Abstract
MntH is the only high-affinity manganese transporter identified in Brucella. A previous study showed that MntH is required for the wild-type virulence of Brucella abortus 2308 in mice (Anderson ES, et al., Infect. Immun. 77:3466–3474, 2009) and indicated that the mntH gene is regulated in a manganese-responsive manner in this strain by a Mur homolog. In the study presented here, the transcriptional start site for mntH in B. abortus 2308 was determined by primer extension analysis. Specific interactions between Mur and the mntH promoter region were demonstrated in an electrophoretic mobility shift assay (EMSA), and a Mur binding site was identified in the −55 to −24 region of the mntH promoter by DNase I footprint analysis. The specificity of the interaction of Mur with the putative Mur box was further evaluated by EMSA employing oligonucleotides in which the consensus nucleotides in this region were substituted. These studies not only confirm a direct role for Mur in the Mn-responsive regulation of mntH expression in Brucella abortus 2308 but also identify the cis-acting elements upstream of mntH that are responsible for this regulation.
INTRODUCTION
A substantial number of bacterial proteins require metal ions for their activity and proper function. However, the accumulation of metals beyond the level at which they are needed can be toxic due to incorrect metal-protein interactions (39) and the capacity of certain metals, such as iron and copper, to participate in the production of toxic oxygen radicals (38). To ensure that they only accumulate the levels of metals they need to meet their physiologic requirements, bacteria produce transporters that mediate both the influx and efflux of specific metal ions (39). The expression of the genes encoding these transport systems typically is tightly controlled by transcriptional regulators whose activities respond to the levels of specific metal ions in the bacterial cell (11). This specific and differential regulation of metal transport genes enables bacteria to actively adapt to different and sometimes rapidly changing levels of available metals in the external environment (39).
Manganese is an essential cofactor for a variety of bacterial proteins (25). Bacterial genes encoding manganese transporters typically are regulated by MntR- or Mur-type transcriptional regulators (12, 30). Mur is a structural homolog of the iron-responsive ferric uptake regulator (Fur) (13, 14), which controls the expression of iron uptake genes in many bacteria (20). Although Mur was first described in Rhizobium leguminosarum (7) as a suspected iron-responsive regulator (40), genetic and biochemical studies have clearly shown that Mur is a manganese-responsive transcriptional regulator of manganese uptake genes (7) in R. leguminosarum and other alphaproteobacteria (5, 28). Mur does not, however, appear to directly participate in the regulation of iron-responsive genes in these bacteria. Instead, the iron-responsive transcriptional regulators Irr and RirA control the expression of the iron metabolism genes in the alphaproteobacteria (18, 30, 33).
The Brucella spp. are members of the alphaproteobacteria and are the causative agents of brucellosis (32). Brucellosis causes sterility and abortion in wild and domestic animals and a severe febrile illness in humans (24). Brucella strains rely upon MntH as their sole high-affinity manganese transporter (1), and MntH plays a critical role in the virulence of B. abortus 2308 in experimentally infected mice. The expression of the mntH gene is regulated in a manganese-responsive manner in this strain, and genetic studies have implicated Mur in this regulation. The purpose of the studies described in this report was to determine whether or not Mur plays a direct role in the manganese-responsive regulation of mntH expression in B. abortus 2308, and if so, to identify the nucleotide sequences to which Mur binds in the mntH promoter region.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Brucella strains were cultivated on Schaedler agar supplemented with 5% defibrinated bovine blood (SBA) at 37°C with 5% CO2 or in brucella broth at 37°C with shaking. Low-manganese minimal medium was prepared as previously described (1). Ampicillin (25 μg/ml) and kanamycin (45 μg/ml) were included in these growth media as appropriate for the selection of Brucella strains carrying antibiotic resistance markers. Brucella stock cultures were stored at −80°C in brucella broth supplemented with 25% glycerol. Escherichia coli strains were grown at 37°C on LB agar or in LB broth or these media supplemented with 100 μg/ml ampicillin or 45 μg/ml kanamycin as needed. E. coli stock cultures were maintained in LB supplemented with 25% glycerol at −80°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | Laboratory cloning strain | Invitrogen |
| BL21(DE3) | Laboratory cloning strain | 37 |
| Brucella abortus | ||
| 2308 | Virulent challenge strain | Laboratory stock |
| Plasmids | ||
| pUC19 | ColE1-based cloning vector; Apr | 41 |
| pASK-IBA6 | ColE1-based expression plasmid for production of Strep-tagged recombinant proteins; Apr | IBA GmbH |
| pEAM2 | pASK-IBA6 derivative containing a cloned copy of the mur gene from B. abortus 2308 | This study |
| pMRS | Derivative of pUC19 containing the hybridized Murbox control F and R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
| pMRS1 | Derivative of pUC19 containing the hybridized Murbox mutation 1 F and 1 R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
| pMRS2 | Derivative of pUC19 containing the hybridized Murbox mutation 2 F and 2 R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
| pMRS3 | Derivative of pUC19 containing the hybridized Murbox mutation 3 F and 3 R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
| pMRS4 | Derivative of pUC19 containing the hybridized Murbox mutation 4 F and 4 R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
| pMRS5 | Derivative of pUC19 containing the hybridized Murbox mutation 5 F and 5 R oligonucleotides cloned into the EcoRI/BamHI sitea | This study |
The sequences of the Murbox control and Murbox mutation oligonucleotides are listed in Table S1 in the supplemental material.
