Abstract
Mycobacterium tuberculosis is a pathogen of major global importance. Validated drug targets are required in order to develop novel therapeutics for drug-resistant strains and to shorten therapy. The Clp protease complexes provide a means for quality control of cellular proteins; the proteolytic activity of ClpP in concert with the ATPase activity of the ClpX/ClpC subunits results in degradation of misfolded or damaged proteins. Thus, the Clp system plays a major role in basic metabolism, as well as in stress responses and pathogenic mechanisms. M. tuberculosis has two ClpP proteolytic subunits. Here we demonstrate that ClpP1 is essential for viability in this organism in culture, since the gene could only be deleted from the chromosome when a second functional copy was provided. Overexpression of clpP1 had no effect on growth in aerobic culture or viability under anaerobic conditions or during nutrient starvation. In contrast, clpP2 overexpression was toxic, suggesting different roles for the two homologs. We synthesized known activators of ClpP protease activity; these acyldepsipeptides (ADEPs) were active against M. tuberculosis. ADEP activity was enhanced by the addition of efflux pump inhibitors, demonstrating that ADEPs gain access to the cell but that export occurs. Taken together, the genetic and chemical validation of ClpP as a drug target leads to new avenues for drug discovery.
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis, is responsible for immense global suffering; over 9 million new cases of tuberculosis are reported each year, and it is estimated that there are 0.5 million cases of multidrug-resistant strains (37, 38). An urgent need for therapeutics acting on novel pathways is evident (7). Current drug discovery efforts are hampered by the lack of suitable drug targets against which to develop inhibitors (39); genetic approaches to identifying genes essential during infection or in culture have been successful but cannot predict whether inhibition of the target protein will be effective (1). Cellular pathways which are central to the survival of the bacterium during the infection process are attractive candidates for rational drug design.
The Clp protease is one of the major cellular proteases responsible for degrading misfolded or damaged proteins and plays a central role in maintaining protein function, especially under stress conditions (17). Clp proteases have a broad substrate range and are implicated in many processes, including stationary-phase survival (36), stress responses (11, 27), and virulence (11, 12, 18, 27). Since Clp activity is required for the removal of damaged proteins, which are likely to occur during infection and exposure to hostile conditions, it is likely to be a key component of the bacterial response to stress.
The Clp protease complex is composed of a proteolytic subunit (ClpP) and a regulatory, ATPase subunit (ClpC or ClpX); ClpP forms two rings of heptamers enclosing a central pore, which associates with a hexameric ring of ClpC or ClpX (17). The ClpC/ClpX ATPase unfolds proteins which are transported into the ClpP chamber where degradation by ClpP occurs. Control of proteolysis is achieved by the specificity and activity of ClpC/ClpX and other accessory proteins.
M. tuberculosis has two homologs of the ClpP proteolytic core (ClpP1, Rv2461; ClpP2, Rv2460c) and three potential Clp ATPases (ClpC1, Rv3596c; ClpC2, Rv2667; ClpX, Rv2457c). It is likely that ClpP1 and ClpP2 have different substrate specificities, as in other bacteria with multiple ClpP subunits (10). The substrate specificity or interacting partners of each ClpP protease are not fully understood, although interactions between ClpP2 and ClpC1 have been demonstrated (2, 31) and ClpC1 ATPase activity has been demonstrated in vitro (15).
ClpP1 and ClpP2 are arranged as an apparent operon and presumed to be coexpressed (6). Regulation of ClpP1-ClpP2 expression is mediated by the ClgR regulator (8, 9, 30); both clpP genes are highly expressed during aerobic and hypoxic growth (24) and are further upregulated during reaeration from anaerobic conditions (30), suggesting a key role in maintaining protein function during changes in oxygen availability. In addition, ClgR-mediated upregulation of Clp genes is required during bacterial infection of macrophages (9).
The ClpP proteolytic subunit is an unusual drug target in that both inhibition and activation are lethal events. Inhibition of ClpP using cyclic peptides results in cell death in Caulobacter crescentus where ClpP is essential (5), whereas activation of uncontrolled proteolysis by acyldepsipeptides (ADEPs) leads to death in several Gram-positive bacterial species (Bacillus subtilis, Streptococcus, and Enterococcus) (4). Resistance to ADEPs is mediated by the loss of ClpP activity in bacteria in which the protease is nonessential (4). Since at least one of the ClpP proteins (ClpP2) is predicted to be essential in M. tuberculosis (28), targeting its activity using the ADEP activators is an attractive option (32). The mode of action of ADEPs has been determined using structural and biochemical studies; binding of ADEP to ClpP leads to the activation of its proteolytic activity independent of the normal regulatory control of the ATPase subunits (19, 21) and uncontrolled proteolysis leads to filamentation and ultimately cell death (4).
