Abstract
Streptococcus pneumoniae and probably most other members of the genus Streptococcus are competent for natural genetic transformation. During the competent state, S. pneumoniae produces a murein hydrolase, CbpD, that kills and lyses noncompetent pneumococci and closely related species. Previous studies have shown that CbpD is essential for efficient transfer of genomic DNA from noncompetent to competent cells in vitro. Consequently, it has been proposed that CbpD together with the cognate immunity protein ComM constitutes a DNA acquisition mechanism that enables competent pneumococci to capture homologous DNA from closely related streptococci sharing the same habitat. Although genes encoding CbpD homologs or CbpD-related proteins are present in many different streptococcal species, the genomes of a number of streptococci do not encode CbpD-type proteins. In the present study we show that the genomes of nearly all species lacking CbpD encode an unrelated competence-regulated murein hydrolase termed LytF. Using Streptococcus gordonii as a model system, we obtained evidence indicating that LytF is a functional analogue of CbpD. In sum, our results show that a murein hydrolase gene is part of the competence regulon of most or all streptococcal species, demonstrating that these muralytic enzymes constitute an essential part of the streptococcal natural transformation system.
INTRODUCTION
In streptococcal species belonging to the mitis phylogenetic group, such as Streptococcus gordonii, Streptococcus pneumoniae, Streptococcus mitis, Streptococcus oralis, Streptococcus infantis, Streptococcus cristatus and Streptococcus sanguinis, competence for natural genetic transformation is regulated by a signaling peptide of the competence-signaling peptide (CSP) type (17, 23). Streptococcal strains and species belonging to this group produce a number of distinct CSPs with different primary structures and specificities (24). The CSP precursor, ComC, is ribosomally synthesized and contains a double-glycine-type leader that is removed concomitant with export by a dedicated ABC transporter, termed ComA (14, 15, 22). In the case of S. sanguinis, the ComC leader peptide is not of the GG type, but the mature secreted peptide pheromone is clearly related to the CSPs produced by the other mitis group streptococci (17). The protein that translocates the CSP peptide across the cytoplasmic membrane of S. sanguinis has not been identified. The extracellular concentration of CSP is monitored by the ComDE two-component regulatory system (16, 37). At the critical external concentration of CSP, phosphorylated ComE activates expression of the alternative sigma factor ComX and other early competence genes. ComX then activates transcription of the late competence genes, some of which encode proteins involved in DNA processing, uptake, and recombination (32).
In S. pneumoniae only 23 of more than 100 CSP-inducible genes are required for transformation (6, 39). Two of the dispensable genes, cbpD and comM, are key players in a competence-associated mechanism called fratricide (13, 18, 26). When pneumococci develop the competent state, they synthesize and secrete the late gene product CbpD. This murein hydrolase functions as a weapon, termed fratricin, that lyses and kills noncompetent cells. In other words, the competent pneumococci act as predators that attack and lyse noncompetent target cells present in the same culture. The competent cells that secrete CbpD protect themselves by producing the immunity protein ComM, a membrane-embedded protein that neutralizes the muralytic activity of CbpD by an unknown mechanism (18). Since ComM is the product of an early competence gene, noncompetent pneumococci lack this immunity protein and are therefore susceptible to CbpD. Pneumococcal CbpD consists of one catalytic domain and two types of cell wall binding domains. The N-terminal CHAP (cysteine histidine-dependent aminohydrolase/peptidase) domain most likely functions either as an amidase that disrupts the N-acetylmuramyl-l-Ala bond or as an endopeptidase that cleaves within the peptide part of peptidoglycan (1, 30, 44). At the C-terminal end of CbpD, there are four choline-binding repeats. These repeats, which together constitute a choline-binding domain (CBD), anchor CbpD noncovalently to choline-decorated teichoic acid present in the pneumococcal cell wall (46). In the middle of the protein, between the CHAP and CBD domains, CbpD contains two Src homology 3 (SH3)-binding (SH3b) domains. These domains bind to the peptidoglycan part of the cell wall (8).
As fratricide is coregulated with natural transformation, we speculated that it evolved to facilitate acquisition of homologous donor DNA from other pneumococcal strains and related streptococcal species. Recently, we obtained strong evidence that this is indeed the case. In experiments carried out in vitro, we showed that the fratricide mechanism enhances gene exchange between pneumococci and also demonstrated that it has a large positive impact on the efficiency of gene transfer from the commensals S. mitis and S. oralis to S. pneumoniae (25). The fact that pneumococcal CbpD stimulates interspecies gene exchange in vitro shows that it is able to lyse S. mitis and S. oralis. This cross-species activity was expected as CbpD proteins from S. mitis and S. oralis are highly homologous to pneumococcal CbpD (Fig. 1). In addition, all three species possess choline-decorated wall teichoic acid (WTA) and lipoteichoic acid (LTA), designated C-polysaccharide and F-polysaccharide, respectively (12). Recently, an in silico survey revealed that genes encoding CbpD-like proteins are present in the genomes of many species belonging to the genus Streptococcus (5). The CbpD-like proteins were identified on the basis of their highly conserved N-terminal CHAP domains. Interestingly, their C-terminal halves are not conserved, suggesting that the cell wall binding specificities of the various CbpD-like proteins differ. In all cases ComX-binding motifs (often referred to as com- or cin-boxes [3, 38]) were present in the promoter regions of the genes (5). Consequently, it is reasonable to assume that the CbpD-like proteins serve the same function as pneumococcal CbpD, namely, to kill and lyse noncompetent streptococci to provide homologous donor DNA for the competent attacker cells. Many of the species harboring these CbpD-like genes have never been demonstrated to be competent for natural transformation. However, recently it has been reported that natural transformation is probably much more widespread in the genus Streptococcus than previously recognized (19, 34).
Fig 1.