Production of recombinant Brucella Mur.
The oligonucleotide primers rMur Forward and rMur Reverse (see Table S1 in the supplemental material), which encode BsaI restriction sites, were used with Pfx DNA polymerase (Invitrogen) to amplify a 426-bp DNA fragment containing the mur gene from B. abortus 2308 genomic DNA by PCR, and this fragment was cloned into the expression vector pASK-IBA6 (IBA GmbH, Göttingen, Germany). The resulting plasmid, designated pEAM2, which contains a gene fusion encoding an N-terminal Strep-tag II version of the Brucella Mur, was introduced into E. coli DH5α by chemical transformation for screening purposes and the propagation of the plasmid. Once the authenticity of the gene fusion was confirmed by nucleotide sequence analysis, this plasmid was introduced into E. coli strain BL21(DE3) by electrotransformation. For recombinant protein production, E. coli cultures were grown overnight at 37°C in LB broth, and 10 ml of these cultures was used to inoculate 1 liter of LB in a 4-liter Erlenmeyer flask. These cultures were grown at 37°C with shaking at 250 rpm to an optical density at 600 nm of approximately 0.5, at which time anhydrotetracycline (AHTC) was added to the cultures to obtain a final concentration of 0.2 μg/ml and the incubation continued. Three to 4 h after the addition of AHTC, bacterial cells were harvested by centrifugation at 2,700 × g for 15 min at 4°C and lysed with CelLytic B-cell lysis reagent (Sigma) in the presence of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) on ice for 30 min. The cell lysates were subjected to centrifugation at 17,510 × g for 15 min at 4°C, and the supernatant was collected and passed through a Strep-Tactin gravity flow column (IBA GmbH, Göttingen, Germany) following the manufacturer's instructions. Protein fractions were pooled and dialyzed in electrophoretic mobility shift assay (EMSA) binding buffer (10 mM Tris-HCl, 40 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, and 5% glycerol [pH 7.5]) and concentrated by centrifugation through an Amicon Ultra-15 centrifugal filter (molecular weight cutoff of 3,000) following the manufacturer's instructions. Total protein was quantified by the Bradford assay (4), and the degree of purity was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Aliquots of the recombinant Brucella Mur protein were maintained at −80°C in the presence of 10% glycerol.
Identification of the mntH transcriptional start site by primer extension analysis.
Total RNA was isolated from B. abortus 2308 grown in low-manganese medium and this medium supplemented with 50 μM MnCl2 employing the methods described by Mohanty et al. (23). The primer mntH-PE (see Table S1 in the supplemental material) was annealed to 50 μg of total RNA, and the primer extension reaction was carried out using a modification of the methods described by Sambrook et al. (34). Briefly, the reaction was performed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) in 5× first-strand buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 100 mM dithiothreitol) plus 0.5 mM deoxynucleoside triphosphates (dNTPs). The 5′ end of the mntH transcript was identified by electrophoresis in a denaturing 6% (wt/vol) acrylamide and 7 M urea sequencing gel in glycerol-tolerant buffer (0.089 M Tris base, 0.0285 M taurine, and 0.5 mM Na2EDTA) alongside a DNA sequence ladder generated using the same primer. The PCR product used as a template for the sequencing ladder was a 1-kb DNA fragment encompassing the coding region of mntH and upstream sequence amplified from B. abortus 2308 genomic DNA with the primers mntH-Forward and mntH-Reverse (see Table S1 in the supplemental material). DNA sequence analysis was performed using the SequiTherm Excel II DNA sequencing kit following the instructions provided by the manufacturer (Epicentre, Madison, WI).
