Abstract
Iron is one of the crucial elements required for the growth of Mycobacterium tuberculosis. However, excess free iron becomes toxic for the cells because it catalyzes the production of reactive oxygen radicals, leading to oxidative damage. Hence, it is essential for the pathogen to have the ability to store intracellular iron in an iron-rich environment and utilize it under iron depletion. M. tuberculosis has two iron storage proteins, namely BfrA (Rv1876; a bacterioferritin) and BfrB (Rv3841; a ferritin-like protein). However, the demonstration of biological significance requires the disruption of relevant genes and the evaluation of the resulting mutant for its ability to survive in the host and cause disease. In this study, we have disrupted bfrA and bfrB of M. tuberculosis and demonstrated that these genes are crucial for the storage and supply of iron for the growth of bacteria and to withstand oxidative stress in vitro. In addition, the bfrA bfrB double mutant (H37Rv ΔbfrA ΔbfrB) exhibited a marked reduction in its ability to survive inside human macrophages. Guinea pigs infected with H37Rv ΔbfrA ΔbfrB exhibited a marked diminution in the dissemination of the bacilli to spleen compared to that of the parental strain. Moreover, guinea pigs infected with H37Rv ΔbfrA ΔbfrB exhibited significantly reduced pathological damage in spleen and lungs compared to that of animals infected with the parental strain. Our study clearly demonstrates the importance of these iron storage proteins in the survival and pathogenesis of M. tuberculosis in the host and establishes them as attractive targets for the development of new inhibitors against mycobacterial infections.
INTRODUCTION
Iron is an essential nutrient for almost all microbes, including pathogens such as Mycobacterium tuberculosis (8, 15, 21). It is an indispensable cofactor for proteins involved in critical cellular processes, such as electron transfer, oxygen transport, DNA synthesis, etc. (21). Although iron is essential, excess free iron is potentially toxic for the cells because it catalyzes the production of reactive oxygen radicals by a Fenton reaction, leading to oxidative damage (3). Thus, all living organisms tightly regulate the cellular levels of iron by employing efficient iron acquisition and storage mechanisms. Microorganisms have evolved two types of proteins for storing iron, ferritins (Ftn) and bacterioferritins (Bfr) (3); these are distinguishable by the presence of heme in the latter. The primary function of bacterioferritins and ferritins is to store iron during iron adequacy and supply it to the cell for various functions. It has been observed that prokaryotes possess a homolog of either an Ftn or Bfr; however, some microorganisms, such as Escherichia coli, Vibrio cholerae, Clostridium acetobutylicum, and M. tuberculosis, have evolved with the presence of both Ftn and Bfr. Although a close structural similarity exists between Ftn and Bfr proteins, their amino acid sequences exhibit little homology with no immunological cross-reactivity, suggesting different origins. These proteins, which exist as macromolecular assemblies, characteristically are composed of 24 identical subunits of 18 to 20 kDa. Once assembled into a spherical protein shell, they can contain ∼600 to 2,400 iron atoms per molecule. These subunits are assembled into a complex with 4-, 3-, and 2-fold symmetry axes (6). Even though the exact mechanism of iron storage in vivo remains elusive, there is enough evidence to demonstrate that the formation of iron core requires the binding of ferrous iron to ferritin/bacterioferritin protein followed by migration to the ferroxidase catalytic site, where ferrous (Fe2+) iron is oxidized to the ferric (Fe3+) state.
The sequencing of the M. tuberculosis H37Rv genome revealed the presence of two putative iron storage proteins, namely, BfrA (Rv1876), a bacterioferritin, and BfrB (Rv3841), a ferritin-like protein (7). The expression of both bfrA and bfrB is regulated by the binding of iron-activated IdeR (iron-dependent regulator) to the tandem operator sites present upstream of these iron storage genes. The regulation of the expression of bfrA in response to iron levels perhaps serves as a crucial mechanism for the adaptation and survival of M. tuberculosis in the host (22). Moreover, the expression of bfrB was found to be upregulated during adaptation to stationary phase and low-oxygen conditions (22, 24, 27). In the past, there have been several suggestive pieces of evidence for the role of these proteins in iron storage and release, such as (i) the induction of BfrA and BfrB production in high-iron culture medium (22) and (ii) their reduced expression in iron-starved cultures of M. tuberculosis (10). However, recently we have published the crystal structures of BfrA and BfrB to elucidate the structural aspects related to iron storage and have provided evidence to show that BfrA is bound to iron atoms at the ferroxidase center (12). Similarly, BfrB has been experimentally shown to take up iron and carry out ferroxidase activity as well as the release of stored iron (14).