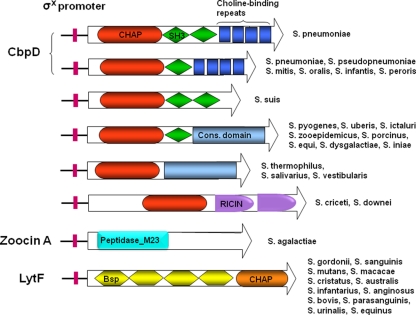
Domain organization of σX-controlled murein hydrolases from different species of streptococci: CHAP, cysteine, histidine-dependent amidohydrolases/peptidases; SH3, binds peptidoglycan (8), choline-binding repeats, bind choline residues linked to teichoic acid; conserved domain, uncharacterized domain that probably mediates binding to the cell wall of target cells; RICIN, carbohydrate binding domain; peptidase M_23, zinc metallopeptidase with a range of specificities. The gene from each streptococcal species that corresponds to the depicted gene product has the following locus tag: spr2006 (S. pneumoniae R6), SPPN_11215 (S. pseudopneumoniae IS7493), SM12261_0760 (S. mitis NCTC 12261), SOR_1962 (S. oralis Uo5), HMPREF9967_0542 (S. infantis SK 1076), HMPREF9180_0030 (S. peroris ATCC 700780), SSUBM407_1976 (S. suis BM 407), SPy_0031 (S. pyogenes M1 GAS), SUB0048 (S. uberis 0140J), Sict7_010100010455 (S. ictaluri 707-05), SeseC_00035 (S. equi subsp. zooepidemicus ATCC 35246), STRPO_1436 (S. porcinus strain Jelinkova 176), SEQ_0031 (S. equi subsp. equi 4047), SDD27957_00205 (S. dysgalactiae subsp. dysgalactiae ATCC 27957), no locus tag assigned (S. iniae), stu0039 (S. thermophilus LMG 18311), STRSA0001_1425 (S. salivarius SK126), HMPREF9425_1781 (S. vestibularis ATCC 49124), ScriH_010100000025 (S. criceti HS-6),HMPREF9176_1241 (S. downei F0415), SAG0031 (S. agalactiae 2603V/R), SGO_2094 (S. gordonii strain Challis), SSA_0036 (S. sanguinis SK 36), SMU.836 (S. mutans UA159), SmacN1_010100005870 (S. macacae NCTC 11558), HMPREF9422_1401 (S. cristatus ATCC 51100), HMPREF9421_0380 (S. australis ATCC 700641), STRINF_01688 (S. infantarius subsp. infantarius ATCC BAA-102), HMPREF9966_1370 (S. anginosus SK52), HMPREF9319_1268 (S. bovis ATCC 700338), HMPREF0833_11509 (S. parasanguinis ATCC 15912), Suri2_010100004140 (S. urinalis 2285-97), and HMPREF0819_0723 (S. equinus ATCC 9812).
Although many streptococcal species possess genes encoding CbpD-like proteins, homologues of cbpD genes are not found in naturally transformable streptococci such as S. gordonii, S. sanguinis, and S. mutans. Does this mean that these species lack the fratricide mechanism? To address this question we searched the genomes of these bacteria for possible CbpD substitutes. Here, we report the identification of a novel murein hydrolase, termed LytF, which is unrelated to CbpD but apparently serves the same function (Fig. 1). Intriguingly, most streptococci lacking a cbpD gene seem to have a lytF gene instead.
MATERIALS AND METHODS
Bacterial growth conditions and storage.
Bacterial species, strains, and plasmids used in this work are described in Table 1. Bacterial cultures were grown at 37°C in Todd-Hewitt medium (Becton, Dickinson and Co.) (for S. gordonii, S. thermophilus, S. mitis, S. oralis, and S. infantis), C medium (28) (for S. pneumoniae), M17 supplemented with 0.5% glucose (GM17) (Oxoid) (for Lactococcus lactis), or brain heart infusion broth (Becton, Dickinson and Co.) supplemented with 5% horse serum (PAA Laboratories Inc.) (for Streptococcus anginosus [Streptococcus milleri], S. cristatus, Streptococcus constellatus, and Streptococcus pyogenes and for Staphylococcus aureus). S. sanguinis was grown in pH-regulated Todd-Hewitt medium (pH 6.8) to avoid autoinduction of competence. Precultures of the bacteria were grown to an optical density at 550 nm (OD550) of 0.3 and maintained as glycerol stocks at −80°C.
Table 1.
Bacterial species, strains, and plasmids
| Species, strain, or plasmid | Genotype or relevant feature(s)a | Reference or sourceb |
|---|---|---|
| S. gordonii strains | ||
| NCTC 7865T | NCTC | |
| AH2 | NCTC 7865, but comA::pEVP3 Cmr Rifr through spontaneous conversion | 48 |
| NCTC 3165 | NCTC | |
| NCTC 7868 | Strain Challis | NCTC |
| SGH139 | NCTC 7868, but comA::pEVP3 Cmr | This work |
| SGH24 | SGH139 with a deletion of sgo_2094 by replacement with the aacA-aphD kanamycin resistance gene; Cmr Kanr | This work |
| SGH25 | SGH139, but Smr through spontaneous conversion; Cmr Smr | This work |
| SGH37 | SGH139, but Rifr through spontaneous conversion; Cmr Rifr | This work |
| SGH43 | SGH25, with a deletion of sgo_2094 by replacement with the aacA-aphD kanamycin resistance gene; Cmr Smr Kanr | This work |
| SGH141 | SGH25 with a deletion of celB by replacement with the aacA-aphD kanamycin resistance gene; Cmr Smr Kanr | This work |
| SGH142 | SGH25 with a deletion of comD by replacement with the aacA-aphD kanamycin resistance gene; Cmr Smr Kanr | This work |
| Other species or strains | ||
| E. coli DH5α | Invitrogen | |
| E. coli BL21 | Invitrogen | |
| E. coli ECH12 | DH5α harboring pHOG2 | This work |
| E. coli ECH13 | BL21 harboring pHOG2 | This work |
| E. coli ECH142 | DH5α harboring pAH1 | This work |
| S. sanguinis SK36 | M. Kilian | |
| S. sanguinis SSH605 | SK36 with a deletion of ssa_0036 by replacement with the aacA-aphD kanamycin resistance gene; Kanr | This work |
| S. anginosus NCTC 10713T | NCTC | |
| S. anginosus NCTC 10708 | NCTC | |
| S. constellatus NCTC 11325T | NCTC | |
| S. cristatus NCTC 12479T | NCTC | |
| S. infantis DSM 12492T | DSMZ | |
| S. mitis NCTC 12261 | NCTC | |
| S. oralis SK155 | M. Kilian | |
| S. pneumoniae RH14 | 7 | |
| S. pyogenes NCTC 8198T | NCTC | |
| S. thermophilus LMG 18311 | ATCCd BAA-250 | |
| Staphylococcus aureus RN 4220 | S. J. Foster | |
| Lactococcus lactis NZ9000 | 27 | |
| Plasmids | ||
| pEVP3 | Carries chloramphenicol resistance gene (cat) | 4 |
| pAH1 | pEVP3 with comA fragment; Cmr | This work |
| pFW13 | Carries the aacA-aphD kanamycin resistance gene | 40 |
| pRSET A | Invitrogen | |
| pRSET/EmGFP | Carries the gfp gene; Ampr | Invitrogen |
| pHOG2 | pRSET A containing lytF-gfp fusion | This work |
Amp, ampicillin; Cm, chloramphenicol; Kan, kanamycin; Rif, rifampin; Sm, streptomycin.
NCTC, National Collection of Type Cultures; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; ATCC, American Type Culture Collection.
Construction of S. gordonii mutants.
The sequences of all primers used are given in Table 2. DNA was introduced into recipient strains by natural transformation. Bacterial cultures were grown to an OD550 of ∼0.2 and induced to the competent state by addition of 250 ng ml−1 synthetic Challis strain CSP (NH2-DVRSNKIRLWWENIFFNKK-COOH). Then, the transforming DNA was added, and the cultures were further incubated for 120 min at 37°C. Selection of transformants was carried out on Todd-Hewitt agar containing the appropriate antibiotic at the following concentrations: kanamycin (Kan) at 350 μg ml−1 or spectinomycin (Spc) at 150 μg ml−1. Resistance to streptomycin (Sm) or rifampin (Rif) was obtained through spontaneous conversion by selection on agar plates containing 1 mg ml−1 streptomycin or 20 μg ml−1 rifampin, respectively. Transformation of S. sanguinis was performed in the same way, by adding 250 ng ml−1 synthetic CSP13b (NH2-DLRGVPNPWGWIFGR-COOH).