EMSA.
The methods described by Platero et al. (28) were employed for the EMSAs, with the exception that the nonionic detergent NP-40 was excluded from the reaction mixture. The DNA fragment representing the mntH promoter region was amplified by PCR from B. abortus 2308 genomic DNA using Pfx polymerase and the primers designated mntH-MurboxF and mntH-MurboxR (see Table S1 in the supplemental material) and end labeled with [γ-32P]ATP (Perkin Elmer, Waltham, MA) using T4 polynucleotide kinase (Promega, Madison, WI). An unlabeled version of this DNA fragment was used as a specific competitor in this reaction mixture, and an unlabeled version of a 150-bp DNA fragment, representing the upstream region and 20 bp of the coding region of the recA gene (BAB1_1224) amplified from B. abortus 2308 genomic DNA by PCR with the primers recAF and recAR (see Table S1 in the supplemental material), was used as a nonspecific competitor.
EMSA analysis employing nucleotide fragments with mutated forms of the Mur binding site was performed using the methods described by Stojilkkovic et al. (36). Synthetic and complementary oligonucleotides (35 nucleotides in length), within which selected nucleotides in the putative Mur binding site in the mntH promoter region were substituted, were synthesized by Integrated DNA Technologies (Coralville, IA). EcoRI and BamHI sites were also incorporated at the ends of these oligonucleotides, which are labeled Murbox mutations 1 to 5 F and Murbox mutations 1 to 5 R in Table S1 in the supplemental material. Complementary oligonucleotides were annealed, phosphorylated, and cloned into BamHI-EcoRI-digested pUC19, resulting in the construction of plasmids designated pMRS1 to pMRS5 (Table 1). Double-stranded DNA fragments containing the mutated Mur binding sites were removed from the pUC19 derivatives by digestion with BamHI and EcoRI and used for EMSA analysis using the procedures described in the previous paragraph.
DNase I footprint analysis.
The DNase I footprint assay was performed as described previously (8), with slight modifications. Briefly, the oligonucleotide primers mntH-MurboxF and mntH-MurboxR (see Table S1 in the supplemental material) were individually labeled with [γ-32P]ATP (Perkin Elmer) using the T4 polynucleotide kinase reaction (Promega, Madison, WI) prior to their use in PCRs with Pfx polymerase to generate 110-bp DNA fragments encompassing the putative Mur box and flanking regions of the mntH promoter. The resulting PCR products were subjected to agarose electrophoresis and purified by gel extraction (Fermentas, Glen Burnie, MD). DNA probes corresponding to 15,000 cpm of the forward labeled and reverse labeled templates were incubated separately in EMSA binding buffer supplemented with 100 ng/ml bovine serum albumin (BSA) and 50 ng/ml salmon sperm DNA (nonspecific competitor) in the presence of 100 μM MnCl2 and increasing concentrations of the recombinant Brucella Mur protein. The reaction mixture was incubated at room temperature for 30 min prior to treatment with 0.05 U of DNase I freshly diluted in 10× DNase I buffer (400 mM Tris-HCl [pH 8.0], 100 mM MgSO4, 10 mM CaCl2) for 1 min. This reaction was stopped by the addition of 5 mM EDTA and heating at 65°C for 10 min. Reaction mixtures were ethanol precipitated and resuspended in 4 μl of formamide loading buffer (98% formamide, 10 mM EDTA [pH 8.0], 1 mg/ml xylene cyanol FF, 1 mg/ml bromophenol blue). Digested DNA fragments were separated on a denaturing 6% (wt/vol) acrylamide and 7 M urea sequencing gel in glycerol-tolerant buffer (as described above). Gels were dried in a vacuum dryer and subjected to autoradiography. The sequence protected by Mur was determined by comparing the nucleotide sequences generated for a 100-bp region of the mntH promoter region using the SequiTherm Excel II DNA sequencing kit (Epicentre, Madison, WI) and B. abortus 2308 DNA preparations exposed to DNase I treatment with and without recombinant Mur as templates.
RESULTS
Mur directly binds to the mntH promoter in B. abortus 2308.
