. 1D).
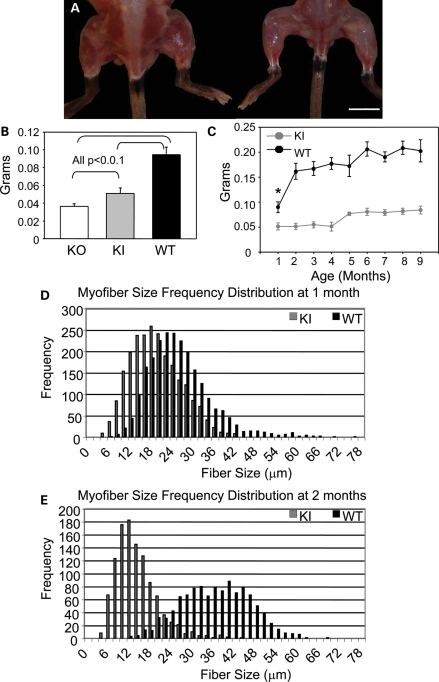
Necropsy showed that the difference in the body mass was mostly due to diffuse muscle atrophy (Fig. 2A), so we examined the masses of various skeletal muscles individually. At 1 month, quadriceps muscle mass was 64% of what was seen in wild-type animals, while in KO animals the quadriceps mass was 38% of wild-type values (Fig. 2B). Mtm1 p.R69C animals showed consistently reduced quadriceps masses over a 9-month period compared with wild-type littermates (Fig. 2C). Morphometric techniques were used to record minimum Feret (MinFeret) diameters of individual myofibers in Mtm1 p.R69C and wild-type animals at 1 and 2 months of age. At 1 month, when Mtm1 p.R69C animals are not overtly weak, MinFeret fiber diameters were 20.9 ± 3.8 µm (coefficient of variation (Cv) = 18.2%) in Mtm1 p.R69C and, 29.7 ± 8.4 µm (Cv = 28.3%) in wild-type animals (P = 0.006) (Fig. 2D). In contrast, in 1-month-old Mtm1 KO animals, MinFeret diameters were 12.4 ± 3.2 µm (Cv = 25.8%), which is ∼60% that of Mtm1 p.R69C diameters. By 2 months, at the clinically detectable onset of weakness, a dramatic reduction in myofiber diameter was noted in Mtm1 p.R69C animals [9.7 ± 3.3 µm (Cv = 34.0%) versus 36.7 ± 5.9 µm (Cv = 16.1%) in wild-type animals (P < 0.001)] (Fig. 2E). Other major muscles, including the gastrocnemius, tibialis anterior and triceps, from Mtm1 p.R69C animals also showed significant reductions in mass relative to the wild-type, but not to the same degree as noted for quadriceps. Interestingly, the gastrocnemius was significantly larger in Mtm1 p.R69C animals relative to Mtm1 KO animals, while tibialis anterior and triceps muscle masses did not differ significantly between the two MTM models (Supplementary Material, Fig. S2A–D).
Figure 2.
Significant but stable muscle atrophy occurs in Mtm1 p.R69C mice. (A) Gross photograph of 3-month-old Mtm1 p.R69C (right) shows significant muscle atrophy compared with a wild-type littermate (bar = 1 cm). (B) Bar plot showing that Mtm1 p.R69C quadriceps mass (0.05 ± 0.006 g) is intermediate to that of Mtm1 KO (0.036 ± 0.003 g) and wild-type littermates (0.095 ± 0.01 g) at 1 month of age [analysis of variance (ANOVA) P < 0.001; F = 213.21]. The P-values reported in B were obtained from post hoc testing. (C) Mtm1 p.R69C quadriceps mass is 30–40% that of wild-type littermates over a 9-month period (n = 8 for each group). Histograms of MinFeret diameters from Mtm1 p.R69C and wild-type littermate quadriceps at 1 (D) and 2 (E) months show that Mtm1 p.R69C myofibers (20.9 ± 3.8 µm) were significantly smaller than the wild-type (29.7 ± 8.4 µm) at 1 month (P = 0.006), but the difference in fiber size at 2 months is even more pronounced (Mtm1 p.R69C: 9.7 ± 3.3 µm versus wild-type: 36.7 ± 5.9 µm; P < 0.001) and this coincides with the onset of weakness. Abbreviations: KI, Mtm1 p.R69C; KO, Mtm1 knockout; WT, wild-type.
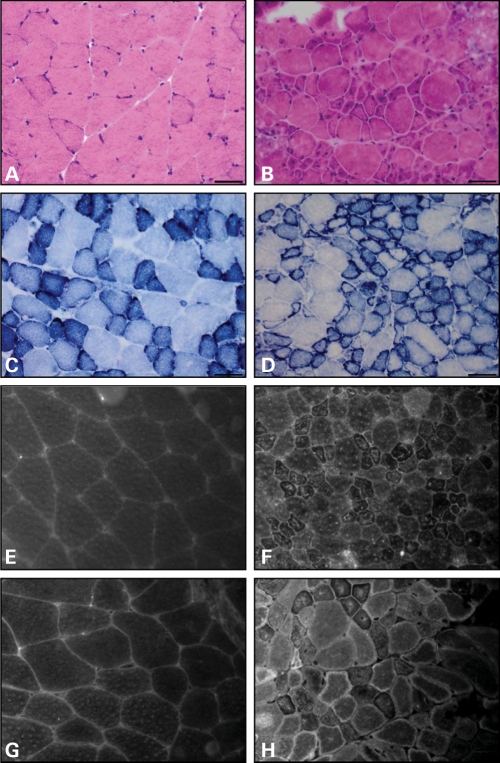
Hematoxylin and eosin-stained frozen cross-sections of Mtm1 p.R69C gastrocnemius muscles showed increased variability in fiber size with numerous small, rounded, hypotrophic fibers admixed with scattered, larger fibers (Fig. 3A and B). There was no observed increase in endomysial fibrous tissue, and neither necrotic nor regenerating myofibers were evident. Nicotinamide adenine dinucleotide dehydrogenase-tetrazolium reductase (NADH-TR) staining showed no fibers with central accumulation of staining product in Mtm1 p.R69C animals, but many small fibers had a peripheral rim of intense staining, which is characteristic of MTM and sometimes referred to as ‘necklace fibers’ (Fig. 3C and D) (5,17–19). No fiber type predominated (Supplementary Material, Fig. S3).
Figure 3.
Mtm1 p.R69C muscle develops structural changes of MTM. Hematoxylin and eosin-stained frozen cross section of quadriceps from 3-month-old wild-type (A) and Mtm1 p.R69C (B). Note the increased number of small myofibers, many of which show central nuclei in Mtm1 p.R69C animals. NADH-TR stain shows the expected delicate staining pattern in wild-type animals (C). In Mtm1 p.R69C muscle, (D) increased NADH-TR staining is noted at the periphery of small fibers. Quadriceps from a 3-month-old wild-type littermate (E, G) and an Mtm1 p.R69C mouse (F, H) stained with anti-DHPR antibodies to highlight T-tubules, and anti-RyR antibodies to mark sarcoplasmic reticulum. DHPR and RyR immunolocalization in wild-type animals show the expected delicate reticular staining pattern, while numerous fibers in Mtm1 p.R69C animals display alterations in the staining pattern including increase in central and peripheral staining, especially of small fibers. Bars = 50 μm.
Even though Mtm1 p.R69C animals exhibit little or no overt muscle weakness at 1 month of age, their muscles show an increased proportion of fibers with central nuclei (3.6 ± 1.8%), compared with wild-type littermates (0.9 ± 1.2%) (P = 0.007). By 2 months, when Mtm1 p.R69C animals become noticeably weak, 10.0 ± 4.8% of fibers have central nuclei, whereas only 1.5 ± 0.8% of wild-type littermate fibers exhibit this finding (P = 0.006). The proportion of fibers with central nuclei increases in Mtm1 p.R69C animals over time. At 9 months, 18.3 ± 10.8% of Mtm1 p.R69C muscle fibers have central nuclei, but only 2.1 ± 3.1% of wild-type fibers have central nuclei (P < 0.001).