We investigated the essentiality of ClpP1 for M. tuberculosis growth in culture as a first step to establishing the validity of the Clp protease as a drug target. We demonstrate that ClpP activators are effective against M. tuberculosis and that overexpression of ClpP1 and ClpP2 have different consequences.
MATERIALS AND METHODS
Growth assays.
Escherichia coli and Staphylococcus aureus were grown in LB broth or LB agar plates. M. tuberculosis H37Rv was cultured in Middlebrook 7H9 medium supplemented with 0.05% (wt/vol) Tween 80 and 10% (vol/vol) OADC (oleic acid, albumin, dextrose, catalase; MTA; Becton Dickinson) or on Middlebrook 7H10 agar supplemented with 10% (vol/vol) OADC. Aerobic growth was measured in 16 mm diameter tubes containing a 2-mm stir bar; cultures were incubated with stirring at 250 rpm. Reserpine was used at 12 μg/ml and verapamil at 40 μg/ml. The Wayne model of hypoxia was used as described previously (33); briefly, seed cultures were grown in Dubos medium supplemented with 0.05% (wt/vol) Tween 80 and 10% (vol/vol) Dubos medium supplement (DTA; Becton Dickinson) aerobically. Seed cultures were used to inoculate 17 ml DTA in 20-mm-diameter glass tubes containing a 2-mm stir bar to a theoretical optical density at 590 nm (OD590) of 0.004; cultures were incubated with stirring at 150 rpm. The starvation model was as described previously (3); briefly, strains were cultured in 100 ml MTA plus 0.2% glycerol for 7 days, washed twice, and resuspended in phosphate-buffered saline (PBS). Cells were incubated standing, and a single tube was taken for each time point. MICs on solid medium were determined using the agar proportion method; briefly, 104 CFU was plated onto medium containing ADEPs or drug-free controls. CFU were scored after 4 weeks of incubation at 37°C. The MIC was defined as the lowest concentration at which viability was reduced by 99% (MIC99). MICs in liquid medium were determined using a 10-point serial dilution in 96-well plates. Growth was measured after 5 days and plotted against compound concentration; curves were fitted using GraphPad Prism, and MICs were defined as the minimum concentration required to inhibit growth completely.
Construction of deletion strains.
The deletion delivery vector pNIPPY40G was constructed as follows: the upstream and downstream regions of clpP1 were amplified and joined using PCR. The primer pairs were F1 (CCA AGC TTC GTC TCG ATC GAC TTG TCT G) and R1 (CGA ACG CAT GTC AGT CAC TT) for the upstream region, F2 (TTC GTC GAT CAC ATC ATC ACC) and R2 (CCA AGC TTG CCA GGA CAC CGG ATA ACT) for the downstream region, and BR1 (AAG TGA CTG ACA TGC GTT CG TTC GTC GAT CAC ATC ATC ACC) and BR2 (GGT GAT GAT GTG ATC GAC GAA CGA ACG CAT GTC AGT CAC TT) for the bridging PCR. The final product was cloned into the HindIII site of p2NIL (25); the marker gene cassette (hyg, sacB, lacZ) from pGOAL19 (25) was cloned in as a PacI fragment.
The complementation expression vector pDIAL3 (Pmyc1-tetO-clpP1) was constructed by amplifying the Pmyc1-tetO promoter from pSE100 (13) and cloning into pGEM-T (Promega); the L5 integration cassette carrying the integrase, attP site and gentamicin resistance was added as a HindIII fragment (22). ClpP1 was amplified using primers containing PacI restriction sites and cloned downstream of Pmyc1-tetO in the correct orientation for expression.