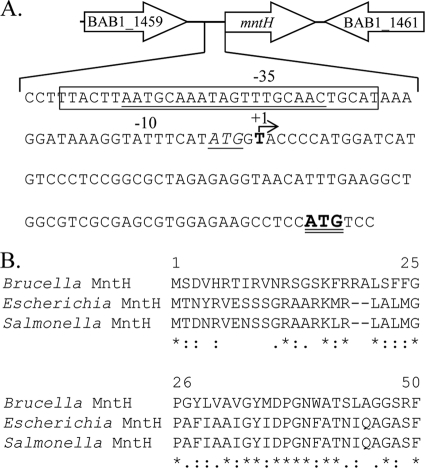
A previous study showed that Mur is required for the manganese-responsive repression of mntH expression in B. abortus 2308 (1), but a direct interaction between Mur and the mntH promoter has not been demonstrated. A putative Mur recognition site (MRS) (8), 5′-AATGCAAATAGTTTGCAAT-3′, lies upstream of the mntH coding region in the Brucella melitensis 16M genome sequence (32), and this same genomic arrangement occurs in B. abortus 2308 (Fig. 1A). To evaluate the functionality of this MRS, primer extension analysis was first used to determine the transcriptional start site for mntH in B. abortus 2308 and define the promoter for this gene. As shown in Fig. 1A, the putative MRS overlaps the −35 region of the mntH promoter, which would be consistent with the predicted function of Mur as a repressor of mntH expression. Interestingly, the thymidine (T) residue identified as the transcriptional start site for mntH represents the middle nucleotide in the second codon of the mntH coding region annotated in the B. abortus 2308 and B. melitensis 16M genome sequences. The proposed start codon for the annotated mntH coding region is presented in italics and underlined in Fig. 1A. Because we were unable to detect a transcriptional start site for the mntH gene upstream of the T residue depicted in Fig. 1A, we propose that an ATG start codon 81 nucleotides downstream of the annotated start codon (shown in boldface with a double underline in Fig. 1A) is the authentic start codon for the Brucella mntH gene. This would produce a Brucella MntH protein that matches the N terminus of the E. coli and Salmonella MntH proteins better than those annotated in the B. abortus 2308 and B. melitensis 16M genome sequences (Fig. 1B).
Fig 1.
(A) Genetic organization of the mntH gene and surrounding genes in B. abortus 2308. The transcriptional start site for mntH identified by primer extension is indicated in boldface with an arrow and a +1 designation above it. The predicted Mur binding site (or MRS) is underlined, and the Mur binding region identified by DNase I footprint analysis in this study is enclosed in a box. The predicted mntH start codon annotated in the B. abortus 2308 genome sequence in GenBank is denoted in italics with a single underline, and the ATG that is proposed to be the authentic mntH start codon based on the experimental findings presented in this report is depicted in boldface with a double underline. (B) ClustalW alignment of the N-terminal amino acids of the Brucella abortus MntH with the corresponding amino acids of the MntH proteins of Escherichia coli (GenBank accession no. NP_416893.1) and Salmonella enterica serovar Typhimurium (GenBank accession no. NP_461349.1). The revised annotation of the Brucella mntH coding region described in the text was used for this analysis.
The Brucella Mur binds in a specific manner to a 110-bp DNA fragment representing the mntH promoter region in an EMSA (Fig. 2A). The addition of the chelator EDTA alleviates the capacity of Mur to bind to the mntH promoter (Fig. 2B), and the inhibitory effect of EDTA can be overcome by adding increasing concentrations of MnCl2 to the reaction mixtures. These experimental findings show that Mn2+ can modulate the DNA binding activity of the Brucella Mur and further support its proposed role as a manganese-responsive transcriptional repressor.
Fig 2.