A number of medical complications, including pyloric stenosis, gall stones, kidney stones, hepatic peliosis and others have been associated with long-term (over 1 year) survivorship in MTM (8,9). No evidence for any of these was detected in Mtm1 p.R69C mice of various ages following complete necropsies on 64 animals.
These results demonstrate that the MTM phenotype of the Mtm1 p.R69C mouse shows less severe muscle atrophy, a delayed onset of weakness and a considerably longer lifespan than that of the Mtm1 KO mouse. Myofibers in the Mtm1 p.R69C mouse show structural changes of MTM, including central nucleation and hypotrophy, even before the overt onset of weakness. Myofiber hypotrophy remains stable, while the proportion of myofibers with central nuclei increases over time.
Mtm1 p.R69C animals show alterations in triad and sacroplasmic reticulum structure and in the expression of genes involved in Ca2+ handling
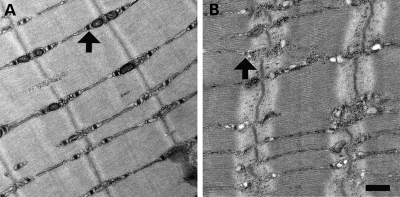
Myotubularin deficiency is associated with T-tubule abnormalities, both in MTM patients and in animal models (17,18,20,21). To determine whether this is also the case in the Mtm1 p.R69C mouse, we used antibodies specific to the dihydropyridine receptor (DHPRa1), a T-tubule marker, and the ryanodine receptor (RyR), a sarcoplasmic reticulum marker, in immunofluorescence staining of frozen cross-sections of Mtm1 p.R69C quadriceps. Wild-type littermate animals showed the expected delicate cytoplasmic staining pattern and some sarcolemmal staining for both antibodies (Fig. 3E and G). Mtm1 p.R69C animals showed an abnormal staining pattern with intense DHPRa1 and RyR staining in the center of many small fibers, while many of the larger fibers showed intense RyR staining at the periphery but no increased central staining, suggestive of necklace fibers (Fig. 3F and H). Ultrastructural evaluation of extensor digitorum longus (EDL) muscles revealed a near-absence of T-tubules and triads and occasional longitudinal (L) tubules (Fig. 4A and B), consistent with previously reported findings in other myotubularin-deficient mice (17), fish (20), dogs (18) and humans (20).
Figure 4.
Ultrastructural evaluation of Mtm1 p.R69C mice. Electron microscopy of EDL muscles at 6 months of life reveals the presence of numerous triad structures (arrows) in the muscles of wild-type animals (A), with only occasional and poorly defined triads seen in Mtm1 p.R69C animals (B). Bar = 500 nm.
Alterations in the expression of genes associated with functions in calcium handling and excitation–contraction coupling have been described in the Mtm1 KO mouse, so we examined the expression of these genes in Mtm1 p.R69C mouse quadriceps using real-time PCR (17). The expression of Cacng1 (1.7-fold), Camk2D (1.8-fold) and Chrna1 (3.9-fold) were all upregulated in Mtm1 p.R69C mouse quadriceps, but to lesser degrees than has been reported in KO mice (17). Rrad and Sln expression were upregulated 60-fold and 22.4-fold, respectively, in Mtm1 p.R69C mice (17). Homer1 expression was decreased 0.77-fold in Mtm1 p.R69C mice. These results indicate that the Mtm1 p.R69C mouse shows the expected structural alterations in the triad and sacroplasmic reticulum and in the expression of muscle calcium handling genes.
Mtm1 p.R69C animals are weak but not as physically compromised as the Mtm1 KO mouse
We were interested in comparing general muscle function of Mtm1 p.R69C animals to KO and wild-type animals, and in following Mtm1 p.R69C muscle function over time. A dynamometer was used to measure the force generated by the hindlimbs and forelimbs, and an open field was used to test overall locomotive function and rearing ability. Given the short lifespan of KO animals, their performance could only be compared with Mtm1 p.R69C and wild-type animals at 1 month (n = 12 for each group). A cohort of 12 Mtm1 p.R69C mice and 12 wild-type littermates was followed over 12 months to test if weakness was stable or progressive over time.
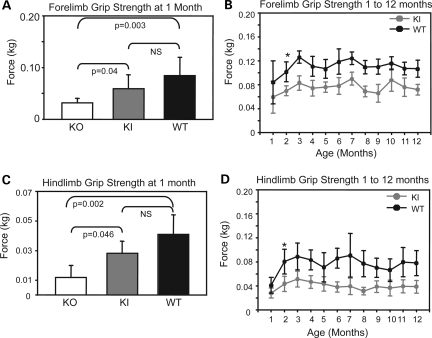
KO animals generated significantly less force (0.032 ± 0.01 kg) with their forelimbs than either Mtm1 p.R69C (0.059 ± 0.02 kg) or wild-type (0.085 ± 0.03 kg) animals at 1 month of age (Fig. 4A). Mtm1 p.R69C animals tended to generate less force with their forelimbs than wild-type animals, but the difference was not significant at 1 month. Mtm1 p.R69C mice generated significantly less forelimb force than wild-type littermates at 2 months (P < 0.001), and it remained 60–70% that of wild-type animals over a 12-month period (Fig. 5B). At 1 month of age, KO animals produced significantly less force (0.01 ± 0.008 kg) with the hindlimbs than Mtm1 p.R69C (0.028 ± 0.01 kg) or wild-type animals (0.041 ± 0.01 kg), and the difference between Mtm1 p.R69C and wild-type animals was not significant at this time point (Fig. 5C). At 2 months of age, Mtm1 p.R69C mice generated significantly less force with their hindlimbs than wild-type mice (P < 0.001), and it remained 40–60% that of wild-type animals over 12 months (Fig. 5D).
Figure 5.
Forelimb and hindlimb grip strength was assessed in Mtm1 KO, Mtm1 p.R69C and wild-type animals (all n = 12) using a dynamometer. (A) KO animals (0.032 ± 0.01 kg) generated significantly less force with their forelimbs than Mtm1 p.R69C (0.059 ± 0.02 kg) or wild-type (0.085 ± 0.03 kg) animals (ANOVA P < 0.01; F = 24.2). The difference between Mtm1 p.R69C and wild-type animals was not significant at 1 month. (B) Mtm1 p.R69C animals generate significantly less forelimb force than wild-type littermates starting at 2 months of age, which persists over a 12-month interval (*P < 0.001). (C) KO animals generated significantly less force with their hindlimbs at 1 month (0.01 ± 0.01 kg) than Mtm1 p.R69C (0.028 ± 0.01 kg) or wild-type (0.041 ± 0.01 kg) animals (ANOVA P < 0.006; F = 7.5). The difference between Mtm1 p.R69C and wild-type animals was not significant at 1 month. (D) Mtm1 p.R69C animals generate significantly less hindlimb force than wild-type animals starting at 2 months and this remains the case over a 12-month interval (*P < 0.001). The P-values reported in (A) and (C) were obtained from post hoc testing. Abbreviations: KI, Mtm1 p.R69C; KO, Mtm1 knockout; NS, not significant; WT, wild-type.
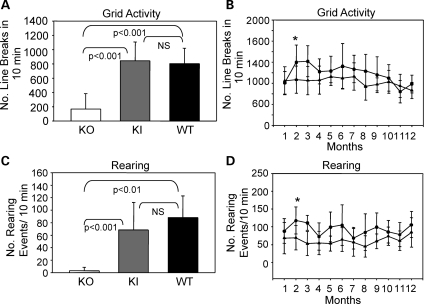
Mtm1 p.R69C mice were far more active in the open field (977 ± 221 line breaks/10 min) than KO mice (168 ± 217 line breaks/10 min) at 1 month of age (P < 0.001) (Fig. 6A). Mtm1 p.R69C and wild-type animals (747 ± 213 line breaks/10 min) showed no significant difference in number of line breaks/10 min at 1 month (Fig. 6A). Open field activity of Mtm1 p.R69C animals over a 12-month period showed a trend towards decreased activity relative to the wild-type, but the range was wide, from 72% at 2 months to 102% at 11 months (Fig. 6B). At 1 month of age, KO animals showed only 3.3 ± 5.4 rearing events/10 min with 6 of the 12 animals studied not even recording a single rearing event. This differed significantly from wild-type animals (88.3 ± 34.5 rearing events/10 min) (P < 0.001) and Mtm1 p.R69C animals (68.7 ± 43.8 rearing events/10 min) (P < 0.001) (Fig. 6C). The difference between Mtm1 p.R69C and wild-type animals was not significant at 1 month (Fig. 6C). Mtm1 p.R69C animals tended to show fewer rearing events, which ranged from 60 to 85% that of wild-type animals when studied over a 12-month period (Fig. 6D).