Table 2.
Primersa
| Primer | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| a1 | ATTAGGATCCAT8AT8GAYAC8TAYGT8CC8GA |
| a2 | TTAAGGTACC8CC8GA8AT8CC8GC8CCRTC |
| hso36 | AGCAATGGCTAGCAATGAACAGTTTAGCCCATCCCAAC |
| hso38 | CTAATTTTCAAGTAACACGCCCAACGGTAGTGAGCAAGGGCGAGGAG |
| hso39 | AGCAATCTCGAGTTACTTGTACAGCTCGTCCATGC |
| hso50 | CTTGATAATCGTCTCAACTGACT |
| hso51 | AGCTTAAGATCTAGAGCTCGAGGATCAAGGTTGCTGAAAGCAGTAAGAA |
| hso52 | GGAAATATTCATTCTAATTGGTAAGCGCCCCTATCATTTAGCACCTCCCT |
| hso53 | AATGCTACGATTGCTTCACAAAAT |
| kana-f | ATCCTCGAGCTCTAGATCTTAAGCTT |
| kana-r | ACTCCGGATGCATATGCATGCT |
| khb113 | CTACAGGGAAGAGACCAAGG |
| khb114 | CAAGCTTAAGATCTAGAGCTCCTTGGATAATGCCAGATTAGG |
| khb115 | GAGCTCTAGATCTTAAGCTTG |
| khb116 | CTCCGGATGCATATGCATGC |
| khb117 | GCATGCATATGCATCCGGAGGCTGTGTGGTATAATATAGAGAAC |
| khb118 | GTCAAGTCAAAAATATTGTCCTG |
| khb121 | GTGGTAGTTTAGGTTTGTCTG |
| khb122 | CAAGCTTAAGATCTAGAGCTCTGATATTACCATTATTAAACCCC |
| khb123 | GAAAAGAAATTTATAAGAGGACTTAAGTAGGCTTCTCAACAAAAGGAG |
| khb124 | CTGCTATTGTCCCATAAAATTC |
| ss1 | CACCTGTATTGGGTCCCTTG |
| ss2 | AGCTTAAGATCTAGAGCTCGAGGATCTGGGCCCACCTGTATTATTC |
| ss3 | GGAAATATTCATTCTAATTGGTAAGCGCAAGTCGCTTTCGCACTTCTC |
| ss4 | TTTCTATCAGACGGCCTTGC |
All primers were designed in the course of this study. 8, inosine.
Strain SGH139 was derived from Streptococcus gordonii NCTC 7868 by insertion duplication mutagenesis targeting the comA gene (sgo_2097). An ∼1,300-bp region of comA was amplified from NCTC 7868 using the primers a1 and a2, containing BamHI and KpnI sites at their 5′ ends, respectively. The fragment was digested with BamHI and KpnI and ligated into the pEVP3 vector, precleaved with the same enzymes. The resulting plasmid (pAH1) was transformed into electrocompetent Escherichia coli DH5α cells, according to standard procedures (45), resulting in ECH142. pAH1 was purified from ECH142 and transformed into S. gordonii NCTC 7868 by natural transformation, resulting in the insertion duplication mutant SHG139 that contains a disrupted comA gene.
Strains SGH25 and SGH37 were derived from SGH139. Spontaneous resistance to streptomycin (SGH25) or rifampin (SGH37) was obtained by selection on agar plates containing 1 mg/ml streptomycin or 20 μg/ml rifampin, respectively.
Strain SGH43 was derived from SGH25 by replacing lytF (sgo_2094) with a kanamycin resistance gene (aacA-aphD). The kanamycin resistance gene was amplified from pFW13 (40) using the primers kanaF and kanaR. Fragments corresponding to the ∼1,000-bp upstream and downstream regions of sgo_2094 were amplified using the primer pairs hso50/hso51 and hso53/hso54, respectively. The upstream and downstream fragments were then fused to the 5′ and 3′ ends of the kanamycin cassette by overlap extension PCR (21), using the primers hso50 and hso54. The resulting PCR product was transformed into the SGH25 strain by natural transformation, giving rise to the SGH43 strain.
Strain SGH141 was constructed by replacing the celB gene (sgo_1601) in SGH25 with the kanamycin resistance gene referred to above. The primer pair khb115/khb116 was used to amplify the kanamycin resistance gene from pFW13. Fragments corresponding to the ∼1,000-bp upstream and downstream regions of celB were amplified using the primer pairs khb113/khb114 and khb117/khb118, respectively. These two fragments were subsequently fused to the 5′ and 3′ ends of the kanamycin resistance gene using the primers khb113 and khb118. The resulting PCR product was transformed into the SGH25 strain by natural transformation, giving rise to the SGH141 strain.
Strain SGH142 was derived from SGH25 by replacing the comD gene (sgo_2146) with the pFW13 kanamycin resistance gene, as described above. Fragments corresponding to the ∼1,000-bp upstream and downstream regions of comD were amplified using the primer pairs khb121/khb122 and khb123/khb124, respectively. These two fragments were subsequently fused to the kanamycin resistance gene using the primers khb121 and khb124. The resulting PCR product was transformed into the SGH25 strain by natural transformation, giving rise to the SGH142 strain.
The pHOG2 plasmid was derived from the pRSETA expression vector (Invitrogen). The S. gordonii sgo_2094 gene, except for the region encoding the CHAP domain, was amplified using the primers hso36 and hso37. Similarly, the emerald green fluorescent protein (EmGFP) gene was amplified from pRSET/EmGFP (Invitrogen) with the primer pair hso38/hso39. The two fragments were subsequently fused by overlap extension PCR using the primers hso36 and hso39. The resulting PCR construct was cleaved with the restriction enzymes XhoI and NheI and ligated into the pRSETA vector precleaved with the same enzymes. The resulting plasmid, pHOG2, was transformed into competent E. coli DH5α cells, giving rise to ECH12. Finally, pHOG2 was purified from the ECH12 strain and transformed into E. coli BL21, resulting in ECH13.
S. sanguinis strain SSH605 was constructed by replacing the ssa_0036 gene in S. sanguinis SK36 with the kanamycin resistance gene described above. Fragments corresponding to the ∼1,000-bp upstream and downstream regions of ssa_0036 were amplified using the primer pairs ss1/ss2 and ss3/ss4, respectively. The upstream and downstream fragments were then fused to the 5′ and 3′ ends of the kanamycin cassette, using the primers ss1 and ss4. The resulting PCR product was transformed into S. sanguinis SK36 by natural transformation, giving rise to the S. sanguinis SSH605 strain.
Zymogram analysis.