Single crossover (SCO) strains were generated by electroporation of the delivery vector pNIPPY40G and isolated on hygromycin, kanamycin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates (14). A blue colony was selected and streaked onto plates containing no antibiotics to allow the second recombination event to occur. Double crossover (DCO) strains were isolated by plating onto sucrose, X-Gal plates; white colonies were selected and patch tested to confirm kanamycin and hygromycin sensitivity. The complementation vector pDIAL3 was transformed into the SCO strain to generate a merodiploid; DCO strains were isolated in the same way except that gentamicin was included for selection. Colony PCR was used to distinguish the presence of the deletion and wild-type alleles. Southern analysis was carried out to confirm the expected genotype of selected strains. Gene switching was carried out as described previously (26).
Construction of overexpression plasmids.
The clpP1 gene was first PCR amplified from H37Rv using primer pair clpP1-C1 (CCG GAT CCT TAA TTA AGA ACT CGA GGA AAG CAG GTG; BamHI [underlined]) and clpP1-C2 (GGA AGC TTA ATT AAG GGG CTG GAT CTG AGA ATT T; HindIII) and cloned into pSC-A (Agilent Technologies) and sequence verified; clpP1 was subcloned into pSMT3 as a BamHI/HindIII fragment to generate pOPPY1. The clpP2 gene was PCR amplified using primer pair clpP2-C1 (GGC TGC AGT TAA TTA ACA CGT CAA TGG AGA AGC ACA; PstI) and clpP2-C2 (GGA AGC TTA ATT AAC CGC CAG CTA AGT GGT CTC; HindIII) and cloned into pSC-A and sequence verified. clpP2 was subcloned into pSMT3 as a PstI/HindIII fragment to generate pOPPY2.
Quantification of mRNA.
Extraction of total RNA was carried out as previously described (3); total RNA was purified using the RNeasy minikit (Qiagen). Samples were treated with RQ1 DNase for 1 h at 37°C in the presence of RNasin Plus RNase inhibitor (Invitrogen) and purified by phenol-chloroform extraction. The RNA was precipitated in 0.3 M sodium acetate and 2.5 volumes of ice-cold 100% ethanol and stored at −80°C. A two-step quantitative reverse transcription (qRT)-PCR was performed using the Transcriptor High Fidelity cDNA synthesis kit (Roche), PCR master mix (Roche), and the Roche LightCycler 480 instrument according to manufacturer's instructions. To quantify genomic DNA contamination, cDNA reactions excluding reverse transcriptase were analyzed. DNA standard curves using M. tuberculosis H37Rv chromosomal DNA were generated for each gene. Each qRT-PCR was performed in duplicate. Transcript levels of target genes were normalized to the transcript levels of the housekeeping sigma factor sigA (endogenous control). The transcript levels of sigA were similar for all samples tested (data not shown). TaqMan primers and TaqMan MGB probes for clpP1, clpP2, and sigA were designed using Primer Express 2.0 software (Applied Biosystems). The sequences for the primers and probes were as follows (where TAMRA is 6-carboxytetramethylrhodamine and FAM is 6-carboxyfluorescein): clpP1-TaqManF1, 5′-TCG CCG TGA TCA AGA AAG AA-3′; clpP1-TaqManR1, 5′-GGC TGG CCG GTG AAT TC-3′; clpP1-MGBprobe, FAM-GGC TGG CCG GTG AAT TC-TAMRA; clpP2-TaqManF1, 5′-GCG ATA TCA CCA TGT ACA TCA ACT C-3′; clpP2-TaqManR1, 5′-GGC CCG CAC GTA TTG C-3′; clpP2-MGBprobe, FAM-TGG CGA TCT ACG ACA CC-TAMRA; sigA-TaqManF1, 5′-CCG ATG ACG ACG AGG AGA TC-3′; sigA-TaqManR1, 5′-GGC CTC CGA CTC GTC TTC A-3′; sigA-MGBprobe, FAM-CCT CCG GTG ATT TC-3′.
Synthesis of ADEPs.
The reference acyldepsipeptide compounds and additional analogs, belonging to the enopeptin class of natural product antibiotics, were prepared according to literature procedures (29). Briefly, pentapeptolide macrocyclization precursors were prepared by standard peptide coupling chemistry and converted to the corresponding depsipeptide intermediates through a macrolactamization procedure (29). Side chain elaboration via acylation of the serine nitrogen with various phenylalanine derivatives gave the title compounds which were characterized by high-pressure liquid chromatography-mass spectrometry (LCMS) and proton nuclear magnetic resonance (NMR) spectral analysis.
RESULTS
ClpP1 is essential for M. tuberculosis growth in culture.