Figure 6.
Open field grid and rearing activity in Mtm1 KO, Mtm1 p.R69C and wild-type animals. (A) Open field activity in 1-month-old KO, Mtm1 p.R69C and wild-type animals (n = 12 for each group). KO animals (168 ± 217) have significantly fewer line breaks in 10 min than Mtm1 p.R69C (977 ± 221) or wild-type animals (747 ± 213) (ANOVA P < 0.001; F = 22.77). Mtm1 p.R69C and wild-type animals show no significant difference in activity at 1 month. (B) Mtm1 p.R69C animals tend to show less activity than wild-type littermates when studied over a 12-month period, and although this difference is significant at 2 months (*P = 0.01), it did not remain so over the entire period. (C) Rearing activity at 1 month is significantly reduced in KO (3.3 ± 5.4) relative to Mtm1 p.R69C (68.7 ± 43.8) and wild-type (88.3 ± 34.5) (ANOVA P < 0.001; F = 21.22). The difference between Mtm1 p.R69C and wild-type was not significant. (D) Mtm1 p.R69C animals tend to show less rearing activity than wild-type littermates over a 12-month interval and although this difference is significant at 2 months (*P = 0.003), it did not remain so over the entire period. The P-values reported in (A) and (C) were obtained from post hoc testing. Abbreviations: KI, Mtm1 p.R69C; KO, Mtm1 knockout; NS, not significant; WT, wild-type.
These data demonstrate that Mtm1 p.R69C animals have significantly better overall muscle function than KO animals at 1 month, that Mtm1 p.R69C animals become significantly weaker than wild-type animals at 2 months of age, and that weakness remains relatively stable and is not progressive in Mtm1 p.R69C animals. The locomotive and rearing activity of Mtm1 p.R69C animals is considerably better than it is in KO animals and this also likely contributes to their overall health and longer lifespan.
The c.205C>T base pair change in exon 4 of Mtm1 results in skipping of exon 4 and undetectable myotubularin protein levels
The c.205C>T change in Mtm1 exon 4 is predicted to introduce the R69C missense change in the PH-GRAM domain of myotubularin. Prior in vitro studies found that myotubularin bearing the R69C mutation has ∼60% of the phosphoinositide-binding capacity of wild-type myotubularin (15), so we expected R69C myotubularin to be hypofunctional. However, myotubularin protein was undetectable in immunoblots of Mtm1 p.R69C mouse quadriceps tissue (Fig. 7A). Since the anti-myotubularin antibody used was raised against the COOH terminus of myotubularin and the R69C change occurs near the NH2-terminus, we considered it unlikely that an alteration in epitope binding was occurring (22). Although previous work indicated that missense changes may destabilize myotubularin and, in turn, render it undetectable in MTM patient tissues (23), we considered the possibly that the introduced point mutation somehow altered myotubularin mRNA structure, which effectively reduced the amount of myotubularin that was translated.
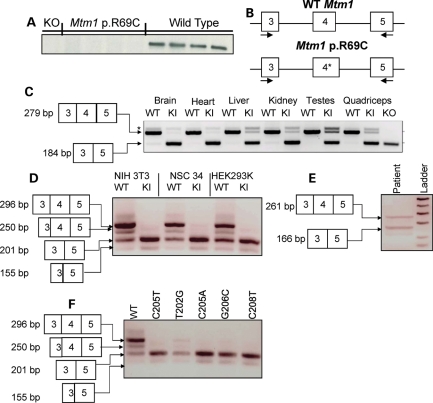
Figure 7.
Myotubularin protein expression and mRNA structure in Mtm1 p.R69C mice. (A) Immunoblot showing no detectable myotubularin in Mtm1 KO and Mtm1 p.R69C quadriceps. (B) Diagram of the strategy behind the splicing assay, depicting primers that anneal in exons 3 and 5 and flank exon 4. The c.205C>T change in Mtm1 p.R69C is denoted with an ‘*’. (C) cDNA was generated from a variety of tissues and subjected to PCR analysis and PCR products were resolved on a gel and sequenced. A 279 bp band predominated in wild-type animals and it includes Mtm1 exons 3, 4 and 5, whereas the predominate PCR product in Mtm1 p.R69C tissue was 184 bp, which included exons 3 and 5, but not exon 4. The 279 bp product was detected in the Mtm1 p.R69C brain, heart, liver, kidney, testes and quadriceps. KO was used as a positive control since the mouse was generated by deleting Mtm1 exon 4, which yields a premature stop codon in exon 5. *Denotes a non-specific band that occurs in both wild-type and Mtm1 p.R69C tissues. (D) Wild-type and Mtm1 p.R69C minigene plasmids were generated and transfected into a variety of cell lines. The minigene plasmids have FLAG sequence 5′ to Mtm1 exons 3, 4 and 5 and intervening sequences. The Mtm1 p.R69C plasmid bears the c.205C>T mutation in exon 4. Total RNA was isolated and cDNA was synthesized and subjected to PCR analysis. The resulting products were resolved on an agarose gel and sequenced. The 296 bp product with exons 3, 4 and 5 predominated in all cell lines with the wild-type minigene. All cells with wild-type minigene also had the 201 bp fragment that contained exons 3 and 5, but not exon 4. A 250 and 155 bp product was also noted in the cells with wild-type minigene, which is the result of the activation of a cryptic slice site in exon 3. In all three cell lines with the Mtm1 p.R69C minigene, the predominant product was the 201 bp fragment that skipped exon 4, but the 155 bp product due to the utilization of the cryptic splice site in exon 3 was also evident. PCR performed on untransfected cells showed no product (data not shown). (E) cDNA was prepared from quadriceps tissue from an MTM patient with c.205C>T. PCR was performed and skipping of exon 4 is noted as a 166 bp product. (F) Additional human point mutations in Mtm1 exon 4 (indicated above) were generated and tested using the minigene system and exon 4 skipping is noted in all of them. Abbreviations: KI, Mtm1 p.R69C; KO, Mtm1 knockout; WT, wild-type.
To test the impact of the point mutation on mRNA structure, PCR primers that flank exon 4 (Fig. 7B) were used in RT–PCR experiments with muscle cDNAs as a template (5). In wild-type animals, a 279 bp PCR product that includes Mtm1 exons 3, 4, and 5 was expected, while in Mtm1 KO animals a 184 bp product is expected as the mouse was generated by removing exon 4. Splicing of exons 3 and 5 together produces an out of frame mRNA with a premature stop codon in exon 5, and no functional myotubularin is translated (5). Therefore, KO cDNA served as a positive control in this experiment.
Quadriceps muscle from wild-type animals yielded a 279 bp product, while a 184 bp product that was missing exon 4 was detected in KO animals (Fig. 7C). The predominant PCR product in the Mtm1 p.R69C animals was 184 bp, and sequencing revealed that it was missing exactly exon 4 (data not shown). Mtm1 p.R69C muscle also contained a less prominent 279 bp product that includes exons 3, 4, 5 and the c.205C>T change (Fig. 7C). Splicing is often tissue-dependent, so cDNA was generated from other tissues and subjected to the same PCR analysis. In the brain, heart, liver, kidney and testes, the predominant PCR product in wild-type animals was 279 bp, while in Mtm1 p.R69C animals the 184 bp product predominates, with variable levels of the 279 bp product among the various tissues (Fig. 7C).