Zymogram analysis was used to detect muralytic activity in whole-cell extracts from competent and noncompetent cultures of S. gordonii and S. sanguinis. To make extracts from competent cells, bacterial cultures were first grown at 37°C until they reached an OD550 of ∼0.2. Then, 250 ng ml−1 of the appropriate CSP was added. After further incubation at 37°C for 15 min, cultures were harvested, and cells from 10-ml samples were pelleted by centrifugation. Prior to electrophoresis, pellets were dissolved in 50 μl of SDS sample buffer and 50 μl of MilliQ-water and heated to 95°C for 10 min. Extracts from noncompetent cells were made in the same way, except that no CSP was added.
The zymogram assay was performed according to the method described by Leclerc and Asselin (31). Whole-cell extracts were separated by SDS-PAGE using a 4% stacking gel and a 10% resolving gel as described by Laemmli (29). Heat-killed substrate cells were mixed into the resolving gel solution before it polymerized. To prepare the substrate cells, 300-ml cultures were grown at 37°C to an OD550 of 0.2. Then, the cells were immediately harvested by centrifugation (10,000 × g) at 4°C for 10 min. The resulting pellet was washed once in 5 ml of ice-cold Tris-HCl (20 mM, pH 7.4) buffer supplemented with 100 mM NaCl. After cells were washed, they were suspended in 1.25 ml of Tris-HCl (1.5 M, pH 8.8) and heat treated for 10 min at 95°C.
Following electrophoresis at 2.5V cm−1, the gels were washed in distilled H2O (dH2O) two times for 30 min each before the refolding buffer (50 mM NaCl, 20 mM MgCl2, 0.5% Triton X-100, and 20 mM Tris-HCl, pH 7.4) was added. The gels were incubated in refolding buffer until lytic activity was observed as bands of clear zones in the opaque gel.
Cocultivation and transformation assays.
Attacker (SGH25 and SGH43, Smr) and target (AH2, Rifr) strains were grown at 37°C to an OD550 of ∼0.2. SGH25 or SGH43 cells (volume, 0.5 ml) were then mixed with an equal volume of AH2 cells and 250 ng ml−1 of Challis CSP. Cocultivation of attacker and target cells was carried out at 37°C for 2 h, after which the samples were put on ice. Next, samples were serially diluted in 10-fold steps and spread on Todd-Hewitt agar plates supplemented with streptomycin (100 μg ml−1) or streptomycin (100 μg ml−1) and rifampin (2 μg ml−1). The agar plates were incubated at 37°C for 15 to 17 h before the number of Smr and Smr Rifr colonies was determined. Uninduced samples were run in parallel as negative controls. Cocultivation experiments involving the attacker strain SGH37 and the target strains SGH140 or SGH141 were performed as described above.
Purified genomic DNA was used to determine transformation efficiency of strains SHG25 and SGH43. Bacterial cultures of these strains were grown at 37°C until they reached an OD550 of 0.2. Next, 250 ng of Challis CSP was added to 1-ml samples of cells together with saturating amounts of genomic DNA (0.5 μg ml−1) isolated from the rifampin-resistant AH2 strain. The cultures were further incubated at 37°C for 2 h and immediately put on ice. Then, the samples were serially diluted and spread on Todd-Hewitt agar plates containing streptomycin (100 μg ml−1) or streptomycin (100 μg ml−1) and rifampin (2 μg ml−1). Uninduced samples were run in parallel as negative controls. Colonies were counted after anaerobic incubation of the plates at 37°C for 15 to 17 h.
Preparation of cell wall sacculi.
Cell wall sacculi from S. gordonii strain Challis were prepared essentially as described by Reusch (43). In brief, cells were harvested from a Challis culture at an OD550 of 0.37, pelleted by centrifugation, and suspended in 3.3 ml of freshly prepared extraction fluid consisting of 1% SDS (wt/vol)–0.5% β-mercaptoethanol (vol/vol) (SDS-ME). Next, the cells were incubated at 85°C for 15 min, followed by dilution in dH2O to a final volume of 5 ml. The cells were dispersed with an Ultra-Turrax T25 basic instrument (Ika-Werke) for 15 s at maximum speed and harvested by centrifugation at 17,500 × g at 4°C for 30 min. The resulting pellet was dissolved in 3.3 ml of hot (85°C) SDS-ME solution. Extraction of the pellet with hot SDS-ME solution was repeated for a total of five times. The extracted material was washed five times with dH2O, once with 2.0 M NaCl, once with dH2O, once with 2.0 M NaCl, and four times with dH2O. The washed sacculi were then dissolved in 200 μl of phosphate-buffered saline (PBS), pH 7.4, and treated with trypsin (1.5 μg ml−1) overnight at 37°C. Sacculi were harvested at 16,000 × g at 4°C, dissolved in 300 μl of 4% SDS, and incubated at 85°C for 15 min. After centrifugation of the sacculi suspension for 30 min at 16,000 × g at 4°C, the sacculi were resuspended, washed three times in dH2O, and suspended in 300 μl of PBS. To remove covalently attached wall teichoic acid (WTA), sacculi were treated with 48% hydrofluoric acid for 48 h at 4°C, as described by Eldholm et al. (8). Purified sacculi were stored at −20°C.
Expression and purification of LytF-GFP.
Construction of the pHOG2 expression vector, which contains a gene encoding the His-tagged LytF-GFP fusion protein under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter, is described above. An overnight culture of the ECH13 strain carrying the pHOG2 vector was diluted to an OD600 of ∼0.1 in 100 ml of Luria-Bertani broth (1% Bacto tryptone [wt/vol], 1% NaCl [wt/vol], and 0.5% Bacto yeast extract [wt/vol]). The ECH13 culture was incubated at 37°C until it reached an OD600 of 0.5. The culture was then induced with 1 mM IPTG and incubated for 2 h with shaking (200 rpm) at 30°C. Next, the cells were pelleted by centrifugation at 4°C, resuspended in 8 ml of PBS and lysed by repeated freezing and thawing in liquid nitrogen and a 42°C water bath. After cell debris was removed by centrifugation (70,000 × g for 10 min at 4°C), the supernatant was filtered through a 0.45-μm-pore-size filter. His-tagged LytF-GFP was purified from the resulting supernatant by the use of Protino Ni-TED columns (Macherey Nagel) as described by the manufacturer.
Epifluorescence microscopy.
The bacterial culture used to study LytF-GFP binding was harvested at an OD550 of ∼0.4. The pellet from 1 ml of culture was washed once in 1 ml of PBS and, after centrifugation, dissolved in 200 μl of PBS. Five microliters of this suspension was transferred to a glass slide and mixed with purified fusion protein dissolved in PBS containing 0.05% Tween 20. The suspension was spread thinly over the slide and incubated at room temperature for 8 min. Then, the slide was washed three times with PBS containing 0.05% Tween 20. Finally, after a coverslip was positioned over the sample, the slide was imaged using a Zeiss LSM 700 confocal microscope. The same protocol was used to study binding of purified LytF-GFP to sacculi.
RESULTS
Identification of LytF, a novel competence-induced murein hydrolase.