We are interested in the Clp proteolytic systems of M. tuberculosis. Unusually, M. tuberculosis encodes two ClpP subunits (ClpP1 and ClpP2) colocated on the chromosome and presumed to be coexpressed as a bicistronic operon (6). clpP2 is predicted to be an essential gene using high-density transposon mutagenesis, but no data are available for clpP1 (28).
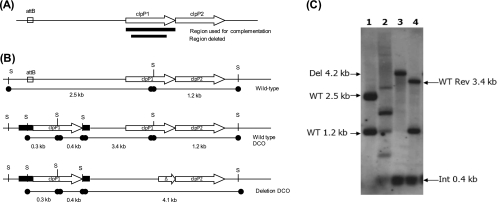
We attempted to construct an unmarked, in-frame deletion of clpP1 on the chromosome using a two-step homologous recombination method (25). A deletion delivery vector carrying a deletion of 525 bp of clpP1 flanked by 1,000 bp was transformed into M. tuberculosis, and recombinant strains (single crossovers) were identified; double crossovers were isolated from this strain and analyzed by PCR. Since we used an unmarked deletion allele, DCOs could carry the deletion allele or the wild-type allele; all DCOs obtained were wild type (40/40), suggesting that the gene is essential for growth. In order to confirm this, we constructed a merodiploid strain containing a functional copy of clpP1 expressed under the control of the tetracycline-inducible promoter (13) integrated into the chromosome at the mycobacteriophage L5 attB site (20). In this background, we were able to delete the chromosomal copy—19/40 DCOs had the deletion allele, 21/40 had the wild-type allele (Fig. 1)—demonstrating that the gene is essential for growth in culture (P < 0.0001 using Fisher's exact test). We further confirmed essentiality by demonstrating that the integrated vector carrying the only functional copy of the gene could not be replaced by a vector lacking clpP1 using gene switching (26).
Fig 1.
ClpP1 is essential for in vitro growth of M. tuberculosis. (A) Chromosomal organization of the probable clpP1-clpP2 operon. The region used in the complementation vector to generate the merodiploid and the region used as the probe for Southern blotting are indicated. (B) Schematics of the merodiploid and Del-Int (deletion, integration) strain showing the SphI restriction sites and expected sizes for Southern analysis. The attB site utilized by the integrating vector is marked. (C) Southern analysis of strains. Genomic DNA from wild-type (WT) H37Rv (lane 1), merodiploid (lane 2), Del-Int DCO (lane 3), and WT revertant (Rev) DCO (lane 4) strains was digested with SphI and hybridized with the clpP1 probe. The expected bands (and sizes) for WT, Del, WT Rev DCO, and Int alleles are indicated by arrows.
Activators of ClpP proteolysis are effective against M. tuberculosis.
Genetic validation of essentiality is often the first step in selecting new drug targets for further investigation, and thus, the essentiality of ClpP1 suggests its potential as a drug target in M. tuberculosis. However, chemical validation of a target is preferable to demonstrate that modulation directly leads to bacterial death. The Clp proteolytic systems are unusual targets in that activation rather than inhibition is cidal, and a series of acyldepsipeptides (ADEPs) which lead to uncontrolled proteolytic activity have been characterized (4).
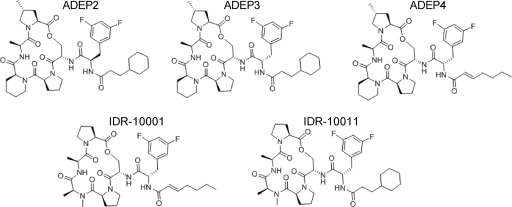
We synthesized three of the ADEP series (ADEP2 and ADEP4 were previously reported to have antibacterial activity, while ADEP3, a side chain epimer of ADEP2, was reported devoid of similar activities) for testing against M. tuberculosis and S. aureus (Fig. 2). We also synthesized two other derivatives, IDR-10001 and IDR-10011, which incorporate N-methylalanine instead of the more rigid homoproline in the depsipeptide core structure to begin to explore structure-activity relationships in the chemical series (Fig. 2).
Fig 2.
Structures of the ADEP compounds and derivatives. Three previously described ADEP compounds were synthesized (ADEP2, ADEP3, ADEP4); two additional new analogs (IDR-10001 and IDR-10011) were also synthesized.