Although the c.205C>T mutation does not involve the consensus splice sites for exon 4 of the Mtm1 gene, cis-acting regulatory elements in pre-mRNA have been shown to influence pre-mRNA splicing through the creation of an exonic splicing enhancer (ESE) or the destruction of an exonic splicing silencer (ESS). To examine the impact of c.205C>T on Mtm1 mRNA, we used ESEFinder 3.0, PESX and ESRsearch applications that can be used to predict the binding of various RNA-binding proteins and identify sequence motifs that have the potential to alter splicing by modulating possible ESE or ESS sites located in exon 4 of Mtm1 (24–30). ESEfinder revealed no predicted changes in binding sites of splicing regulatory proteins; however, both PESX and ESRsearch predicted that the introduced c.205C>T base pair alters a putative ESS. This altered ESS could explain the observed change in splicing transcripts generated in the Mtm1 p.R69C mice.
We used an in vitro minigene system to further test that the c.205C>T mutation led to improper splicing. Wild-type genomic DNA was PCR amplified and a product containing Mtm1 exons 3, 4 and 5 and the intervening sequences were cloned into the minigene plasmid pCMV-Tag 2B, which incorporate a 5′ FLAG sequence. PCR mutagenesis was performed to generate the c.205C>T base pair change in the wild-type construct. The constructs were confirmed by sequencing and transfected into NIH3T3 cells (mouse fibroblasts), NSC34 cells (mouse motor neuron) and HEK293 cells (human kidney), total mRNA was isolated and cDNA was synthesized and used as template in PCR. Primers used in the minigene assay included: X3FWD, which anneals in the 5′ FLAG sequence of the minigene construct, and MTM1 cDNA 3′, which anneals in exon 5. Reactions were resolved on an agarose gel and the resulting bands were excised and confirmed by sequencing.
Cells transfected with the wild-type Mtm1 minigene construct showed one predominant, 296 bp band that contained exons 3, 4 and 5 (Fig. 7D). All of the cell lines also had a 250 bp band that includes only a part of exon 3 due to the activation of a cryptic splice site, and all of exons 4 and 5 (Fig. 7D). Due to the activation of the same cryptic splice site, a 201 bp product missing exon 4 and a 155 bp product missing exon 4 and a part of exon 3 were also detected to variable degrees in all of the cell lines with the wild-type construct (Fig. 7D). The Mtm1 p.R69C minigene construct yielded a 201 bp band that included exons 3 and 5, but not exon 4 in all of the tested cell lines (Fig. 7D). A less prominent band of 155 bp that was missing part of exon 3 and all of exon 4 also occurred in all of the cell lines (Fig. 7D). No resolvable full-length PCR product containing exons 3, 4 and 5 was present in any cell line transfected with the c.205C>T minigene. The activation of the exon 3 cryptic splice site, as well as exon 4 skipping in cells with the wild-type minigene, suggests that some dysregulation of splicing is occurring in the minigene system and supports the findings in Mtm1 p.R69C mouse tissues.
To determine whether exon skipping occurs in humans, cDNA was prepared from the quadriceps tissue of an MTM patient with the c.205C>T mutation. The patient was weak and hypotonic at birth, and required intubation due to respiratory distress. He is now 21 years old, has severe scoliosis and uses a wheelchair. His motor function has remained stable since 6 years of age. A similar PCR strategy yielded a 261 bp product predicted to include exons 3, 4 and 5 and a 166 bp fragment expected to include exons 3 and 5, but not exon 4 (Fig. 7E).
We also tested other known human point mutations that have been described in MTM1 exon 4 to determine whether they cause skipping. PCR site-directed mutagenesis was used to introduce the c.T202G (p.Y68D), c.C205A (p.R69S), c.G206C (p.R69P) and c.C208T (p.L70F) point mutations in minigene vectors (10,12). Minigene vectors were transfected into HEK293 cells, and cDNA was prepared and used as a template in a similar PCR assay. All four-point mutations led to exon 4 skipping with variable degrees of activation of the cryptic splice site in exon 3 (Fig. 7F). No PCR product was generated in untransfected cells (data not shown).
Overall, these data show that the c.205C>T base change and other exon 4 missense changes lead to exon 4 skipping in many, but not all, Mtm1 mRNA molecules. Because skipping exon 4 generates an out of frame message, a reduction in myotubularin protein levels would be expected. This likely accounts for the inability to detect myotubularin in the Mtm1 p.R69C mouse, and may explain why myotubularin protein cannot be detected in some MTM patients with missense mutations (23). Some full-length Mtm1 mRNA, albeit with the c.205C>T base change, is detected in Mtm1 p.R69C mice and in humans with the c.205C>T mutation. Myotubularin protein translated from this message would contain the R69C amino acid change, and likely retains enough residual activity to account for the relatively mild phenotype of the mouse and patients who bear this mutation.
PI3P levels are elevated in both Mtm1 p.R69C and Mtm1 KO skeletal muscle
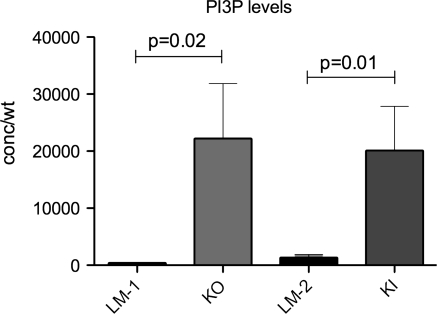
Myotubularin is a lipid phosphatase that removes the 3-phosphate group from PI3P so a loss of function, as seen in MTM, is predicted to result in increased levels of this phosphoinositide. Given the less severe MTM phenotype in Mtm1 p.R69C mice, we were interested in determining the levels of PI3P in skeletal muscle from 5-month-old Mtm1 p.R69C and 2-month-old Mtm1 KO mice. These ages were chosen because myopathy is well established in each model at these time points. To do this, we performed lipid-protein overlay enzyme-linked immunosorbent assays (ELISAs) on lipid extracts from quadriceps. As predicted, and in agreement with previous data from the zebrafish model of MTM (20), muscle from affected 2-month-old KO mice had significantly higher levels of PI3P than age-matched wild-type littermates (22 219 ± 9638 versus 419 ± 57 pmol/g, P = 0.02, n = 8) (Fig. 8). PI3P levels in skeletal muscle from 5-month Mtm1 p.R69C animals were also significantly elevated when compared with age-matched wild-type siblings (20 113 ± 7720 versus 1348 ± 540 pmol/g, P = 0.01, n = 8) (Fig. 8). These data suggest that PI3P levels are elevated in both Mtm1 p.R69C and Mtm1 KO mouse muscle and suggests that other functions of myotubularin besides its lipid phosphatase activity may contribute to MTM pathogenesis.
Figure 8.
PI3P levels are increased in skeletal muscle from Mtm1 KO and Mtm1 p.R69C animals. Total lipids were extracted from quadriceps muscle from 2-month-old Mtm1 KO (KO) and 5-month-old Mtm1 p.R69C (KI) animals and from age-matched littermate controls (LM) (n = 8 per condition). These ages were selected because weakness is well established at these times. PI3P levels were then measured using a lipid-antibody overlap enzyme-linked immunosorbent assay (ELISA). Levels from Mtm1 KO (22 219 ± 9638 pmol/g) and Mtm1 p.R69C animals (20 113 ± 7720 pmol/g) were significantly higher than those of littermate control animals (419 ± 57 and 1348 ± 540 pmol/g, respectively).
DISCUSSION
The Mtm1 p.R69C mouse models a less severe MTM phenotype
Perhaps the most notable difference between Mtm1 KO and Mtm1 p.R69C mice is the significantly longer lifespan of the latter, supporting the notion that Mtm1 p.R69C mice have some residual myotubularin activity (5). Weakness developed postnatally in both Mtm1 p.R69C and KO mice, although the onset of weakness occurs later and is not progressive in Mtm1 p.R69C mice (1,5). No Mtm1 p.R69C mouse developed paralysis due to the myopathy, as can occur in Mtm1 KO mice (5) and the weakness is stable with no significant decline over a 12-month period. Open field activity and rearing ability of Mtm1 p.R69C mice is dramatically better than that of KO mice, and the ability to access food and water clearly assist in preventing dehydration or caloric deficiency. Wild-type animals developed obesity at ∼5 months, due to the high-fat diet these animals were maintained on, but this did not occur in Mtm1 p.R69C littermates. The C57BL/6 strain is prone to diet-related obesity (31) which suggests that the Mtm1 p.R69C animals may utilize the extra calories or fail to properly absorb nutrients from the diet. We believe that the former possibility is more likely correct because the gastrointestinal tract is histologically unremarkable. It is possible that additional calories are spent in increased respiratory activity or in increased energy demands of muscle calcium handling. These hypotheses will be tested in future studies.
