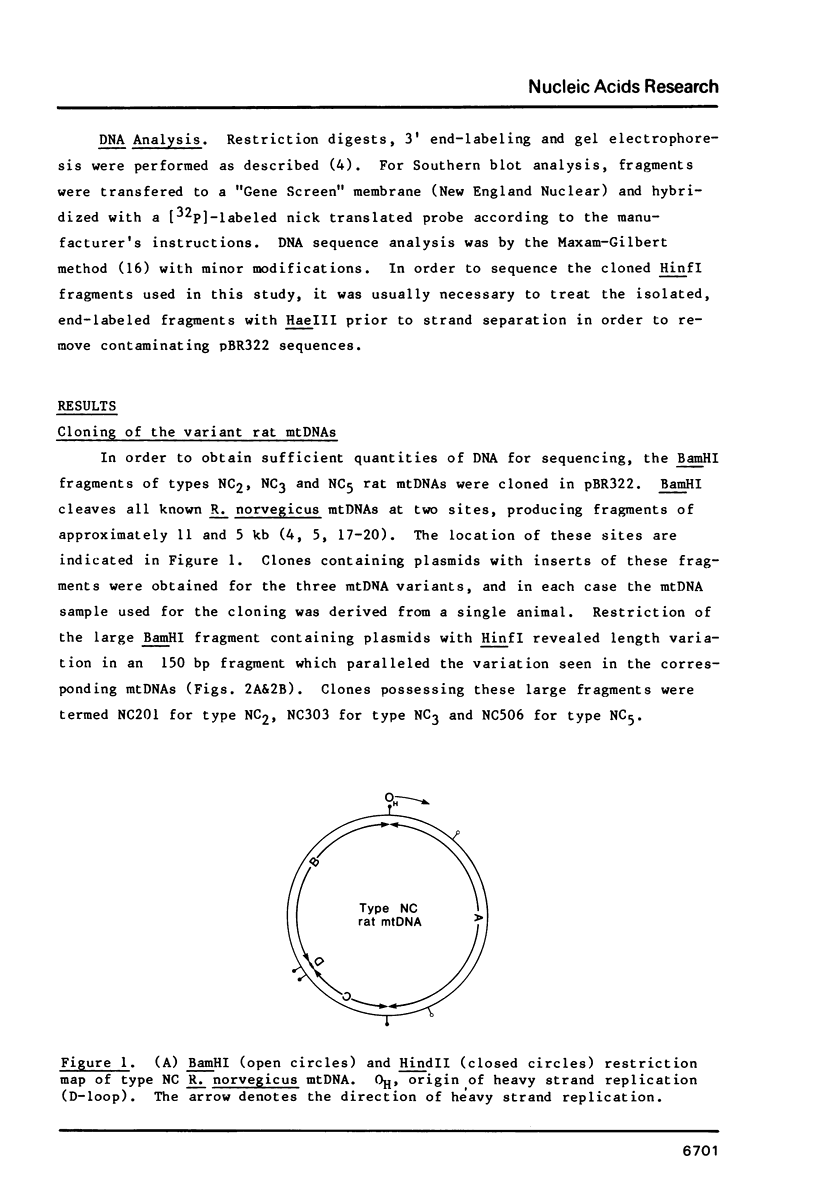
Abstract
Three closely related variants of rat (Rattus norvegicus) mtDNA have been shown to differ in the number of T residues found in a run of Ts (light strand) which spans the junction between the tRNACys and tRNATyr genes. The number of Ts in the repeat varies from 6 to 8 in these DNAs. Another, less closely related, R. norvegicus variant has a run of 5 Ts at this site and in the related species, Rattus rattus, a run of 4 Ts is found. In R. norvegicus mtDNA runs of 5 As and 5 Gs are found just to the 3' side of the variable T repeat, and it is suggested that the three runs of repeated nucleotides may stabilize heteroduplexes which result from strand slippage and which give rise to the insertions and/or deletions. Among 17 mtDNA clones derived from an individual with the 8T repeat, one clone was found which possessed a 9T repeat. This variant may represent an additional DNA type originally present within the individual.