In view of the well-established importance of iron for M. tuberculosis, the role of BfrA and BfrB in iron storage and supply as well as in protection against iron-mediated oxidative stress and their overexpression during hypoxic conditions, which is often associated with the latent phase (20, 23, 27), these proteins represent attractive targets for the development of new therapeutic molecules against tuberculosis (4, 10). However, the biological significance of these iron-storing proteins for M. tuberculosis has not been genetically proven. In this study, we have disrupted both bfrA and bfrB in M. tuberculosis H37Rv to evaluate their importance in the survival and pathogenesis of M. tuberculosis in the host.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Mycobacterial strains were grown on Middlebrook 7H11 (MB7H11) solid medium or in MB7H9 medium supplemented with 10% albumin dextrose catalase (ADC), 0.2% glycerol, and 0.05% Tween 80 at 37°C and with spinning at 200 rpm. M. tuberculosis H37Rv was transformed with pJV53 to overexpress recombineering proteins that enhance the recombination frequency for the generation of M. tuberculosis mutants (25). Thus, for all of the experiments carried out in this study, we have used M. tuberculosis H37Rv/pJV53 as the wild-type strain. E. coli strains XL-1 Blue (Stratagene) and HB101 (Life Technologies) were used for cloning. Kanamycin, hygromycin, and chloramphenicol were used at a concentration of 25, 50, and 30 μg/ml, respectively, for mycobacteria or at 25, 150, and 30 μg/ml, respectively, for E. coli. 2′2′-Dipyridyl (DPI), an iron chelator, was added to MB7H9 medium at a final concentration of 100 μM for growing M. tuberculosis in iron-depleted conditions.
Table 1.
Bacterial strains, plasmids, and cell line used in this study
| Strains, plasmids, and cell line | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli XL-1 Blue | endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44F′[::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK− mK+) | Stratagene, Heidelberg, Germany |
| E. coli HB101 | F− (gpt-proA)62 leuB6 glnV44 ara-14 galK2 lacY1 (mcrC-mrr) rpsL20 (Strr) xyl-5 mtl-1 recA13 | Life Technologies, CA |
| M. tuberculosis | M. tuberculosis H37Rv expressing recombineering proteins gp60 and gp61 | This study |
| H37Rv ΔbfrA | M. tuberculosis H37Rv bfrA mutant | This study |
| H37Rv ΔbfrB | M. tuberculosis H37Rv bfrB mutant | This study |
| H37Rv ΔbfrAΔbfrB | M. tuberculosis H37Rv bfrA bfrB double mutant | This study |
| H37Rv ΔbfrA Comp | H37Rv ΔbfrA complemented with wild-type bfrA | This study |
| H37Rv ΔbfrB Comp | H37Rv ΔbfrB complemented with wild-type bfrB | This study |
| Plasmids | ||
| pYUB854 | Cloning vector with hygromycin resistance gene cassette flanked by two multiple cloning sites | 3a |
| pSD7 | Mycobacterial promoter cloning vector carrying kanamycin resistance gene and promoterless chloramphenicol resistance gene | 7a |
| pJV53 | Mycobacterium-E. coli shuttle vector encoding recombineering proteins gp60 and gp61 | 25 |
| pVRΔA | pYUB854 with bfrA::hyg | This study |
| pVRΔB | pYUB854 with bfrB::hyg | This study |
| pVRΔAB | pYUB854 with bfrB::CATtrrn | This study |
| pVR.BfrA | pSD7 carrying bfrA gene with native promoter | This study |
| pVR.BfrB | pSD7 carrying bfrB gene with native promoter | This study |
| Cell line | ||
| THP-1 | Human acute monocytic leukemia cell line | NCCS, Pune, India |
Disruption of bfrA and bfrB genes in M. tuberculosis.