Muscle atrophy is noted prior to the onset of overt weakness in Mtm1 p.R69C mice, but by 2 months of age, body mass in Mtm1 p.R69C animals becomes significantly less than that of wild-type animals, and this coincides with the onset of weakness in Mtm1 p.R69C animals. Morphometric studies of myofiber size suggests that diffuse muscle atrophy largely accounts for the decrease in body mass, similar to the case in Mtm1 KO mice (5). The weakness was relatively stable in Mtm1 p.R69C mice, consistent with the relative stability of symptoms seen in Mtm1 p.R69C patients. Interestingly, although a number of medical complications have been described in MTM patients who are long-term survivors (>1 year of age) (8,9), none of these complications was observed in Mtm1 p.R69C mice, even at advanced ages.
Mtm1 p.R69C mouse muscle displayed all of the histopathologic changes expected of centronuclear myopathy. The proportion of fibers harboring central nuclei increased over time in both Mtm1 p.R69C and KO mice (5). This progressive increase in the proportion of fibers with central nuclei has also been reported in an Mtm1 p.R69C patient who had two muscle biopsies taken ∼3 years apart (14). In the Mtm1 p.R69C mouse, the increase in proportion of fibers with central nuclei in the presence of relatively stable muscle weakness over time suggests that central nuclei, while diagnostically useful, may not be critical to the pathogenic mechanism underlying weakness in MTM. No significant type 1 fiber predominance was noted, as has been described in some MTM patients (2), but muscle fiber type patterns are quite different in mice, most of whose muscles contain predominantly fast fiber types. The noted alterations in NADH-TR staining and in DHPR and RyR immunofluorescence staining are consistent with observations made in the patient biopsy material and the Mtm1 KO mouse (3,5), and they likely correspond to the disorganized T-tubules and sacroplasmic reticulum that have been previously observed in MTM animal models and in patients (17,18,20,21).
Mtm1 p.R69C and Mtm1 KO mice show alterations in the expression of the same subset of genes involved in Ca2+ homeostasis, which suggests that this exquisitely regulated aspect of muscle biology is defective in both models (17). Defects in excitation–contraction coupling are now thought to be a major component underlying the pathogenesis of MTM and possibly all centronuclear myopathies (17,21). The Mtm1 p.R69C and KO mouse models represent different ends of the MTM phenotypic spectrum, and when used together may provide considerable insight into the role of myotubularin in the excitation–contraction coupling. A more detailed analysis of Mtm1 p.R69C muscle physiology and calcium handling is in progress.
The longer lifespan of the Mtm1 p.R69C mouse makes it an excellent preclinical model of MTM to develop and optimize new therapeutic approaches. Muscle atrophy appears before the onset of weakness in the Mtm1 p.R69C mouse, so it seems reasonable to test therapeutic interventions that enhance muscle mass in MTM. A recent study using a soluble ActRIIB receptor in Mtm1 KO mice described selective myofiber hypertrophy and a moderate increase in lifespan in treated animals (32). Similar studies using Mtm1 p.R69C mice are currently in progress. Also, because the onset of weakness occurs uniformly at 2 months, it can be easily monitored, along with body mass and muscle atrophy, as early markers of an intervention's therapeutic effectiveness. The proportion of centrally nucleated myofibers is an additional parameter that could be readily used to monitor the effect of a therapeutic intervention in the Mtm1 p.R69C mouse.
The c.205C>T base change in Mtm1 induces exon 4 skipping
We predicted that modeling the c.205C>T mutation would generate a hypofunctional myotubularin, as the R69C missense change occurs in the PH-GRAM domain of myotubularin and reduces phosphoinositide-binding capacity of this domain (15). The milder symptoms and longer lifespan of the Mtm1 p.R69C model are consistent with hypofunctional myotubularin; however, myotubularin, which is a low abundance protein to begin with, was undetectable on immunoblots. Some missense mutations are thought to destabilize myotubularin protein (23), which may contribute to our inability to detect myotubularin in Mtm1 C205T mice, but examination of Mtm1 mRNA structure provided an alternative, or more likely, complementary explanation. Unexpectedly, the introduced c.205C>T base change induced exon 4 skipping in the majority of Mtm1 mRNA, which is expected to generate an out of frame message that produces no functional myotubularin due to the generation of a stop codon in exon 5. Yet some Mtm1 mRNA containing the c.205C>T base change is produced, and the myotubularin translated from this mRNA appears to provide enough activity to produce the relatively mild phenotype observed in Mtm1 p.R69C mice. The inability to detect myotubularin could therefore be due to protein instability caused by the missense mutation, and/or because the point mutation alters the mRNA structure. Exon 4 skipping observed in the mouse appears to be relevant to human MTM, as we confirmed this phenomenon in quadriceps from a patient with the c.205C>T mutation.
We also tested four other human exon 4 point mutations that occur at or near nucleotide position 205 in a minigene system, and all induced exon skipping. These data indicate that this region of exon 4 may be sensitive to single base pair alterations and our bioinformatic analysis suggests that the function of a putative ESS may be altered by these mutations. Even though all of the tested mutations induce similar degrees of exon 4 skipping in the minigene system, patients show some phenotypic differences as c.205C>T (p.R69C) and c.C208T (p.L70F) are associated with a mild clinical course while c.C205A (p.R69S) is associated with severe disease (10). The clinical course of c.T202G (p.Y68D) is moderate in nature (10), and our findings using the minigene system suggest that this mutation may produce more full-length Mtm1 mRNA than the others. While missense changes likely alter the stability and activity of myotubularin, the effect of missense changes on mRNA structure may also need to be taken into account when formulating genotype–phenotype correlations. Some missense mutations may not induce exon skipping at all, while others may significantly enhance skipping and dramatically reduce the amount of full-length MTM1 mRNA available for translation.
How c.205C>T and other base changes promote skipping is not clear. The base change may alter the function of regulatory sequences or it may induce changes in pre-mRNA secondary structure that somehow promote skipping. These findings indicate that antisense oligonucleotide therapy, a previously untested approach to MTM treatment, may be a suitable strategy to prevent exon skipping and restore the expression of full-length Mtm1 mRNA in a subset of MTM patients (27,33,34). We are currently exploring the use of antisense oligonucleotide therapy and we are searching for other human exonic MTM1 point mutations that may also induce skipping.
Muscle from Mtm1 p.R69C and Mtm1 KO mice with weakness showed similarly elevated PI3P levels, suggesting that other biologic functions of myotubularin besides its lipid phosphatase activity may account for some differences in phenotype noted in these models. These other biologic functions are currently not clear but may relate to the interactions of myotubularin with other proteins, such as desmin (35). This is a subject that requires further study.
Conclusions
The MTM1 c.205C>T base change is associated with relatively mild MTM clinical course in patients. We modeled this mutation in mice, with the goal of generating a less severely affected MTM model to complement the Mtm1 KO mouse. We believed such a model would expand opportunities for preclinical research. The Mtm1 p.R69C mouse recapitulates the pathologic and clinical features of a less severe human MTM phenotype and, notably, it has a considerably longer lifespan than the Mtm1 KO mouse. Unexpectedly, introducing the c.205C>T base change in Mtm1 exon 4 induced skipping of exon 4, suggesting that antisense oligonucleotide therapy may be a viable treatment strategy in a subset of MTM patients.
MATERIALS AND METHODS
Gene targeting
The targeting vector was generated at InGenious Gene Targeting (Stony Brook, NY, USA) and is shown in Supplementary Material, Figure S1A. An 8.4 kb genomic DNA fragment used in the targeting vector was subcloned from an identified B6 BAC clone (RP23-125F4). The long homology arm extends 6.1 kb 5′ to the point mutation. The Lox/FRT-Neo cassette is situated 345 bp downstream of the point mutation in exon 4. The short homology arm extends 1.9 kb 3′ to the floxed Neo cassette. Restriction digests and sequencing were performed following each modification of the targeting vector. The boundaries of each homology arm were confirmed by sequencing with P6 and T7 primers (sequence of all primers appears in Supplementary Material) that read through both sides of the backbone vector. The floxed Neo cassette was confirmed by sequencing with N1 and N2 primers that read through both sides of the Neo cassette and into the genomic sequences. The point mutation was confirmed by sequencing with PT4 and SQ1 primers. Sequencing results confirmed that no additional mutations were introduced into the exons.