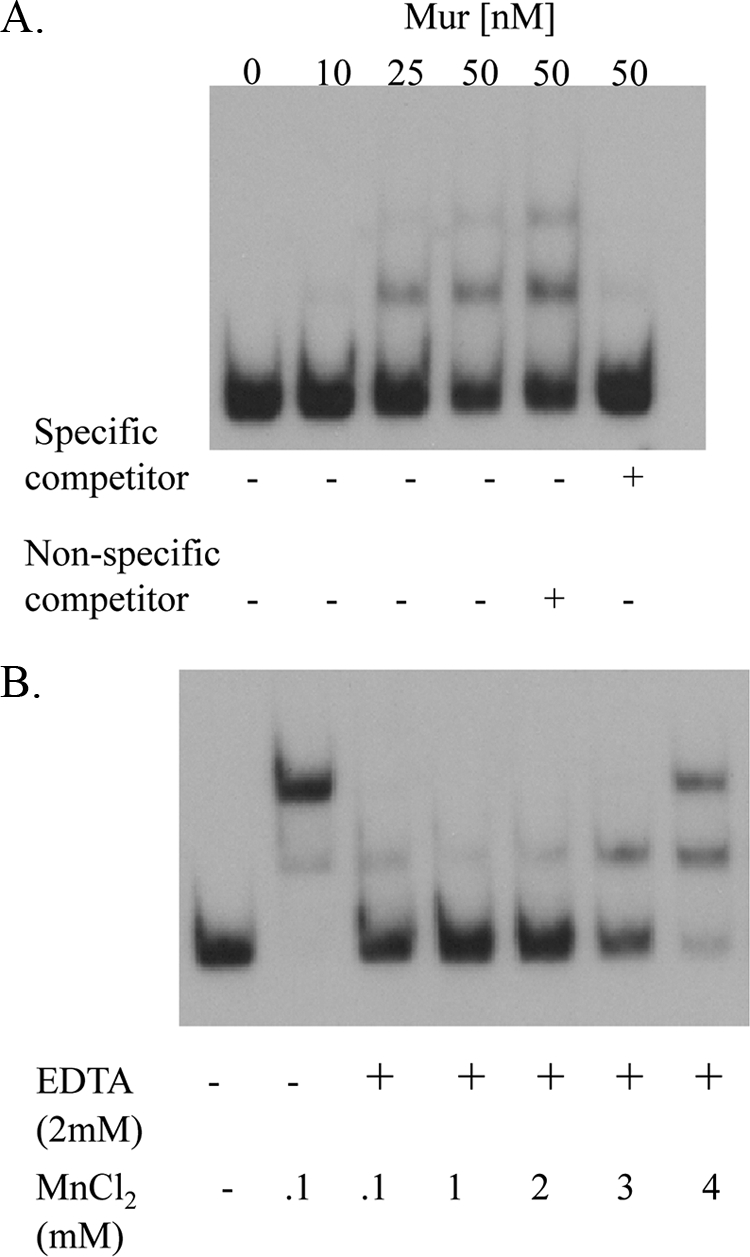
Mur binds to the mntH promoter region in B. abortus 2308 in a manganese-responsive fashion in an EMSA. (A) 0.25 nM labeled DNA probe was used in these reactions; the concentration of recombinant Mur added to the reaction mixtures is shown above the lanes, and the plus-or-minus symbols below the lanes indicate whether the reaction mixtures contain a 100× concentration of an unlabeled DNA fragment representing the mntH promoter region (specific competitor) or an unlabeled DNA fragment representing sequences upstream of the Brucella recA gene (nonspecific competitor). (B) 0.1 pM labeled DNA probe was incubated with 50 nM recombinant Mur in these reaction mixtures. The concentration of MnCl2 present in the reaction mixture is shown below the lanes, and the plus symbols indicate that the reaction mixtures contain 2 mM EDTA.
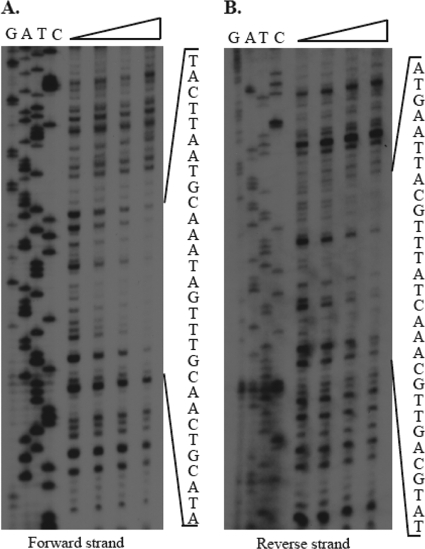
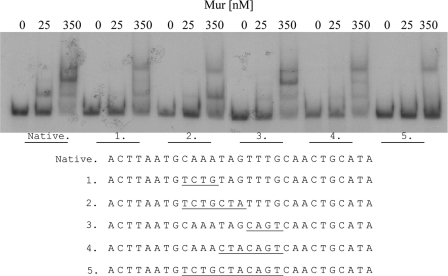
A DNase I footprint analysis determined that the Mur binding site in the mntH promoter is comprised of the 28-nucleotide sequence 5′-TACTTAATGCAAATAGTTTGCAACTGCA-3′ (Fig. 3), which overlaps the predicted MRS in the mntH promoter (Fig. 1A). In an attempt to identify specific subsets of nucleotides in the MRS that are required for Mur binding, five synthetic double-stranded DNA fragments representing the mntH promoter, each with a mutated version of a portion of the CAAATAGTTTG core region of the MRS, were evaluated for their capacity to bind to Mur in an EMSA. As shown in Fig. 4, the synthetic mntH promoter fragments with mutations affecting the CAAATAG residues in the core region of the MRS (e.g., fragments 1, 2, 4, and 5) did not show the same altered mobility patterns in response to increasing concentrations of Mur in an EMSA, as did a fragment containing the native core region of the MRS or one in which only the TTTG component had been mutated (fragment 3). These experimental findings indicate that the CAAATAG region of the MRS plays an important role in the specificity of Mur binding to the mntH promoter in B. abortus 2308.