Primers were designed to amplify (i) an ∼700-bp amplicon comprised of ∼200 bp of the 5′-proximal end of the bfrA and bfrB genes and ∼500 bp of the immediate upstream regions of bfrA and bfrB (amplicon I), and (ii) an ∼700-bp amplicon comprised of ∼200 bp of the 3′-distal end of bfrA and bfrB genes and ∼500 bp of the immediate downstream region of bfrA and bfrB genes (amplicon II). The amplicons I and II were PCR amplified and cloned into the vector pYUB854 flanking the hygromycin cassette at KpnI/XbaI and XhoI/SpeI restriction sites, respectively, to generate pVRΔA and pVRΔB. The linear allelic exchange substrate (AES) of ΔbfrA::hyg and ΔbfrB::hyg was excised from pVRΔA and pVRΔB by using KpnI/SpeI and electroporated into M. tuberculosis strains separately as described earlier (25) to generate the mutant bfrA (H37Rv ΔbfrA) and bfrB (H37Rv ΔbfrB) strains of M. tuberculosis. For the generation of the double mutant of M. tuberculosis (mutated in bfrA and bfrB genes), the bfrB gene was disrupted in the H37Rv ΔbfrA strain. Briefly, a hygromycin resistance cassette in pVRΔB was removed by using XbaI and XhoI and was replaced with the chloramphenicol resistance gene expressed under the mycobacterial Trrn promoter to generate pVRΔAB, which was digested with SpeI to generate ΔbfrB::CATtrrn AES. The AES then was electroporated into H37Rv ΔbfrA to generate the double mutant of M. tuberculosis, namely, H37Rv ΔbfrA ΔbfrB.
Disk diffusion assay.
To evaluate the ability of H37Rv ΔbfrA ΔbfrB to withstand oxidative stress, wild-type and mutant strains were subjected to oxidative stress-inducing agents, such as cumene hydroperoxide and plumbagin, by employing the disk diffusion assay as described previously (22). Briefly, all of the mycobacterial strains were grown to an A600 of 0.7 to 0.8 in MB7H9 supplemented medium. The cells were washed once with equal volumes of phosphate-buffered saline (PBS) (pH 7.4), followed by dilution to an A600 of 0.5. Two hundred fifty μl (1 × 106 cells) was plated on MB7H11 agar supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC). The plates were allowed to dry for 10 min, and a 5-mm sterile disk was placed on the plate. The oxidative agents were added to the disk at various concentrations, and the plates were incubated at 37°C for 15 days to measure the zone of inhibition.
Comparison of the growth of H37Rv ΔbfrA ΔbfrB and the parental strain in human macrophages.
Human monocytic THP-1 cells were cultured in complete RPMI-glutamax medium (containing 10% heat-inactivated fetal bovine serum [FBS] and 1% antibiotic-antimycotic mix) (Gibco) and were differentiated to macrophages by the addition of 30 nM phorbol 12-myristate 13-acetate (PMA) (Sigma) for 16 h at 37°C in 5% CO2. Cells were washed with complete RPMI medium and rested for 2 h in fresh medium before infection. For infection, 5 × 105 macrophages were infected with 1 × 105 mycobacteria to achieve a multiplicity of infection (MOI) of 1:5 (1 bacterium per 5 macrophages) in 24-well plates for 4 h in triplicates (17). Following infection, extracellular bacteria were removed by treatment with 200 μg/ml amikacin for 2 h. For the determination of the intracellular growth of M. tuberculosis strains, macrophages were harvested and lysed by the addition of 0.025% SDS (Sigma) at 2, 4, and 8 days postinfection. Appropriate dilutions of the lysates then were plated on MB7H11 agar. Colonies were counted after 4 weeks of incubation at 37°C, and the data were expressed as CFU/ml. The same protocol was followed at day 0 to determine the percent infection.