The c.205C>T point mutation was introduced into Mtm1 cDNA within exon 4 by three-step PCR mutagenesis. Four mutagenesis primers (PT1–4) were used to amplify a 0.6 kb product that includes the point mutation in the desired location (Supplementary Material, Fig. S1B). The point mutation was engineered into primer PT3 (Supplementary Material, Fig. S1B). The PCR product bearing the point mutation was used to replace the wild-type sequence using conventional subcloning techniques.
Ten micrograms of the targeting vector was linearized by NotI and electroporated into C57BL/6 × 129/SvEv hybrid ES cells. Clones were selected in G418 and survivors expanded for PCR analysis to identify recombinant ES clones. The PCR screening strategy consisted of primers (A1, A2) designed to anneal downstream of the short homology arm outside of the 3′ region used to generate the targeting construct and a primer (F7) designed to anneal within the Neo cassette, thus allowing positive recombinant clones to be identified by a 2.1 kb PCR product. Control PCR using primers (AT1 and AT2) designed to anneal at the 3′ and 5′ ends of the short homology arm resulted in a 1.7 kb product. PCR using SQ1 and LAN1 primers was used to confirm the point mutation in the long homology arm, and the product was sequenced using the PT1 primer. Five clones with the introduced point mutation were reconfirmed for short homology arm integration by sequencing (Supplementary Material, Fig. S1C).
Positive clones identified by the PCR screening strategy were further confirmed by Southern blotting. DNA was digested with BamHI, electrophoretically separated, transferred to nylon membrane and hybridized with a probe designed to target the short homology arm. Wild-type C57BL/6 and 129/SvEv and hybrid strains were used as positive controls.
Generation and genotyping of mice
The Institutional Animal Care and Use Committee approved of all animal procedures in accordance with the principles delineated in the NIH Guide for the Care and Use of Laboratory Animals. Positive ES cell clones were microinjected into C57BL/6 blastocysts at InGenious Gene Targeting. Chimeras with a high percentage agouti coat color were mated to wild-type C57BL/6 mice to produce F1 heterozygous offspring. F1 tail DNA was analyzed using PCR and the A2/F7 primer set to generate a 2.2 kb product. F7 anneals inside the Neo cassette and A2 anneals 3′ to the short homology arm, outside the region used to create the targeting construct. The expanded positive ES cell clone was used as a positive control. Tail DNA samples from properly targeted F1 mice were PCR amplified by primer SQ1 and LAN1 and the resulting 1 kb product was sequenced using primer PT1 to confirm the introduced c.205C>T point mutation (Supplementary Material, Fig. S1D). The F1 heterozygous female mice were mated to wild-type C57BL/6 male mice to yield the F2 generation that was genotyped using the SQ1 and LAN1 primers with the following thermocycling parameters: 95°C for 5 min, 95°C for 30 s, 55°C for 30 s, 72°C for 2 min for 35 cycles followed by an extension period of 72°C 5 min. Heterozygous female mice were mated with wild-type male C57BL/6 mice for 10 generations with PCR genotyping performed at weaning.
Animals used
Survival time was determined by comparing a cohort of 57 Mtm1 p.R69C animals to 54 wild-type littermates. Twelve Mtm1 p.R69C and 12 wild-type littermates were used to test muscle performance using a dynamometer, and activity using an open field, over a 12-month period. The Mtm1 p.R69C and wild-type data at 1 month were compared with twelve 1-month-old Mtm1 KO animals. Quadriceps, gastrocnemius, tibialis anterior and triceps muscle masses, as well as liver, kidney, heart and lung masses were determined following dissection from Mtm1 p.R69C and wild-type littermates (n = 8 each group) each month for 9 months and the masses were recorded. These organ masses were compared with eight 1-month-old Mtm1 KO mice. Mtm1 p.R69C and wild-type animals were maintained on a high-fat diet with ad libitum access to food and water, which led to significant obesity at 5 months in the later and impacted the normalization of individual muscle mass to body mass. At some time points, it even suggested that Mtm1 p.R69C organ masses exceeded that of the wild-type, while the actual masses of these tissues showed no significant change between 1 and 9 months. Therefore, the muscle performance data reported are not normalized to body mass. During the course of this study, 64 Mtm1 p.R69C animals that died underwent a necropsy that included a detailed gross examination of all organs and microscopic evaluation of grossly evident lesions.
Histopathology and morphometry
Cross-sections (8 μm) cut from quadriceps muscle that was frozen in liquid nitrogen-chilled isopentane were stained with hematoxylin and eosin or NADH-TR and visualized using a Zeiss Axioscope 2 Plus microscope. Non-overlapping fields of muscle were photographed using Zeiss AxioCam MRC 5 camera. All the fibers in the field were individually selected using the AxioVs40 V 4.6.3.0 morphometry software and the minimum Feret (MinFeret) diameter was recorded. The MinFeret diameter is the smallest diameter across an ellipse that contains the true center (36,37). Since a large number of myofibers were measured to ensure that the entire specimen was represented, in general a larger number of fibers were counted in the Mtm1 p.R69C (range: 793–1107 fibers at 1 month; 1219–2095 fibers at 2 months) and Mtm1 KO animals (range: 586–856 fibers at 1 month) relative to controls (range: 301–879 fibers at 1 month; 219–453 fibers at 2 months). This was due to the relatively small size of myofibers in the affected animals. Histograms were generated in Excel using pooled MinFeret diameters and the mean and standard deviations calculated for statistical analysis. The coefficient of variation (Cv) was calculated as the ratio of the standard deviation to the mean.
Immunofluorescence
Cross-sections (8 μm) of isopentane frozen quadriceps were cut using a cryostat, washed and blocked in blocking buffer [poshphate buffered saline (PBS) with 10% fetal bovine serum and 0.1% Triton X-100] for 1 h at room temperature. Fiber types were stained using anti-myosin heavy chain type 2A (SC-71-S) at 5 μm/ml, anti-myosin heavy chain type 2B (BF-F3-S) at 10 μm/ml (both from the Developmental Hybridoma Bank) or anti-myosin heavy chain, slow, undiluted (NCL-MHCS, Novacastra) antibodies. Monoclonal anti-RyR 1 (Clone: R129) and anti-dihydropyradine receptor (Clone: D218) antibodies (Sigma, St Louis, MO, USA) were diluted 1:50 in blocking buffer and applied overnight at 4°C. The next day, slides were washed in PBS and alexa-fluor-conjugated anti-mouse secondary antibody was then applied at 1:50 or 1:100 in blocking buffer for 1 h at room temperature. Slides were washed in PBS and mounted in medium containing 4′,6-diamidino-2-phenylindole (Vectorshield, Burlingame, CA, USA). Slides were imaged using a Leica epiflourescence microscope and images captured using SPOT Advanced imaging software.
Electron microscopy
Fixed tissue was subjected to osmication, stained using uranyl acetate, dehydrated in alcohols and embedded in TAAB epon (Marivac Ltd., Halifax, Nova Scotia, Canada). Subsequently, 1 μm scout sections were stained with toluidine blue and evaluated. Areas of interest were cut at 95 nm thickness using an ultracut microtome (Leica Camera AG, Solms, Germany) picked up on 100-μm formvar-coated copper grids, stained with 0.2% lead citrate and viewed and imaged using a Tecnai BioTwin Spirit Electron Microscope (FEI Co., Hillsboro, OR, USA).
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting
Muscle tissue was homogenized in extraction buffer (22) supplemented with Halt protease inhibitor (Pierce Biotechnology, Rockford, IL, USA). Protein concentrations were determined using the bicinchoninic acid method (Pierce Biotechnology). Fifty micrograms of total protein was separated on a 4–20% gradient Bis–Tris gel (Invitrogen) and transferred to polyvinylidene fluoride membrane (Bio-Rad). Membranes were probed with polyclonal anti-myotubularin antibody (r1947) (22) for 2 h followed by donkey anti-rabbit antibody conjugated to horseradish peroxidase (Jackson Immunoresearch). Chemiluminescent detection was performed using ECL (Pierce Biotechnology) followed by exposure to photographic film (Pierce Biotechnology).