We examined the activity of the ADEPs against M. tuberculosis on solid medium. MICs were determined for each of the five synthetic compounds (Table 1). All five compounds were active against M. tuberculosis, including compound ADEP3, previously shown to be inactive against S. aureus. ADEP2 was the most active, with an MIC of 25 μg/ml. We determined MICs against S. aureus as a control; our data were in agreement with that previously reported (4), showing good activity of ADEP2 and ADEP4, with ADEP3 being inactive up to 100 μg/ml.
Table 1.
Activity of ADEPsa
| Compound | E. coli | S. aureus |
M. tuberculosis |
|
|---|---|---|---|---|
| Wild type | ClpP1 overexpressor | |||
| IDR-10001 | >100 | 1 | 100 | 100 |
| ADEP2 | NT | 1.25 | 25 | 25 |
| ADEP3 | NT | >100 | 100 | 100 |
| ADEP4 | NT | 0.125 | 50 | 50 |
| IDR-10011 | >100 | 5 | 50 | 50 |
MIC99 (μg/ml) was determined against each bacterial species on solid medium.
NT, not tested.
These data suggest for the first time that ClpP activation is an effective means of controlling the replication of M. tuberculosis and therefore that ClpP is a valid drug target for pursuit.
Overexpression of ClpP1 is tolerated, but overexpression of ClpP2 is toxic in M. tuberculosis.
Since M. tuberculosis has two ClpP homologs, ADEPs could act via activation of either one or both. In order to investigate this, we attempted to generate recombinant strains of M. tuberculosis in which either ClpP1 or ClpP2 was overexpressed. Overexpression of the target of ADEP binding could lead to increased sensitivity to the compounds.
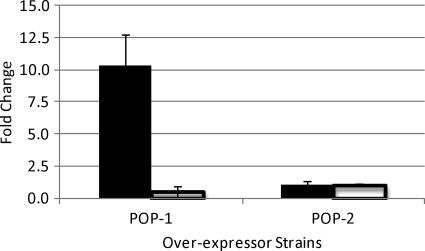
We expressed either ClpP1 or ClpP2 under the control of the strong, constitutive promoter from Mycobacterium bovis BCG hsp60 on a multicopy plasmid. Overexpression of clpP1 was measured at the mRNA level and resulted in approximately 9-fold upregulation of expression (Fig. 3); clpP2 mRNA levels were unchanged in this strain. In contrast, no upregulation of clpP2 mRNA was seen in recombinant strains carrying the clpP2 overexpression plasmid.
Fig 3.
Overexpression of clpP1 and clpP2 in recombinant strains of M. tuberculosis. clpP1 and clpP2 were independently overexpressed in the WT background, and levels of transcript were analyzed via qRT-PCR. POP-1, clpP1 overexpressor strain; POP-2, clpP2 overexpressor strain. Transcript levels were standardized to sigA expression and compared to those in WT and are represented as fold change. Solid bars, clpP1 mRNA; white bars, clpP2 mRNA. Results are average of duplicates; error bars are standard deviations.
Instability in mycobacterial expression vectors has been extensively reported, so we analyzed the plasmids in the recombinant M. tuberculosis strains. PCR analysis of plasmid-carrying strains confirmed that the ClpP1 plasmid was intact, whereas the ClpP2 plasmid had undergone deletions in all three transformants tested; the deletion resulted in the loss of Phsp60-clpP2, explaining the lack of overexpression in this strain (data not shown). We repeated the transformation but were unable to obtain strains in which the plasmid maintained the correct insert, suggesting that overexpression of ClpP2 (independently of ClpP1) is toxic and not tolerated in the mycobacterial cells. To determine if overexpression was due to ClpP2 toxicity, or if it was due to competition with ClpP1, we also tried to overexpress both genes together from the same promoter. In this case, we were able to obtain stable transformants in which both genes were overexpressed (data not shown).
Once we had confirmed that overexpression of ClpP1 at the mRNA level occurred, we determined the MIC99 for the five compounds. No shift was seen toward sensitivity (or resistance); all compounds had the same activity against the ClpP1 overexpressor (Table 1).