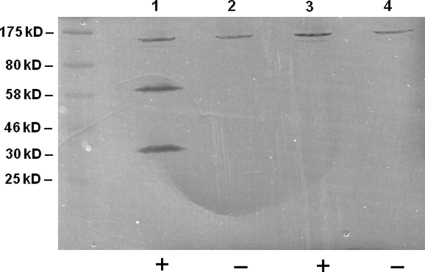
The absence of genes encoding CbpD-like fratricins from the genomes of S. gordonii, S. sanguinis, S. mutans, and several other streptococcal species made us wonder whether these species produce alternative murein hydrolases that carry out the same or a similar function as pneumococcal CbpD. By performing BLASTP searches against the S. gordonii genome with pneumococcal CbpD, we identified a few proteins carrying CHAP domains that were distantly related to the CHAP domain used as a query sequence. Interestingly, the gene encoding one of these proteins, sgo_2094, had a com-box in its promoter region, indicating that it is part of the ComX regulon. By screening the literature we found that sgo_2094 had been identified as a late competence gene by Vickerman et al. (50), who had used microarray analysis to study transcriptional changes in S. gordonii in response to CSP. sgo_2094 encodes a ∼58-kDa protein consisting of an N-terminal signal peptide, three Bsp-like (group B streptococcal secreted protein) domains (42), and a C-terminal CHAP domain (1, 44) (Fig. 1). Proteins with Bsp domains appear to occur only in members of the genus Streptococcus. In all cases described so far, Bsp domains are part of modular proteins that in addition carry a CHAP, amidase, or N-acetylmuramidase domain (30). It follows from these observations that Bsp modules might function as cell wall binding domains that attach the accompanying muralytic domain to its substrate. To determine whether the sgo_2094 gene product functions as a competence-specific murein hydrolase, we performed zymogram analyses with total protein extracts prepared from CSP-induced and noninduced cells of S. gordonii strain Challis. To avoid spontaneous competence induction in cultures used for this experiment, ΔcomA mutants (SGH139 and SGH24) were employed (Table 1). Protein extracts made from SGH139 and SGH24 cells were run on SDS-PAGE gels containing heat-killed cells of the SGH139 strain. The resulting zymogram revealed two clearing zones, corresponding to proteins with molecular masses of ∼60 kDa and ∼30 kDa, which were present only in the protein extract from CSP-induced cells (Fig. 2). Total protein extracts from an S. gordonii strain (SGH24) lacking both the comA and the sgo_2094 genes were analyzed in parallel. In this case, the 60-kDa as well as the 30-kDa clearing zone was missing (Fig. 2). These results show that the sgo_2094 gene product, LytF, encodes a murein hydrolase, and that the 30-kDa band probably represents a degradation product of the full-length LytF protein.
Fig 2.
Murein-hydrolyzing activity in total extracts of S. gordonii strain Challis cells revealed by zymogram analysis. The dark bands represent clearing zones that result from degradation of Challis cells that have been incorporated in the SDS-PAGE resolving gel. Lane 1, extract from competence-induced SGH139 (ΔcomA) cells; lane 2, extract from noncompetent SGH139 (ΔcomA) cells; lane 3, extract from competence-induced SGH24 (ΔcomA ΔlytF) cells; lane 4, extract from noncompetent SGH24 (ΔcomA ΔlytF) cells.
The ssa_0036 gene of S. sanguinis SK36 encodes a protein that is highly similar to Sgo_2094, except that the Ssa_0036 protein contains five instead of three predicted Bsp domains. The properties of the S. sanguinis version of the LytF protein were investigated by zymogram analysis essentially as described for the S. gordonii homologue. A clearing zone corresponding to a protein of ∼70 kDa was detected in the protein extract prepared from CSP-induced S. sanguinis SK36 cells. This zone was absent from extracts prepared from noninduced SK36 cells and a CSP-induced S. sanguinis mutant (SSH605) lacking the ssa_0036 gene (results not shown). The predicted molecular mass of Ssa_0036 is ∼70 kDa, which is in full agreement with the experimental data.
LytF localizes to the equator and septal regions of the S. gordonii cell wall.
To determine whether the Bsp domains target LytF to the cell wall, we made a construct consisting of the Bsp portion of LytF fused to green fluorescence protein (GFP). A His tag was added to the N terminus of the recombinant protein to facilitate purification after overexpression in Escherichia coli. Fluorescence microscopy revealed that the Bsp-GFP fusion protein binds strongly to the S. gordonii cell wall and that it specifically localizes to the equatorial and septal regions (Fig. 3A). In a subfraction of the cells, binding to the polar regions was also observed. As the binding specificity of Bsp-type domains has not been studied before, it was of interest to identify the component of the cell wall that interacts with these domains. To investigate these matters, we prepared cell wall sacculi that consisted only of peptidoglycan and wall teichoic acid. A portion of the sacculus preparation was further treated with hydrofluoric acid to remove the teichoic acid component. Hence, the resulting sacculi consisted of pure peptidoglycan. Interestingly, the Bsp-GFP fusion protein bound to both types of sacculi, displaying a binding pattern identical to that observed with whole S. gordonii cells (Fig. 3B and C). These results demonstrate that Bsp domains recognize the peptidoglycan moiety of the streptococcal cell wall. Furthermore, since the Bsp-GFP fusion protein does not bind to the older section of the cell wall, Bsp domains must recognize structures that are present only in relatively newly synthesized peptidoglycan.
Fig 3.
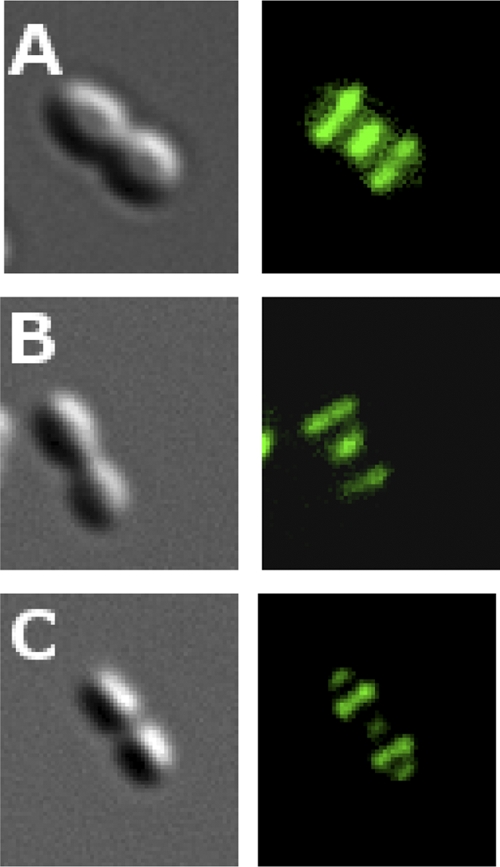
Binding of a LytF-GFP fusion protein to the surface of S. gordonii strain Challis cells. (A) Intact cells. (B) Sacculi. (C) Sacculi lacking teichoic acid. The fusion protein was constructed by exchanging the CHAP domain of LytF with GFP.
What is the biological role of LytF?