Examination of splicing regulatory elements located in Mtm1 exon 4
ESRsearch, ESEFinder and PESX Internet-based programs were used to search for putative splicing regulatory elements that the c.205C>T mutation could disrupt or create (24,25,28–30). All of exon 4 containing either the wild-type or c.205C>T sequence was searched.
Generation of splicing competent minigenes
The pMtm1 wild-type minigene was constructed to contain the wild-type mouse genomic Mtm1 fragment containing exons 3, 4, and 5 and the corresponding intronic sequences. Due to the size of intron 4, only the first 657 nt from the 5′ splice site and 837 nt from the 3′ splice site were used in the creation of the minigene. This was achieved by PCR amplification using Invitrogen Platinum Taq DNA Polymerase High Fidelity (Carlsbad, CA, USA) and cloning each PCR fragment into pCR 2.1-TOPO Vector (Invitrogen). The Mtm1X3BamHIFW primer and Mtm1X4XmaIRV primer pair were used to clone arm 1, while the Mtm1X4XmaIFW and Mtm1X5SalIRV were used to clone arm 2. Each arm was sequenced to verify correct amplification then ligated into Stratagene's (La Jolla, CA, USA) CMV-2B vector. Site-directed mutagenesis using QuikChange XL Site Directed Mutagenesis Kit (Stratagene) was performed to convert the exon 4 c.205C>T and create the Mtm1 p.R69C minigene. The CMV-2B vector was chosen so the minigene would contain the vector-specific 5′ FLAG tag sequence. This unique sequence can be used during PCR experiments to avoid the detection of endogenous Mtm1 transcripts.
Transfection and in vivo splicing assays
All reagents were used according to the manufacturer's recommendations. Transient transfections of cells with plasmid DNA were performed with Lipofectamine 2000 (Invitrogen). Cells were plated 24 h prior to transfection so that their density on the day of transfection was ∼70–90% confluence. Total RNA was isolated 24 h after transfection from cells or tissue using TRIzol (Invitrogen). To generate cDNA, reverse transcription was carried out with Bio-Rad iScript cDNA Synthesis Kit (Hercules, CA, USA). Two micrograms of total RNA was used per 40 µl of reaction mixture. Minigene-specific spliced products were subsequently amplified with Taq polymerase (Sigma) and the FLAG and Mtm1 cDNA 3′ primer pair. Mtm1 cDNA was amplified in mouse using X3 FWD and X5 REV, and in the patient muscle using X3F and X5R. Results were confirmed by a minimum of three independent experiments.
Real-time PCR and data analysis
Total mRNA was isolated from quadriceps muscle by homogenization in TRIzol (Invitrogen) and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). Three independent real-time PCR experiments, with each sample performed in triplicate, were performed for each gene using Taqman gene expression assays (see Supplementary Material for probe names and part numbers) as directed. Real-time PCR data were recorded and analyzed using ABI 7900 HT fast real-time PCR system and 7500 Real-Time PCR System Sequence Detection Software v1.2.3. The relative amount of target gene expression in Mtm1 p.R69C animals was calculated as fold changes relative to wild-type littermate controls that were normalized to glyceraldehyde 3-phosphate dehydrogenase.
Muscle mass and performance studies
Quadriceps, gastrocnemius, tibialis anterior and triceps muscles as well as liver, kidney, heart and lungs were dissected from Mtm1 p.R69C and wild-type littermates (n = 8 each group) each month for 9 months and the masses were recorded. A separate cohort of 12 Mtm1 p.R69C and 12 wild-type littermates were followed to test muscle performance. Grip force strength generated by hindlimb and forelimbs was recorded using a dynamometer with a digital gauge (Columbus Instruments, Columbus, OH, USA). The test was performed by letting the animal grasp a grid and the animal was gently pulled until it released the grid. The recorded force measurement generated for each animal at each time point represents the mean of three successive measurements (38). Open field activity was assessed using the Photobeam Activity System (San Diego Instruments, San Diego, CA, USA). The open field is 42 cm × 42 cm and has photobeams and detectors that are spaced 2.5 cm apart in both X- and Y-planes and 2 cm above the floor of the field. Following a 5 min acclimation period, the Photobeam Activity System Software V.2.0.7.101 recorded the number of beam breaks occurring in a 10 min period due to locomotion in the open field. Rearing was concurrently recorded with the open field activity analysis using a second set of photobeams and detectors that are 6 cm above the floor of the open field. The mouse only breaks these beams when it rears and the number of rearing events that occur in the same 10 min period was recorded.
Statistics
Two group comparisons were performed using a two-tailed t test. Analysis of variance (ANOVA) was used to compare three groups followed by Scheffe post hoc testing. The Kaplan–Meier survival analysis was performed with the log-rank test for significance testing. All tests were performed using SigmaPlot v11 software. Significance was defined as P < 0.05 in all tests.
PI3P ELISA
PI3P Mass ELISAs were performed on lipid extracts from whole quadriceps skeletal muscle preparations according to the manufacturer's recommendations (Echelon Biosciences). Quadriceps were isolated at necropsy, weighed and then incubated in ice cold 5% TCA in order to extract lipids. Extracted lipids were resuspended in PBS-T with 3% protein stabilizer and then spotted on PI3P Mass ELISA plates in triplicate. PI3P levels were detected by measuring absorbance at 450 nm on a plate reader after following the protocol provided with the Mass ELISA kit. Specific amounts were determined by comparison of values to a standard curve generated with simultaneous readings of known amounts of PI3P.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work is supported by the National Institutes of Health (K08 NS049095 to C.R.P., R01 AR044345, P50 NS040828 to A.H.B., K08 AR059750-01 and L40 AR057721-01 to M.W.L.); the Muscular Dystrophy Association (MDA155638, to C.R.P. and MDA201302 to A.H.B.); The Lee and Penny Anderson Family Foundation; The Joshua Frase Foundation; The Foye Family and Mr John Dulin.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Benjamin Bryson, Kristen Roth and Marissa Viola for their scientific contributions and Steven W. Lange for his editorial expertise. Anti-myosin type 2A and 2B antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA, USA.