Overexpression of active proteases could lead to growth defects if it resulted in increased protein turnover or degradation. We characterized the ClpP1 overexpressor strain phenotypically in several growth models. Aerobic growth was slightly reduced during logarithmic growth although the final stationary-phase OD590 reached was the same as for the vector-only control strain (data not shown). Growth was also measured in the Wayne model of hypoxia in which a gradual depletion of oxygen is achieved due to consumption of oxygen during bacterial respiration (33–35). The majority of genes are downregulated in this model, but we have previously shown that ClpP1 and ClpP2 mRNA levels remain high, suggesting a role during hypoxic maintenance (24). The ClpP1 overexpressor showed no defect in its ability to either enter or remain in this state (data not shown). We also looked at the ClpP1 overexpressor during complete nutrient deprivation (3), but the overexpressor showed no compromise in ability to enter and survive this state, with OD590 and CFU over time similar to those of the control strain (data not shown).
Efflux in M. tuberculosis is partly responsible for resistance to ADEPs.
The ADEP compounds showed activity against M. tuberculosis, albeit at a lower level than for S. aureus. This could arise from differences in the cell wall, since the mycobacterial cell wall is very lipid rich and contributes in great part to the intrinsic antibiotic resistance of M. tuberculosis. Alternatively, this could be due to efficient compound efflux from the cell. E. coli is resistant to the action of the ADEPs (Table 1) presumably due to efflux (4).
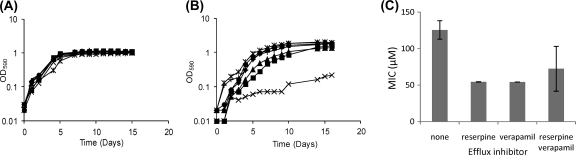
We looked at inhibition of M. tuberculosis growth in liquid medium by ADEP2 in combination with the efflux pump inhibitors reserpine and verapamil (Fig. 4). In the absence of efflux pump inhibitors, no significant growth inhibition was seen at 10 or 15 μg/ml. The addition of efflux pump inhibitors increased the sensitivity of the cells to ADEP2-mediated growth inhibition; at 15 μg/ml, inclusion of reserpine and verapamil with ADEP2 resulted in significant growth retardation, with cells growing very slowly and to a low final OD590 (Fig. 4B). Addition of either reserpine or verapamil singly with ADEP2 resulted in a small effect on growth. The effect of the efflux pump inhibitors was synergistic, suggesting that efflux is mediated by more than one efflux pump. We also tested the effect of efflux pump inhibitors in combination with IDR-10011. MICs were determined in liquid medium (Fig. 4C); again, compound efficacy was increased by the addition of efflux pump inhibitors, although the effect on MICs was seen with either inhibitor, as well as with the combination. These data confirm that efflux is effective against this compound class in M. tuberculosis.
Fig 4.
Increased ADEP activity against M. tuberculosis using efflux pump inhibitors. M. tuberculosis was grown in the presence of two different concentrations of ADEP2 only (■) or in combination with reserpine (▴), verapamil (*), or both (×) at concentrations of 12 μg/ml and 40 μg/ml, respectively. (A) Growth in 10 μg/ml ADEP2. Aerobic growth in stirred culture tubes was monitored over 16 days. (B) Growth in 15 μg/ml ADEP2. Aerobic growth in stirred culture tubes was monitored over 16 days. (C) MICs against compound IDR-10011 were determined against M. tuberculosis in liquid medium (in 96-well plates) in the presence/absence of 12 μg/ml reserpine and/or 40 μg/ml verapamil. The experiment was performed in triplicate, and results are presented as average with standard deviations.
DISCUSSION
Much interest has centered on identifying novel targets for the design of inhibitors against M. tuberculosis, but there remains a dearth of suitably validated targets. Here we show that the Clp protease system is essential for growth in vitro using genetic means; we also demonstrate that activator compounds are effective against mycobacteria. The combination of chemical and genetic validation suggests that the Clp proteases are attractive new targets.
Overexpression of ClpP2, independently of ClpP1, was apparently toxic to the cells; no overexpressing strains could be isolated, and all plasmid transformants had deletions in the vector. This was surprising since ClpP1 and ClpP2 are found in an apparent operon and so are expected to be coexpressed at similar levels. Expression of genes in operons is subject to positional influence, with the downstream gene, in this case clpP2, being expressed to lower levels, although this effect is normally most pronounced in operons with lower expression levels (16). Thus, ClpP2 could be naturally present at lower levels than ClpP1 in the cell.