Fig 3.
DNase I footprinting of the B. abortus mntH promoter with recombinant Brucella Mur. The triangle above the lanes indicates that the corresponding reaction mixtures contain increasing concentrations (0, 1, 5, and 10 μg) of recombinant Brucella Mur.
Fig 4.
Mapping of the subsets of the nucleotides present in the MRS of the B. abortus 2308 mntH promoter that are required for its interaction with Mur in an EMSA. The concentration of Mur added to the reaction mixtures is indicated above the lanes; the specific synthetic oligonucleotides (labeled native, 1, 2, 3, 4, and 5) used in the EMSA reactions are indicated below the lanes, and the nature of the nucleotide substitution in each synthetic oligonucleotide used in these reactions is shown at the bottom of the figure.
DISCUSSION
The gene designated BAB1_1668 in the B. abortus 2308 genome sequence is predicted to encode a homolog of the ferric uptake regulator Fur. This gene was originally cloned during an attempt to identify the iron-responsive regulator of the siderophore biosynthesis genes in B. abortus 2308 by genetic complementation (27). A plasmid-borne copy of BAB1_1668 restores the iron-responsive repression of the fiu-lacZ fusion in the E. coli fur mutant H1780 (15). However, a derivative of B. abortus 2308 from which BAB1_1668 has been deleted exhibits wild-type regulation of its siderophore biosynthesis genes in response to environmental iron levels (31). Moreover, a recombinant version of this Brucella Fur homolog does not bind to the promoter regions of the iron-responsive dhbC (3) or bhuA genes (26) in an EMSA (E. Menscher, unpublished data). These findings suggest that BAB1_1668 does not encode an iron-responsive transcriptional regulator in B. abortus 2308. Instead, data presented in this and a previous report (1) indicate that the BAB1_1668 gene product is serving as a manganese uptake regulator or a Mur.
Although Mur proteins belong to the Fur superfamily of bacterial metalloregulators (20), their activity as transcriptional repressors is modulated in vivo by manganese instead of iron. Mur proteins have been described exclusively in the alphaproteobacteria, where Mur regulates the expression of manganese acquisition genes in Rhizobium leguminosarum (7) and Sinorhizobium meliloti (5, 28). Recent studies have also shown that proteins currently designated Fur are functioning as manganese-responsive, instead of iron-responsive, regulators in Bradyrhizobium japonicum (16, 17) and Agrobacterium tumefaciens (19), and thus they appear to be functional Mur homologs as well.
As demonstrated in this report, Mur binds to the mntH promoter region in B. abortus 2308 and directly regulates the expression of this gene in a manganese-responsive manner. The 30-nucleotide sequence in the mntH promoter protected by Mur in the DNase I protection studies contains a motif (5′-AATGCAAATAGTTGCAA-3′) that matches well with the consensus Mur recognition sequence (MRS) or Mur box defined by Rodionov et al. (30) and MRSs identified upstream of the mntH gene of B. japonicum (16), the sitA gene of R. leguminosarum (8), and the mntA and sitA genes of S. meliloti (5, 28) (Fig. 5). Like the Brucella mntH gene, all of these genes are involved in manganese transport and have been shown to be regulated by Mur. The location of the region of the mntH promoter protected by Mur in B. abortus 2308 (−55 to −24 with respect the transcriptional start site) is very similar to the location of the region protected by Mur in the B. japonicum promoter (−56 to −22 with respect to the transcriptional start site) (16). The selective regional mutagenesis of the MRS in the Brucella mntH promoter sequence identified a 7-nucleotide motif in this region that appears to be critical for the specificity of Mur binding. These findings are similar to those obtained with Fur in E. coli (9) and Caulobacter (6), where groups of nucleotides rather than individual nucleotides have been shown to be responsible for the specific binding of Fur to Fur boxes.