REFERENCES
- 1.Jungbluth H., Wallgren-Pettersson C., Laporte J. Centronuclear (myotubular) myopathy. Orphanet. J. Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. doi:10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierson C.R., Tomczak K., Agrawal P., Moghadaszadeh B., Beggs A.H. X-linked myotubular and centronuclear myopathies. J. Neuropathol. Exp. Neurol. 2005;64:555–564. doi: 10.1097/01.jnen.0000171653.17213.2e. [DOI] [PubMed] [Google Scholar]
- 3.Romero N.B. Centronuclear myopathies: a widening concept. Neuromuscul. Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. doi:10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Laporte J., Hu L.J., Kretz C., Mandel J.L., Kioschis P., Coy J.F., Klauck S.M., Poustka A., Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. doi:10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 5.Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J.F., Mandel J.L. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl Acad. Sci. USA. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. doi:10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman G.E., Kopacz K., Zhao W., Mills P.L., Metzenberg A., Das S. Characterization of mutations in fifty North American patients with X-linked myotubular myopathy. Hum. Mutat. 2002;19:114–121. doi: 10.1002/humu.10033. doi:10.1002/humu.10033. [DOI] [PubMed] [Google Scholar]
- 7.McEntagart M., Parsons G., Buj-Bello A., Biancalana V., Fenton I., Little M., Krawczak M., Thomas N., Herman G., Clarke A., et al. Genotype-phenotype correlations in X-linked myotubular myopathy. Neuromuscul. Disord. 2002;12:939–946. doi: 10.1016/s0960-8966(02)00153-0. doi:10.1016/S0960-8966(02)00153-0. [DOI] [PubMed] [Google Scholar]
- 8.Barth P.G., Dubowitz V. X-linked myotubular myopathy—a long-term follow-up study. Eur. J. Paediatr. Neurol. 1998;2:49–56. doi: 10.1016/1090-3798(98)01004-9. doi:10.1016/1090-3798(98)01004-9. [DOI] [PubMed] [Google Scholar]
- 9.Herman G.E., Finegold M., Zhao W., de Gouyon B., Metzenberg A. Medical complications in long-term survivors with X-linked myotubular myopathy. J. Pediatr. 1999;134:206–214. doi: 10.1016/s0022-3476(99)70417-8. doi:10.1016/S0022-3476(99)70417-8. [DOI] [PubMed] [Google Scholar]
- 10.Biancalana V., Caron O., Gallati S., Baas F., Kress W., Novelli G., D'Apice M.R., Lagier-Tourenne C., Buj-Bello A., Romero N.B., et al. Characterisation of mutations in 77 patients with X-linked myotubular myopathy, including a family with a very mild phenotype. Hum. Genet. 2003;112:135–142. doi: 10.1007/s00439-002-0869-1. [DOI] [PubMed] [Google Scholar]
- 11.de Gouyon B.M., Zhao W., Laporte J., Mandel J.L., Metzenberg A., Herman G.E. Characterization of mutations in the myotubularin gene in twenty six patients with X-linked myotubular myopathy. Hum. Mol. Genet. 1997;6:1499–1504. doi: 10.1093/hmg/6.9.1499. doi:10.1093/hmg/6.9.1499. [DOI] [PubMed] [Google Scholar]
- 12.Laporte J., Biancalana V., Tanner S.M., Kress W., Schneider V., Wallgren-Pettersson C., Herger F., Buj-Bello A., Blondeau F., Liechti-Gallati S., et al. MTM1 mutations in X-linked myotubular myopathy. Hum. Mutat. 2000;15:393–409. doi: 10.1002/(SICI)1098-1004(200005)15:5<393::AID-HUMU1>3.0.CO;2-R. doi:10.1002/(SICI)1098-1004(200005)15:5<393::AID-HUMU1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 13.Laporte J., Guiraud-Chaumeil C., Vincent M.C., Mandel J.L., Tanner S.M., Liechti-Gallati S., Wallgren-Pettersson C., Dahl N., Kress W., Bolhuis P.A., et al. Mutations in the MTM1 gene implicated in X-linked myotubular myopathy. ENMC International Consortium on Myotubular Myopathy. European Neuro-Muscular Center. Hum. Mol. Genet. 1997;6:1505–1511. doi: 10.1093/hmg/6.9.1505. doi:10.1093/hmg/6.9.1505. [DOI] [PubMed] [Google Scholar]
- 14.Pierson C.R., Agrawal P.B., Blasko J., Beggs A.H. Myofiber size correlates with MTM1 mutation type and outcome in X-linked myotubular myopathy. Neuromuscul. Disord. 2007;17:562–568. doi: 10.1016/j.nmd.2007.03.010. doi:10.1016/j.nmd.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. doi:10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., Mruk D.D., Cheng C.Y. Myotubularin phosphoinositide phosphatases, protein phosphatases, and kinases: their roles in junction dynamics and spermatogenesis. J. Cell Physiol. 2005;204:470–483. doi: 10.1002/jcp.20303. doi:10.1002/jcp.20303. [DOI] [PubMed] [Google Scholar]
- 17.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A.H., Allard B., Mandel J.L., et al. T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl Acad. Sci. USA. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. doi:10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beggs A.H., Bohm J., Snead E., Kozlowski M., Maurer M., Minor K., Childers M.K., Taylor S.M., Hitte C., Mickelson J.R., et al. MTM1 mutation associated with X-linked myotubular myopathy in Labrador Retrievers. Proc. Natl Acad. Sci. USA. 2010;107:14697–14702. doi: 10.1073/pnas.1003677107. doi:10.1073/pnas.1003677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevilacqua J.A., Bitoun M., Biancalana V., Oldfors A., Stoltenburg G., Claeys K.G., Lacene E., Brochier G., Manere L., Laforet P., et al. ‘Necklace’ fibers, a new histological marker of late-onset MTM1-related centronuclear myopathy. Acta Neuropathol. 2009;117:283–291. doi: 10.1007/s00401-008-0472-1. doi:10.1007/s00401-008-0472-1. [DOI] [PubMed] [Google Scholar]
- 20.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. doi:10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toussaint A., Cowling B.S., Hnia K., Mohr M., Oldfors A., Schwab Y., Yis U., Maisonobe T., Stojkovic T., Wallgren-Pettersson C., et al. Defects in amphiphysin 2 (BIN1) and triads in several forms of centronuclear myopathies. Acta. Neuropathol. 2011;121:253–266. doi: 10.1007/s00401-010-0754-2. [DOI] [PubMed] [Google Scholar]
- 22.Buj-Bello A., Fougerousse F., Schwab Y., Messaddeq N., Spehner D., Pierson C.R., Durand M., Kretz C., Danos O., Douar A.M., et al. AAV-mediated intramuscular delivery of myotubularin corrects the myotubular myopathy phenotype in targeted murine muscle and suggests a function in plasma membrane homeostasis. Hum. Mol. Genet. 2008;17:2132–2143. doi: 10.1093/hmg/ddn112. doi:10.1093/hmg/ddn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laporte J., Kress W., Mandel J.L. Diagnosis of X-linked myotubular myopathy by detection of myotubularin. Ann. Neurol. 2001;50:42–46. doi: 10.1002/ana.1033. doi:10.1002/ana.1033. [DOI] [PubMed] [Google Scholar]
- 24.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. doi:10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. doi:10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 26.Goren A., Ram O., Amit M., Keren H., Lev-Maor G., Vig I., Pupko T., Ast G. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. doi:10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. doi:10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith P.J., Zhang C., Wang J., Chew S.L., Zhang M.Q., Krainer A.R. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. doi:10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. doi:10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. doi:10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black B.L., Croom J., Eisen E.J., Petro A.E., Edwards C.L., Surwit R.S. Differential effects of fat and sucrose on body composition in A/J and C57BL/6 mice. Metabolism. 1998;47:1354–1359. doi: 10.1016/s0026-0495(98)90304-3. doi:10.1016/S0026-0495(98)90304-3. [DOI] [PubMed] [Google Scholar]
- 32.Lawlor M.W., Read B.P., Edelstein R., Yang N., Pierson C.R., Stein M.J., Wermer-Colan A., Buj-Bello A., Lachey J.L., Seehra J.S., et al. Inhibition of activin receptor type IIb increases strength and lifespan in myotubularin-deficient mice. Am. J. Pathol. 2011;178:784–793. doi: 10.1016/j.ajpath.2010.10.035. doi:10.1016/j.ajpath.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aartsma-Rus A., van Ommen G.J. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. doi:10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Roy F., Charton K., Lorson C.L., Richard I. RNA-targeting approaches for neuromuscular diseases. Trends Mol. Med. 2009;15:580–591. doi: 10.1016/j.molmed.2009.10.005. doi:10.1016/j.molmed.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Hnia K., Tronchere H., Tomczak K.K., Amoasii L., Schultz P., Beggs A.H., Payrastre B., Mandel J.L., Laporte J. Myotubularin controls desmin intermediate filament architecture and mitochondrial dynamics in human and mouse skeletal muscle. J. Clin. Invest. 2011;121:70–85. doi: 10.1172/JCI44021. doi:10.1172/JCI44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooke M.H., Engel W.K. The histographic analysis of human muscle biopsies with regard to fiber types. 4. Children's biopsies. Neurology. 1969;19:591–605. doi: 10.1212/wnl.19.6.591. [DOI] [PubMed] [Google Scholar]
- 37.Brooke M.H., Engel W.K. The histographic analysis of human muscle biopsies with regard to fiber types. 3. Myotonias, myasthenia gravis, and hypokalemic periodic paralysis. Neurology. 1969;19:469–477. doi: 10.1212/wnl.19.5.469. [DOI] [PubMed] [Google Scholar]
- 38.Miller T.M., Kim S.H., Yamanaka K., Hester M., Umapathi P., Arnson H., Rizo L., Mendell J.R., Gage F.H., Cleveland D.W., et al. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2006;103:19546–19551. doi: 10.1073/pnas.0609411103. doi:10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.