Studies on the influence of bfrA and bfrB disruption on the growth of the pathogen in guinea pigs.
Pathogen-free outbred female guinea pigs of the Duncan-Hartley strain in the weight range of 200 to 300 g were obtained from the Disease Free Small Animal House Facility, Chowdhary Charan Singh Haryana Agricultural University, Hisar, India. The animals were maintained in a biosafety level 3 facility and routinely cared for according to the guidelines of the CPCSEA (Committee for the Purpose of Control and Supervision on Experiments on Animals), India. All of the experimental protocols included in this study were reviewed and approved by the animal ethics committee of the institute. To study the influence of bfrA and bfrB mutation on the virulence of the pathogen, guinea pigs (n = 6) were infected by the aerosol route with 5 to 10 bacilli of the wild-type, mutant, or respective complemented strains. Animals were euthanized at 5, 10, and 16 weeks postinfection by CO2 asphyxiation. The survival of the animals was monitored for 21 weeks postinfection. After dissection, lungs, liver, and spleen of the animals were scored for pathological changes as described previously (13). For histopathological evaluation, the right lung and a portion of the left dorsal lobe of the liver were removed and fixed in 10% buffered formalin. The left caudal lung lobe and caudal portion of spleen were aseptically removed for the measurement of the bacillary load. Histopathological changes in lungs and liver were evaluated as described earlier (13). The detection limit in the case of lung as well as spleen CFU was 1 log10 CFU/organ.
Statistical analysis.
For comparisons between the groups, the nonparametric Kruskal-Wallis test followed by the Mann-Whitney U test, one-way analysis of variance (ANOVA) with the Tukey posttest, two-way ANOVA with the Bonferroni multiple comparison test, and Student's t test were employed where appropriate. Differences were considered significant when P < 0.05. For the statistical analysis and generation of graphs, Prism 5 software (version 5.01; GraphPad Software Inc., CA) was used.
RESULTS
Functional disruption of the bfrA and bfrB genes of M. tuberculosis and characterization of the mutants.
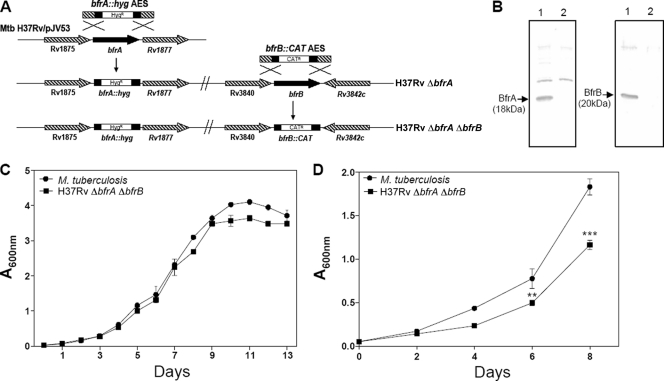
To examine the role of bfrA (Rv1876) and bfrB (Rv3841) in the physiology of M. tuberculosis, we constructed three knockout strains lacking bfrA (H37Rv ΔbfrA), bfrB (H37Rv ΔbfrB), and both genes (H37Rv ΔbfrA ΔbfrB) (Fig. 1A). The expression of BfrA and BfrB proteins was analyzed in the mutants as well as in the parental strain by immunoblotting using antibodies raised against these proteins. It was observed that while M. tuberculosis expressed BfrA and BfrB, as was evident from the presence of 18- and 20-kDa bands, respectively, the mutant strains H37Rv ΔbfrA and H37Rv ΔbfrB showed no commensurate expression of bfrA and bfrB, respectively (see Fig. S1 in the supplemental material). The restoration of BfrA and BfrB expression was observed in M. tuberculosis bfrA and bfrB complemented strains (see Fig. S1). Moreover, H37Rv ΔbfrA ΔbfrB did not show any expression of either bfrA or bfrB (Fig. 1B). Further, we assessed the growth characteristics of these mutants under various growth conditions. No significant differences were observed in the growth of the double mutant compared to that of the parental strain under standard culture conditions by using MB7H11 or MB7H9 medium (Fig. 1C). The growth of H37Rv ΔbfrA and H37Rv ΔbfrB also was similar to that of the parental strain in MB7H9 medium. To find out whether the cell compensates for the absence of BfrB by the overproduction of BfrA and vice versa, we analyzed the expression of BfrA in the H37Rv ΔbfrB mutant and the expression of BfrB in the H37Rv ΔbfrA mutant of M. tuberculosis; however, these studies revealed no compensatory overproduction in any of these cases (data not shown).