Fig 5.
Comparison of the experimentally documented Mur binding sites present in the promoter regions of the mntH and sitA genes of Brucella abortus, Bradyrhizobium japonicum (16), Rhizobium leguminosarum (8), and Sinorhizobium meliloti (28).
Manganese is an essential micronutrient for Brucella strains (10, 35), and the importance of MntH for the efficient acquisition of this divalent cation in vitro and in vivo has been documented experimentally (1). Like most other biologically beneficial metals, manganese can be toxic if accumulated beyond the levels needed by the bacterial cell, although the potential toxicity of this metal appears to be considerably less than that of other beneficial metals, such as iron, zinc, nickel, or copper (25, 39). A direct role for Mur in preventing manganese toxicity in B. abortus 2308 by repressing mntH expression has not been observed in vitro, however, since both the parent strain and the mur mutant exhibit equivalent growth characteristics during cultivation in laboratory media containing up to 10 mM MnCl2. At higher concentrations, both strains exhibit equivalent levels of growth restriction (E. Menscher, unpublished).
The bioinformatics-based study performed by Rodionov et al. (30) suggests that mntH and a gene designated irr1 by this group (BMEI1563 and BAB1_0393 in the B. melitensis 16M and B. abortus 2308 genome sequences, respectively) are the only Brucella genes directly regulated by Mur. It is important to note, however, that these genes do not encode the Brucella iron-responsive regulator Irr that has been characterized by Martínez et al. (21, 22). Instead, the genes encoding Irr are designated BAB1_2175 and BMEI1955 in the B. abortus 2308 and B. melitensis 16M genome sequences, respectively. This distinction is important because Mur regulates irr expression and cellular manganese levels modulate Irr activity in B. japonicum (17, 29), and evidence for indirect effects of manganese on the expression of iron metabolism genes has been reported for A. tumefaciens fur (19) and S. meliloti mur mutants (5).
During their residence in host macrophages, the brucellae must deal with both endogenous reactive oxygen species (ROS) generated by their respiratory metabolism as well as exogenous ROS generated by the NADPH activity of the host phagocyte (32). Efficient manganese transport mediated by MntH has been linked to ROS resistance in B. abortus 2308 (1), and Irr is required for the optimal expression of the siderophore biosynthesis (22) and heme transport genes (2) in this bacterium. Having the capacity to modulate irr expression or Irr activity such that iron acquisition genes are only maximally expressed in Brucella strains when these bacteria possess protective levels of manganese would be beneficial in their intracellular niche in the host, because it could reduce the risk for ROS toxicity due to iron-mediated Fenton chemistry. Thus, to better understand the importance of Mur in the physiology of Brucella strains, it will be important to determine whether or not Mur directly regulates the expression of irr or whether cellular manganese levels influence Irr activity. Likewise, it will be important to define the biological function of Irr1 and determine if the corresponding gene is regulated by Mur.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by a grant (AI-63516) from the NIAID to R.M.R.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Anderson ES, et al. 2009. The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect. Immun. 77:3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson ES, et al. 2011. The iron-responsive regulator Irr is required for wild-type expression of the gene encoding the heme transporter BhuA in Brucella abortus 2308. J. Bacteriol. 193:5359–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bellaire BH, et al. 2003. Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect. Immun. 71:1794–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Chao TC, Becker A, Buhrmester J, Pühler A, Weidner S. 2004. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J. Bacteriol. 186:3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. da Silva Neto JF, Braz VS, Italiani VC, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res. 37:4812–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Díaz-Mireles E, et al. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150:1447–1456 [DOI] [PubMed] [Google Scholar]
- 8. Díaz-Mireles E, et al. 2005. The manganese-responsive repressor Mur of Rhizobium leguminosarum is a member of the Fur-superfamily that recognizes an unusual operator sequence. Microbiology 151:4071–4078 [DOI] [PubMed] [Google Scholar]
- 9. Escolar L, Perez-Martin J, De Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evenson MA, Gerhardt P. 1955. Nutrition of brucellae: utilization of iron, magnesium and manganese for growth. Proc. Soc. Exp. Biol. Med. 89:678–680 [DOI] [PubMed] [Google Scholar]