LytF is encoded by a late competence gene and is expressed only during competence for natural transformation. What role does LytF play in this process? Although LytF and pneumococcal CbpD are only distantly related through their CHAP domains, they have several properties in common. They are both competence-induced murein hydrolases that bind to the equatorial/septal region of their respective species. As mentioned above, previous studies support the hypothesis that the biological function of CbpD is to lyse susceptible cells in order to release DNA that can be taken up by competent attacker cells (25). Considering the similar properties of CbpD and LytF, we speculated that they are functional analogues that serve the same or similar biological roles in their respective species. To test this experimentally, we performed cocultivation experiments between different strains of S. gordonii to compare the efficiency of gene transfer in the presence and absence of LytF. The mutants SGH25 (ΔcomA Strr) and SGH43 (ΔcomA ΔlytF Strr), derivatives of S. gordonii strain Challis, were used as the competence-induced attacker strains. As target strain we used the mutant AH2 (ΔcomA Rifr), a derivative of S. gordonii strain NCTC 7865. The Challis and NCTC 7865 strains produce CSPs with different primary structures. Challis-CSP (NH2-DVRSNKIRLWWENIFFNKK-COOH) did not cross-induce competence in the NCTC 7865 strain, whereas a weak cross-inducing activity was observed when the Challis strain was subjected to 200 ng/ml of the NCTC 7865 CSP (NH2-DIRHRINNSIWRDIFLKRK-COOH). Consequently, when Challis CSP is added to a mixed culture of Challis and NCTC 7865 cells, only the Challis cells will become competent for natural transformation. Cocultivation experiments showed that a Rifr marker was transferred much more efficiently from NCTC 7865 to Challis cells when LytF was expressed by the competent attacker cells. The number of Strr Rifr doubly resistant transformants obtained with mixed cultures of SGH25 and AH2 cells was, on average, 100-fold higher than with corresponding cultures of SGH43 and AH2 cells (Table 3). This strongly indicates that LytF causes lysis of AH2 target cells, followed by release of DNA that can be taken up by competent SGH25 cells. To rule out the possibility that this result was due to decreased transformability of the strain lacking LytF, the SGH25 and SGH43 strains were transformed with genomic DNA purified from the AH2 strain. The obtained transformation efficiencies show that there are no significant differences between the two strains (Table 3).
Table 3.
Impact of LytF on the efficiency of gene transfer from donor to recipient cells during co-cultivation
| DNA recipient strain | DNA source | Transformation efficiency during cocultivation (%)d | Transformation efficiency with genomic DNA (%)d |
|---|---|---|---|
| SGH25a | AH2 | 0.1 ± 0.019 | |
| SGH43a | AH2 | 0.001 ± 0.0002 | |
| SGH25b | AH2 DNA | 0.87 ± 0.32 | |
| SGH43b | AH2 DNA | 0.66 ± 0.16 | |
| SGH37c | SGH25 | 0.16 ± 0.046 | |
| SGH37c | SGH141 | 0.08 ± 0.016 | |
| SGH37c | SGH142 | 0.006 ± 0.001 |
The S. gordonii Challis strains SGH25 (ΔcomA Strr) and SGH43 (ΔcomA ΔlytF Strr) were induced to competence with Challis CSP and cocultivated with the noncompetent strain AH2 (ΔcomA Rifr). AH2, which is derived from S. gordonii strain NCTC 7865 does not respond to Challis CSP.
Transformation of the SGH23 and SGH43 strains with purified genomic DNA from the AH2 strain. Saturating amounts of DNA were used.
To determine if competent target cells are immune against LytF, the Challis attacker strain SGH37 (ΔcomA Rifr) was cocultivated with the Challis target strains SGH25 (ΔcomA Strr), SGH141 (ΔcomA ΔcelB Strr), and SGH142 (ΔcomA ΔcomD Strr) in the presence of CSP. As it lacks the DNA permease CelB, the SGH141 target strain is not able to take up DNA during competence. The SGH142 target strain contains a disrupted comD gene and will therefore remain noncompetent even in the presence of CSP.
Transformation efficiency was estimated by dividing the number (CFU) of doubly resistant transformants by the total number (CFU) of competent recipient cells and multiplying by 100. Means and standard errors were calculated from three independent experiments.
LytF fratricins have a limited target range.
To gain insight into the specificity of LytF-type murein hydrolases, we performed a number of zymogram analyses in which cells from different species were used as substrates. The results showed that LytF from S. gordonii strain Challis was active against all S. gordonii strains tested but otherwise had a narrow muralytic spectrum (Table 4). The different strains and species used as substrates in the zymograms were also tested for their ability to bind the Bsp-GFP fusion protein described above. The fusion protein bound to all LytF-sensitive strains/species according to the pattern shown in Fig. 3A, while all LytF-resistant species tested negative in the Bsp-GFP binding assay (results not shown).
Table 4.
Specificity of LytF from S. gordonii strain Challis and S. sanguinis SK36 determined by zymogram analysis
| Source of cells incorporated in SDS-PAGE gel as a substrate | Activity of LytF from:a |
|
|---|---|---|
| S. gordonii Challis | S. sanguinis SK36 | |
| S. gordonii NCTC 7865T | + | ND |
| S. gordonii NCTC 3165 | + | + |
| S. anginosus NCTC 10713T | + | − |
| S. anginosus NCTC 10708 | − | + |
| S. constellatus NCTC 11325T | − | + |
| S. sanguinis SK36 | − | + |
| S. cristatus NCTC 12479T | − | + |
| S. pneumoniae RH 14 | − | ND |
| S. mitis NCTC 12261 | − | ND |
| S. oralis SK155 | − | − |
| S. infantis DSM 12492T | − | − |
| L. lactis NZ9000 | − | ND |
| S. aureus RN 4220 | − | ND |
| S. thermophilus LMG 18311 | + | ND |
| S. pyogenes NCTC 8198T | + | ND |
ND, not determined.
The muralytic spectrum displayed by LytF from S. sanguinis SK36 was different from, and included more species than, that of Challis LytF (Table 4). As the amino acid sequences of the CHAP domains of the two LytF proteins are 80% identical, the difference in target ranges is probably due to their N-terminal cell wall binding regions. The N-terminal part of Challis LytF contains three predicted Bsp domains, whereas the corresponding part of S. sanguinis LytF contains five such domains. In addition, large parts of the N-terminal regions of these proteins display relatively low homology. Together, our results show that similar to CbpD-type fratricins, the LytF type has a limited target range and is primarily directed against related bacteria.
Are competent S. gordonii cells immune to LytF?