Fig 1.
Characterization of H37Rv ΔbfrA ΔbfrB. (A) Disruption of bfrA and bfrB genes of M. tuberculosis by using the recombineering method. The diagram represents homologous recombination between bfrA::hyg AES and the bfrA gene in M. tuberculosis to generate H37Rv ΔbfrA, followed by recombination between bfrB::CAT AES and the bfrB gene in H37Rv ΔbfrA to generate H37Rv ΔbfrA ΔbfrB. (B) Confirmation of disruption of bfrA and bfrB deletion in M. tuberculosis by immunoblotting. Five μg of cell-free protein extract of M. tuberculosis (lane 1) and H37Rv ΔbfrA ΔbfrB (lane 2) was loaded onto a 12% polyacrylamide gel and subjected to electrophoresis. BfrA and BfrB were detected by immunoblot analysis using anti-BfrA or anti-BfrB polyclonal antiserum. BfrA and BfrB proteins migrated as protein bands corresponding to a molecular mass of 18 and 20 kDa, respectively (lane 1). The disruption of bfrA and bfrB in H37Rv ΔbfrA ΔbfrB was confirmed by the lack of expression of either protein (lane 2). (C) Growth kinetics of M. tuberculosis and H37Rv ΔbfrA ΔbfrB in MB7H9 medium. Cultures were inoculated in duplicate with a starting absorbance (A600) of 0.025, and the growth was monitored for 13 days. (D) Growth curve of M. tuberculosis H37Rv and H37Rv ΔbfrA ΔbfrB in iron-depleted medium. M. tuberculosis H37Rv and H37Rv ΔbfrA ΔbfrB were grown in MB7H9 medium supplemented with 0.05% Tween 80, 10× ADC, and 100 μM 2′2′ dipyridyl. The growth of the strains was monitored by measuring the A600 for 8 days. There was a significant difference in the growth of H37Rv ΔbfrA ΔbfrB compared to that of M. tuberculosis H37Rv under iron-deprived conditions. The values of absorbance are represented as the means (± standard errors) of three independent samples, and the experiment was repeated three times. **, P < 0.01; ***, P < 0.001 (two-way ANOVA).
H37Rv ΔbfrA ΔbfrB shows reduced growth in the iron-depleted medium.
To demonstrate the effect of iron deprivation on the growth of M. tuberculosis deficient in bacterioferritin A and B, we compared the ability of the mutant H37Rv ΔbfrA ΔbfrB to that of the parental M. tuberculosis strain to grow in MB7H9 medium in the presence of 2′2′-dipyridyl, an iron chelator. The growth of both strains was monitored for 8 days. We observed that the growth of H37Rv ΔbfrA ΔbfrB was significantly reduced compared to that of the parental strain in the presence of 100 μM 2′2′-dipyridyl (Fig. 1D). This concentration of iron chelator had no effect on the growth of the parental strain. Our observations indicate that under iron-deprived conditions, H37Rv ΔbfrA ΔbfrB was compromised for growth compared to the ability of the parental strain. Iron is an essential micronutrient that is required by the bacterial cell for its normal functioning and growth. Under iron-depleted conditions, BfrA and BfrB were able to supply iron in the case of the parental strain, thus leading to a similar growth profile for this strain under the presence or absence of an iron chelator. However, the double-mutant strain lacking both iron storage proteins was unable to store and, hence, supply iron to cells under iron-starved conditions, leading to a reduction in its growth rate (by ∼40%) compared to that of the parental strain. In the absence of the chelator, the growth of the mutant and the parental strain was comparable, indicating that iron supply in the medium was sufficient for cell growth. These observations indeed further emphasize the role of these ferritins in the storage and supply of iron.