- 11. Giedroc DP, Arunkumar AI. 2007. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 29:3107–3120 [DOI] [PubMed] [Google Scholar]
- 12. Guedon E, Helmann JD. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48:495–506 [DOI] [PubMed] [Google Scholar]
- 13. Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288–292 [DOI] [PubMed] [Google Scholar]
- 14. Hantke K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197:337–341 [DOI] [PubMed] [Google Scholar]
- 15. Hantke K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135–139 [DOI] [PubMed] [Google Scholar]
- 16. Hohle TH, O'Brian MR. 2009. The mntH gene encodes the major Mn2+ transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol. Microbiol. 72:399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hohle TH, O'Brian MR. 2010. Transcriptional control of the Bradyrhizobium japonicum irr gene requires repression by Fur and antirepression by Irr. J. Biol. Chem. 285:26074–26080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Johnston AWB, et al. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other α-proteobacteria. Biometals 20:501–511 [DOI] [PubMed] [Google Scholar]
- 19. Kitphati W, Ngok-Ngam P, Suwanmaneerat S, Sukchawalit R, Mongkolsuk S. 2007. Agrobacterium tumefaciens fur has important physiological roles in iron and manganese homeostasis, the oxidative stress response, and full virulence. Appl. Environ. Microbiol. 73:4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 21. Martínez M, Ugalde RA, Almirón M. 2005. Dimeric Brucella abortus Irr protein controls its own expression and binds haem. Microbiology 151:3427–3433 [DOI] [PubMed] [Google Scholar]
- 22. Martínez M, Ugalde RA, Almirón M. 2006. Irr regulates brucebactin and 2,3-dihydroxybenzoic biosynthesis, and is implicated in the oxidative stress resistance and intracellular survival of Brucella abortus. Microbiology 152:2591–2598 [DOI] [PubMed] [Google Scholar]
- 23. Mohanty BK, Giladi H, Maples VF, Kushner SR. 2008. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol. 447:3–29 [DOI] [PubMed] [Google Scholar]
- 24. Nicoletti P. 1989. Relationship between animal and human disease, p. 41–51. In Young EJ, Corbel MJ. (ed.), Brucellosis: clinical and laboratory aspects. CRC Press, Boca Raton, FL [Google Scholar]
- 25. Papp-Wallace KM, Maguire ME. 2006. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60:187–209 [DOI] [PubMed] [Google Scholar]
- 26. Paulley JT, Anderson ES, Roop RM., II 2007. Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect. Immun. 75:5248–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phillips RW, et al. 1996. Molecular cloning of the Brucella abortus iron uptake regulation (fur) gene. Abstr. 96th Annu. Meet. Am. Soc. Microbiol., abstr. D-24, p. 245 [Google Scholar]
- 28. Platero R, de Lorenzo V, Garat B, Fabiano E. 2007. Sinorhizobium meliloti Fur-like (Mur) protein binds a Fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl. Environ. Microbiol. 73:4832–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puri S, Hohle TH, O'Brian MR. 2010. Control of bacterial iron homeostasis by manganese. Proc. Natl. Acad. Sci. U. S. A. 107:10691–10695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodionov DA, Gelfand MS, Todd JD, Curson ARJ, Johnston AWB. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in α-proteobacteria. PLoS Comput. Biol. 2:1568–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roop RM, II, Bellaire BH, Anderson E, Paulley JT. 2004. Iron metabolism in Brucella, p 243–262. In López-Goñi I, Moriyon I. (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 32. Roop RM, II, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med. Microbiol. Immunol. 198:221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rudolph G, Hennecke H, Fischer HM. 2006. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol. Rev. 30:631–648. [DOI] [PubMed] [Google Scholar]
- 34. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. [Google Scholar]
- 35. Sanders TH, Higuchi K, Brewer CR. 1953. Studies on the nutrition of Brucella melitensis. J. Bacteriol. 66:294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stojilkkovic I, Bäumler AJ, Hantke K. 1994. Fur regulon in Gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531–545 [DOI] [PubMed] [Google Scholar]
- 37. Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130 [DOI] [PubMed] [Google Scholar]
- 38. Summers A. 2009. Damage control: regulating defenses against toxic metals and metalloids. Curr. Opin. Microbiol. 12:138–144 [DOI] [PubMed] [Google Scholar]
- 39. Waldron KJ, Robinson NJ. 2010. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 6:25–35 [DOI] [PubMed] [Google Scholar]
- 40. Wexler M, et al. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149:1357–1365 [DOI] [PubMed] [Google Scholar]
- 41. Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.