Full text
PDF








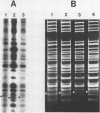
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Aquadro C. F., Greenberg B. D. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. doi: 10.1093/genetics/103.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J. C., Giblin-Davidson C., Laerm J., Patton J. C., Lansman R. A. Mitochondrial DNA clones and matriarchal phylogeny within and among geographic populations of the pocket gopher, Geomys pinetis. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6694–6698. doi: 10.1073/pnas.76.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. G., Castora F. J., Frantz S. C., Simpson M. V. Mitochondrial DNA polymorphism: evolutionary studies of the genus Rattus. Ann N Y Acad Sci. 1981;361:135–153. [PubMed] [Google Scholar]
- Brown G. G., Simpson M. V. Intra- and interspecific variation of the mitochondrial genome in Rattus norvegicus and Rattus rattus: restriction enzyme analysis of variant mitochondrial DNA molecules and their evolutionary relationships. Genetics. 1981 Jan;97(1):125–143. doi: 10.1093/genetics/97.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. G., Simpson M. V. Novel features of animal mtDNA evolution as shown by sequences of two rat cytochrome oxidase subunit II genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3246–3250. doi: 10.1073/pnas.79.10.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Wilson A. C. Length mutations in human mitochondrial DNA. Genetics. 1983 Aug;104(4):699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castora F. J., Arnheim N., Simpson M. V. Mitochondrial DNA polymorphism: evidence that variants detected by restriction enzymes differ in nucleotide sequence rather than in methylation. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6415–6419. doi: 10.1073/pnas.77.11.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B. Evolution of mitochondrial DNA sequences in Xenopus. Dev Biol. 1972 Oct;29(2):139–151. doi: 10.1016/0012-1606(72)90051-6. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Ferris S. D., Wilson A. C., Brown W. M. Evolutionary tree for apes and humans based on cleavage maps of mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2432–2436. doi: 10.1073/pnas.78.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco J. F., Brown G. G., Simpson M. V. Further studies on types A and B rat mtDNAs: cleavage maps and evidence for cytoplasmic inheritance in mammals. Plasmid. 1979 Jul;2(3):426–436. doi: 10.1016/0147-619x(79)90026-x. [DOI] [PubMed] [Google Scholar]
- Griffith O. W., Bridges R. J., Meister A. Transport of gamma-glutamyl amino acids: role of glutathione and gamma-glutamyl transpeptidase. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6319–6322. doi: 10.1073/pnas.76.12.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K., Yourno J. Chemical carcinogens as frameshift mutagens: Salmonella DNA sequence sensitive to mutagenesis by polycyclic carcinogens. Proc Natl Acad Sci U S A. 1974 May;71(5):1612–1617. doi: 10.1073/pnas.71.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon A. M., Pepe G., Bakker H., Holtrop M., Bollen J. E., Van Bruggen E. F., Cantatore P., Terpstra P., Saccone C. The restriction fragment map of rat-liver mitochondrial DNA: a reconsideration. Biochim Biophys Acta. 1977 Sep 20;478(2):128–145. doi: 10.1016/0005-2787(77)90177-0. [DOI] [PubMed] [Google Scholar]
- Lansman R. A., Clayton D. A. Selective nicking of mammalian mitochondrial DNA in vivo: photosensitization by incorporation of 5-bromodeoxyuridine. J Mol Biol. 1975 Dec 25;99(4):761–776. doi: 10.1016/s0022-2836(75)80183-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M. Restriction endonuclease cleavage maps of rat and mouse mitochondrial DNAs. Nucleic Acids Res. 1977;4(5):1291–1299. doi: 10.1093/nar/4.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Shen S. H., Slightom J. L., Smithies O. A history of the human fetal globin gene duplication. Cell. 1981 Oct;26(2 Pt 2):191–203. doi: 10.1016/0092-8674(81)90302-0. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Taira M., Yoshida E., Kobayashi M., Yaginuma K., Koike K. Tumor-associated mutations of rat mitochondrial transfer RNA genes. Nucleic Acids Res. 1983 Mar 25;11(6):1635–1643. doi: 10.1093/nar/11.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]