Competent S. pneumoniae bacteria produce the integral membrane protein, ComM, which protects them against the action of CbpD (18). ComM is encoded by an early competence gene and is consequently expressed about 5 min before CbpD. To address whether competent S. gordonii cells are immune to LytF, we carried out cocultivation experiments with competence-inducible and noninducible mutants of strain Challis. Three different experimental setups were used: experiment 1 using SGH37 (ΔcomA Rifr) and SGH25 (ΔcomA Strr), experiment 2 using SGH37 and SGH141 (ΔcomA ΔcelB Strr), and experiment 3 using SGH37 and SGH142 (ΔcomA ΔcomD Strr). The SGH141 strain lacks the DNA permease CelB (also called ComEC) and is therefore not able to take up DNA during competence, whereas SGH142 contains a disrupted comD gene and will remain noncompetent when subjected to CSP. In experiment 1, transfer of antibiotic resistance markers goes in both directions as both mutant strains become competent when CSP is added. In experiment 2, both mutant strains turn on expression of competence genes upon addition of CSP; however, strain SGH141 is not able to take up donor DNA due to its defective celB gene. As expected, the number of Rif Sm doubly resistant transformants was reduced 2-fold in experiment 2 compared to experiment 1 (Table 3). If competent S. gordonii cells produce an immunity protein that protects them against the muralytic activity of LytF, experiment 3 should have given rise to a higher number of doubly resistant transformants than experiment 2. Our results show that this was clearly not the case (Table 3). On the contrary, the transformation efficiency was found to be 13-fold higher in experiment 2 than in experiment 3. This result strongly indicates that S. gordonii cells lack a competence-induced immunity mechanism that protects them against LytF. The reduced transformation efficiency observed in experiment 3 is presumably due to decreased extracellular levels of LytF, resulting from the fact that LytF is produced by the SGH37 strain only in this cocultivation experiment.
DISCUSSION
Two major types of competence-associated murein hydrolases are present in the genus Streptococcus (Fig. 1). Those belonging to the CbpD-type have closely related N-terminal CHAP domains, while their C-terminal cell wall-targeting regions vary between species. Based on the structure of their C-terminal regions, six CbpD subtypes were identified (the six uppermost variants of CbpD depicted in Fig. 1). One of these, represented by the fratricins produced by Streptococcus criceti and Streptococcus downei, have not been reported before. Alignment of the C-terminal cell wall-targeting regions of fratricins belonging to the same CbpD subtype revealed that their sequences often have diverged considerably at the amino acid level, suggesting that fratricins belonging to the same subtype might possess different binding specificities and target ranges.
The C-terminal CHAP domains of LytF fratricins are highly conserved within the LytF group but are only distantly related to CbpD-type CHAP domains. Interestingly, the CHAP domains carried by LytF-type fratricins are highly similar to the CHAP domains of streptococcal PcsB proteins. Deletion of the pcsB gene or depletion of its gene product generates cells with multiple-division septa, demonstrating that PcsB is required for normal septum formation. It has been proposed that PcsB functions as a division hydrolase, but experimental proof of its muralytic activity is lacking (36, 41). The fact that LytF has murein hydrolase activity suggests that this is also the case for the PcsB proteins. The N-terminal regions of LytF and PcsB are totally unrelated, showing that these proteins serve different biological functions. All LytF-type fratricins have N-terminal regions consisting of two to five successive Bsp domains. The cell wall binding specificities of these regions, which have been shown to vary (Table 4), must be determined by the Bsp domains. Presumably, both the number and structure of Bsp domains play a role in determining their binding properties and target ranges. Until now, proteins with Bsp domains have been found only in streptococci, where they are associated with murein hydrolases that carry enzymatic domains belonging to the PF05257 (CHAP), PF01520 (amidase_3), and PF01183 (Glyco_hydro_25) protein families (29).
BLASTP searches for homologues of CbpD or LytF in the genome of Streptococcus agalactiae were unsuccessful, suggesting that this species produces a different fratricin or lacks a competence-regulated cell lysis mechanism. Similar to many of the streptococcal species listed in Table 1, S. agalactiae has never been demonstrated to be competent for natural transformation. However, since all streptococci seem to possess the core competence genes and regulatory circuits that control their expression, it is reasonable to assume that most or all members of the genus are naturally transformable (19, 34). To investigate the possibility that S. agalactiae produces a competence-induced murein hydrolase that is unrelated to CbpD and LytF, we scanned its genome for alternative candidates. Intriguingly, we found that a gene encoding a close homologue of zoocin A, a protein previously identified as a muralytic bacteriocin, contains a cin-box in its promoter region. The presence of a ComX binding site upstream of the S. agalactiae version of zoocin A strongly indicates that it functions as a fratricin. The zoocin A bacteriocin, which is produced by some strains of Streptococcus equi subsp. zooepidemicus, is a d-alanyl-l-alanine endopeptidase that hydrolyzes the peptidoglycan cross bridges of susceptible streptococci (9, 10, 47). In the S. equi strains carrying the zooA gene, the gene is always accompanied by a neighboring immunity gene termed zif. The zif gene product protects the zoocin A producer strain by inserting an extra alanine residue in the peptidoglycan cross bridges of its cell wall (11). There is no zif immunity gene flanking the S. agalactiae zooA gene, suggesting that the two zoocin A homologues carry out different biological functions in their respective species.
Some investigators might argue that fratricins in reality are bacteriocins that are used by competent cells to eliminate competitors from the common habitat and that the release of transforming DNA from susceptible cells is only a side effect of this chemical warfare. The overwhelming majority of bacteriocins produced by low-GC Gram-positive bacteria are small peptides that have a narrow spectrum of activity. In general, these peptide bacteriocins exert their antibacterial effect by making pores in the cytoplasmic membrane of sensitive bacteria, resulting in leakage of low-molecular-weight cytoplasmic components, destruction of the proton motive force (PMF), and loss of viability (35). In some cases, however, it has been demonstrated that the bactericidal effect of peptide bacteriocins triggers the autolytic system of target bacteria (33). In these cases cell death is followed by cell lysis. In comparison to the ubiquitous peptide bacteriocins, few examples of muralytic antimicrobials are known. Among streptococci, zoocin A, millericin B, and stellalysin, produced by a few strains of Streptococcus equi subsp. zooepidemicus, Streptococcus anginosus, and Streptococcus constellatus, respectively, have been reported to be murein hydrolases that apparently serve as weapons to kill competing bacteria (2, 20, 47). In our view it is too simplistic to regard all bactericidal murein hydrolases solely as antimicrobial compounds. In all likelihood, some of them have evolved primarily for purposes related to their ability to release nutrients and/or DNA from target cells.
S. gordonii strain Challis seems to lack a competence-regulated self-protection mechanism corresponding to the ComM protein produced by S. pneumoniae (18). However, our DNA transfer experiments show that only a fraction of the cell population is lysed during cocultivation, demonstrating that most of the cells are somehow spared from the muralytic activity of LytF. This apparent immunity might be due to the operation of an undiscovered self-protection mechanism or could result from the experimental conditions used. It is, for example, possible that the external concentration of LytF reached in our in vitro cocultivation experiments was not sufficient to cause lysis of a substantial part of the cell population. Regardless of which explanation is correct, some mechanism must exist that controls the activity of LytF under natural conditions. If not, development of the competent state would be suicidal for S. gordonii cells.