ClpP subunits assemble into multimeric complexes with ClpX or ClpC subunits; M. tuberculosis has three potential ATPase subunits (ClpC1, ClpC2, ClpX), and it is not clear which of these can interact with ClpP1 or ClpP2. Protein-protein interaction between ClpP2 and ClpC1 has been demonstrated (31), but this does not exclude interactions being possible between ClpP1 and ClpC1. The ClpP2-ClpC1 complex does constitute a functional protease in vitro, and one substrate has been identified (RseA) (2); in this instance, neither ClpP1 nor ClpX could function, suggesting that the true complex is ClpP2-ClpC and leaving open the possibility that ClpP1 interacts with ClpX. Different ClpP subunits may have different binding affinities or specificities for the ClpC and ClpX ATPase or other accessory subunits; overexpression of ClpP2 might lead to competition for these subunits, thereby blocking normal ClpP1 function. In support of this, we found that coexpression of ClpP1 and ClpP2 from the same strong promoter did allow the upregulation of ClpP2. Additional work to determine the interacting partners and the substrate specificities of the native complexes would shed further light on the different roles of ClpP1 and ClpP2.
ADEP mode of action.
The ADEP series of compounds have an unusual mode of action, since they do not inhibit a cellular function but activate a normal cellular process which becomes detrimental to the cells. This explains why ADEPs are active against organisms in which ClpP is not an essential protein; in such species, ADEP resistance results from deletion of ClpP. However, in M. tuberculosis, ClpP is essential, so that deletion mutants would not be viable; this could be advantageous in that resistance to ADEPs may be more difficult to acquire.
Overexpression of drug targets often leads to resistance when the drug is an inhibitor. In contrast, the ADEP series are activators of proteolytic activity. We did not observe any change in susceptibility to ADEPs in strains of M. tuberculosis which overexpressed clpP1. This may be due to the mode of action, since ADEPs are activators, not inhibitors. If the activation of ClpP is dependent on the concentration of ADEP and if this is the rate-limiting step, then increased levels of ClpP will have no effect on resistance/sensitivity. In addition, ADEPs are efficiently exported from the cell, a process which is likely to be energy and concentration dependent; thus, the intracellular concentration of ADEP would result from a balance between import and export and would be unchanged in the overexpressing strain. Alternatively, ClpP2 and not ClpP1 may be the intracellular target of ADEPs. However, we think that this is unlikely, given the structural similarities between the two homologs, suggesting that ADEPs would activate both ClpP proteins. Alignment of ClpP1 and ClpP2 with the B. subtilis Clp reveals reasonable conservation of residues known to be involved in interaction with the ADEPs; in particular, the Tyr62 residue, which is critical for the interaction, is conserved in both M. tuberculosis proteins (19).
Role of efflux in resistance.
The intrinsic drug resistance of mycobacteria is in part due to the nature of the impermeable cell wall, but increasing evidence points toward efflux as a significant mechanism of resistance in M. tuberculosis (23). Our data confirm that efflux of ADEPs occurs, and this may account wholly for their lower activity against M. tuberculosis than against S. aureus. A combination of efflux inhibition and ADEP was more effective in preventing growth. Further development of ADEPs as inhibitors to reduce efflux or as a combination therapy with an efflux inhibitor could be considered.
Conclusion.
Using genetic and chemical validation approaches, we have demonstrated that the ClpP proteins of M. tuberculosis are exciting new drug targets for exploration. Targeting the Clp series has the added advantage that both inhibitors and activators have the potential to kill the cell. Further work to develop the ADEP activator series or to identify new activators or inhibitors could lead to the development of new therapeutic agents in the long term.
ACKNOWLEDGMENTS
We thank Melanie Ikeh and James Ahn for technical assistance. We thank Gerry Waters, Anna Upton, Takushi Kaneko, Yoann Personne, and Amanda Brown for helpful discussion. We thank David Roberts and Brendan Leeson for critical evaluation of the manuscript.
This work was funded by the Global Alliance for TB Drug Development (TB Alliance).