The competence regulon of all species in the genus Streptococcus seems to include a murein hydrolase, implying that these cell wall-degrading enzymes must serve some important function associated with natural transformation. Based on previous studies in S. pneumoniae, we have proposed that the competence-induced murein hydrolase CbpD together with the immunity protein ComM constitutes a predatory mechanism used by competent pneumococci to acquire homologous DNA from noncompetent target cells sharing the same habitat (25). In the present study we have shown that LytF, a competence-regulated murein hydrolase produced by S. gordonii and at least 11 other streptococcal species, in most respects is a functional analogue of CbpD. This finding strengthens our hypothesis that the biological role of these murein hydrolases is to kill and lyse close relatives of the competent attacker cells in order to release homologous donor DNA. In a very recent report, Stevens et al. (49) show that increasing the error rate during ribosomal decoding promotes competence development in S. pneumoniae. This finding provides strong evidence that a major function of natural transformation in streptococci is repair of damaged DNA. Such repair will work only if competent streptococci possess a mechanism that enables them to capture DNA from closely related bacteria when growing in an environment consisting mostly of distantly related and unrelated bacteria, i.e., in multispecies biofilms in the oral cavity and nasopharynx. In our view, the best candidate for such a mechanism is the fratricide mechanism, which in the present study is shown to be omnipresent in the genus Streptococcus.
ACKNOWLEDGMENT
This work was supported by The Research Council of Norway.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237 [DOI] [PubMed] [Google Scholar]
- 2. Beukes M, Hastings JW. 2001. Self-protection against cell wall hydrolysis in Streptococcus milleri NMSCC 061 and analysis of the millericin B operon. Appl. Env. Microbiol. 67:3888–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell EA, Choi SY, Masure HR. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929–939 [DOI] [PubMed] [Google Scholar]
- 4. Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128 [DOI] [PubMed] [Google Scholar]
- 5. Claverys JP, Martin B, Håvarstein LS. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 64:1423–1433 [DOI] [PubMed] [Google Scholar]
- 6. Dagkessamanskaia A, et al. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071–1086 [DOI] [PubMed] [Google Scholar]
- 7. Eldholm V, Johnsborg O, Haugen K, Solheim Ohnstad H, Håvarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155:2223–2234 [DOI] [PubMed] [Google Scholar]
- 8. Eldholm V, et al. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual cell-envelope binding-specificity to kill target cells during fratricide. Mol. Microbiol. 76:905–917 [DOI] [PubMed] [Google Scholar]
- 9. Gargis AS, O'Rourke ALD, Sloan GL, Simmonds RS. 2009. Prevalence and acquisition of the genes for Zoocin A and Zoocin A resistance in Streptococcus equi subsp. zooepidemicus. J. Mol. Evol. 68:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gargis SR, et al. 2009. Use of 4-sulfophenyl isothiocyanate labeling and mass spectrometry to determine the site of action of the streptococcolytic peptidoglycan hydrolase zoocin A. Appl. Environ. Microbiol. 75:72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gargis SR, et al. 2009. Zif, the Zoocin A immunity factor, is a FemABX-like immunity protein with a novel mode of action. Appl. Environ. Microbiol. 75:6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gillespie SH, et al. 1993. Species of alpha-hemolytic streptococci possessing a C-polysaccharide phosphoryl-containing antigen. Infect. Immun. 61:3076–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of non-competent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102:8710–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229–240 [DOI] [PubMed] [Google Scholar]
- 15. Håvarstein LS, Coomaraswami G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 92:11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863–869 [DOI] [PubMed] [Google Scholar]
- 17. Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationships to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297–1307 [DOI] [PubMed] [Google Scholar]
- 19. Håvarstein LS. 2010. Increasing competence in the genus Streptococcus. Mol. Microbiol. 78:541–544 [DOI] [PubMed] [Google Scholar]
- 20. Heng NCK, et al. 2006. The large antimicrobial proteins (bacteriocins) of streptococci. Int. Congr. Ser. 1289:351–354 [Google Scholar]
- 21. Higuchi R, von Beroldingen CH, Sensabaugh GF, Erlich HA. 1988. DNA-typing from single hairs. Nature 332:543–546 [DOI] [PubMed] [Google Scholar]
- 22. Hui FM, Morrison DA. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnsborg O, Eldholm V, Håvarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767–778 [DOI] [PubMed] [Google Scholar]
- 24. Johnsborg O, Blomqvist T, Kilian M, Håvarstein LS. 2007. Biologically active peptides in streptococci, p 25–59 In Hakenbeck R, Chhatwal S. (ed), Molecular biology of streptococci. Horizon Scientific Press, Norfolk, United Kingdom [Google Scholar]
- 25. Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae. Mol. Microbiol. 69:245–253 [DOI] [PubMed] [Google Scholar]
- 26. Kausmally L, Johnsborg O, Lunde M, Knutsen E, Håvarstein LS. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 28. Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39:508–518 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30. Layec S, Decaris B, Leblond-Bourget N. 2008. Diversity of Firmicutes peptidoglycan hydrolases and specificities of those involved in daughter cell separation. Res. Microbiol. 159:507–515 [DOI] [PubMed] [Google Scholar]
- 31. Leclerc D, Asselin A. 1989. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can. J. Microbiol. 35:749–753 [DOI] [PubMed] [Google Scholar]
- 32. Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martínez-Cuesta MC, et al. 2000. Requirement of autolytic activity for bacteriocin-induced lysis. Appl. Env. Microbiol. 66:3174–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nes IF, Yoon SS, Diep DB. 2007. Ribosomally synthesized antimicrobial peptides (bacteriocins) in lactic acid bacteria: a review. Food Sci. Biotechnol. 16:675–690 [Google Scholar]
- 36. Ng WL, Kazmierczak KM, Winkler ME. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161–1175 [DOI] [PubMed] [Google Scholar]
- 37. Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853–862 [DOI] [PubMed] [Google Scholar]
- 38. Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192–6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peterson SN, et al. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051–1070 [DOI] [PubMed] [Google Scholar]
- 40. Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lütticken R. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137–147 [DOI] [PubMed] [Google Scholar]
- 41. Reinscheid DJ, Gottschalk B, Schubert A, Eikmanns BJ, Chhatwal GS. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reinscheid DJ, et al. 2002. Influence of proteins Bsp and FemH on cell shape and peptidoglycan composition in group B streptococcus. Microbiology 148:3245–3254 [DOI] [PubMed] [Google Scholar]
- 43. Reusch VM. 1982. Isolation and analysis of sacculi from Streptococcus sanguis. J. Bacteriol. 151:1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rigden DJ, Jedrzejas MJ, Galperin MY. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230–234 [DOI] [PubMed] [Google Scholar]
- 45. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Sánches-Puelles JM, Sanz JM, García JL, García E. 1990. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene 89:69–75 [DOI] [PubMed] [Google Scholar]
- 47. Simmonds RS, Simpson WJ, Tagg JR. 1997. Cloning and sequence analysis of zooA, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene 189:255–261 [DOI] [PubMed] [Google Scholar]
- 48. Steinmoen H. 2003. PhD thesis. Agricultural University of Norway, Ås, Norway [Google Scholar]
- 49. Stevens KE, Chang D, Zwack EE, Sebert ME. 2011. Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2(5):e00071–11 doi:10.1128/mBio.00071-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vickerman MM, Iobst S, Jesionowski AM, Gill SR. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 189:7799–7807 [DOI] [PMC free article] [PubMed] [Google Scholar]