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Balganesh TS, Furr NJ. 2007. Molecular approaches to target discovery: evaluating targets for anti-tuberculosis drug discovery programmes. Infect. Disord. Drug Targets 7:120–126 [DOI] [PubMed] [Google Scholar]
- 2. Barik S, Sureka K, Mukherjee K, Basu J, Kundu M. 2010. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75:592–606 [DOI] [PubMed] [Google Scholar]
- 3. Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731 [DOI] [PubMed] [Google Scholar]
- 4. Brötz-Oesterhelt H, et al. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082–1087 [DOI] [PubMed] [Google Scholar]
- 5. Cheng L, et al. 2007. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 16:1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 7. Duncan K. 2003. Progress in TB drug development and what is still needed. Tuberculosis 83:201–207 [DOI] [PubMed] [Google Scholar]
- 8. Engels S, Schweitzer JE, Ludwig C, Bott M, Schaffer S. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigma(H). Mol. Microbiol. 52:285–302 [DOI] [PubMed] [Google Scholar]
- 9. Estorninho M, et al. 2010. ClgR regulation of chaperone and protease systems is essential for Mycobacterium tuberculosis parasitism of the macrophage. Microbiology 156:3445–3455 [DOI] [PubMed] [Google Scholar]
- 10. Fedhila S, Msadek T, Nel P, Lereclus D. 2002. Distinct clpP genes control specific adaptive responses in Bacillus thuringiensis. J. Bacteriol. 184:5554–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frees D, Qazi SNA, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48:1565–1578 [DOI] [PubMed] [Google Scholar]
- 12. Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286–1294 [DOI] [PubMed] [Google Scholar]
- 13. Guo XV, et al. 2007. Silencing essential protein secretion in Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 189:4614–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinds J, et al. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519–527 [DOI] [PubMed] [Google Scholar]
- 15. Kar NP, Sikriwal D, Rath P, Choudhary RK, Batra JK. 2008. Mycobacterium tuberculosis ClpC1: characterization and role of the N-terminal domain in its function. FEBS J. 275:6149–6158 [DOI] [PubMed] [Google Scholar]
- 16. Kovács K, Hurst LD, Papp B. 2009. Stochasticity in protein levels drives colinearity of gene order in metabolic operons of Escherichia coli. PLoS Biol. 7:e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kress W, Maglica Z, Weber-Ban E. 2009. Clp chaperone-proteases: structure and function. Res. Microbiol. 160:618–628 [DOI] [PubMed] [Google Scholar]
- 18. Kwon HY, et al. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence expression and protects against fatal pneumococcal challenge. Infect. Immun. 72:5646–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee BG, et al. 2010. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 17:471–478 [DOI] [PubMed] [Google Scholar]
- 20. Lee MH, Pascopella L, Jacobs WR, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li DHS, et al. 2010. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: a model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 17:959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahenthiralingam E, et al. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 66:3626–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milano A, et al. 2009. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 89:84–90 [DOI] [PubMed] [Google Scholar]
- 24. Muttucumaru DGN, Roberts G, Hinds J, Stabler RA, Parish T. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 84:239–246 [DOI] [PubMed] [Google Scholar]
- 25. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 26. Pashley CA, Parish T. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229:211–215 [DOI] [PubMed] [Google Scholar]
- 27. Robertson GT, Ng WL, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 29. Schmidt U, Lieberknecht A, Griesser H, Talbiersky J. 1982. Synthesis of peptide alkaloids. J. Org. Chem. 47:3261–3264 [Google Scholar]
- 30. Sherrid AM, Rustad TR, Cangelosi GA, Sherman DR. 2010. Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PLoS One 5:e11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh A, Mai D, Kumar A, Steyn AJC. 2006. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U. S. A. 103:11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Socha AM, Tan NY, LaPlante KL, Sello JK. 2010. Diversity-oriented synthesis of cyclic acyldepsipeptides leads to the discovery of a potent antibacterial agent. Bioorg. Med. Chem. 18:7193–7202 [DOI] [PubMed] [Google Scholar]
- 33. Wayne LG. 2001. In vitro model of hypoxically induced nonreplicating persistence of Mycobacterium tuberculosis. In Parish T, Stoker NG. (ed), Mycobacterium tuberculosis protocols. Humana Press Inc, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 34. Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163 [DOI] [PubMed] [Google Scholar]
- 36. Weichart D, Querfurth N, Dreger M, Hengge-Aronis R. 2003. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J. Bacteriol. 185:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. WHO 2010. Global tuberculosis control: WHO report 2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 38. WHO 2010. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response. World Health Organization, Geneva, Switzerland [Google Scholar]
- 39. Williams KJ, Duncan K. 2007. Current strategies for identifying and validating targets for new treatment-shortening drugs for TB. Curr. Mol. Med. 7:297–307 [DOI] [PubMed] [Google Scholar]