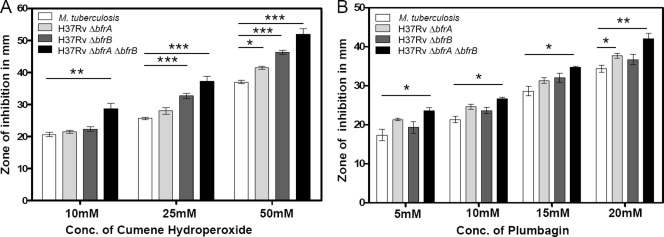
Disruption of bfrA and bfrB genes results in increased sensitivity of M. tuberculosis to oxidative stress.
To evaluate the role of BfrA and BfrB in mediating the response of M. tuberculosis to oxidative stress, H37Rv ΔbfrA, H37Rv ΔbfrB, H37Rv ΔbfrA ΔbfrB, and the parental strain were subjected to various oxidative stress-inducing agents, such as cumene hydroperoxide (an organic peroxide) and plumbagin (a superoxide generator). A concentration range of 10 to 50 mM in the case of cumene hydroperoxide and 5 to 25 mM in the case of plumbagin was employed for the assay. The M. tuberculosis strain lacking both iron storage proteins BfrA and BfrB exhibited a markedly higher sensitivity to both oxidative stress-inducing agents included in this study compared to that of the parental strain (Fig. 2). It was observed that in response to both stress-inducing agents, the zone of inhibition increased with increasing concentrations of the agents. However, a significantly larger zone of inhibition was observed in the case of H37Rv ΔbfrA ΔbfrB compared to that of the parental strain. These observations indicate a crucial role of BfrA and BfrB proteins in protecting the pathogen against oxidative stress. The absence of both iron storage proteins probably enhances the susceptibility of H37Rv ΔbfrA ΔbfrB to Fenton reaction due to the increased availability of free Fe2+ ions inside the cell. The influence of the disruption of a single gene on the ability of M. tuberculosis to withstand the oxidative stresses was variable. H37Rv ΔbfrB was significantly more sensitive to peroxide stress caused by cumene hydroperoxide than the parental strain. However, H37Rv ΔbfrA exhibited a marginal sensitivity to a higher concentration of both cumene hydroperoxide (50 mM) and plumbagin (20 mM). H37Rv ΔbfrA ΔbfrB exhibited a higher sensitivity to oxidative stress induced by both agents than the parental strain (Fig. 2). These observations indicate that the functions of BfrA and BfrB are not entirely overlapping, and the influence of their collective absence on the ability of the cell to withstand oxidative stress could be more prominent than that in the case of a single mutation.
Fig 2.
Influence of disruption of bfrA and bfrB on the ability of M. tuberculosis to withstand oxidative stress. Shown is the range of the inhibition zones observed in the presence of various concentrations of cumene hydroperoxide (A) and plumbagin (B). H37Rv ΔbfrA exhibited a significant sensitivity toward both cumene hydroperoxide and plumbagin, although only at higher concentrations, whereas H37Rv ΔbfrB exhibited a significant sensitivity toward cumene hydroperoxide. H37Rv ΔbfrA ΔbfrB exhibited a significantly higher sensitivity to oxidative stress induced by both agents compared to that of the parental strain. The values of zones of inhibition are represented as the means (± standard errors) of three independent samples, and the experiment was repeated three times. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-way ANOVA).
Further, to demonstrate that the oxidative stress response exhibited by the double mutant was specific and not due to the generally compromised health of the cells leading to nonspecific effects, we compared the effect of the antitubercular drugs isoniazid, rifampin, streptomycin, and ethionamide on the growth of H37Rv ΔbfrA ΔbfrB and the parental strain. The sensitivity of M. tuberculosis and H37Rv ΔbfrA ΔbfrB strains to various concentrations of these antibiotics for 7 and 14 days at 37°C was found to be comparable (data not shown), thus indicating that the oxidative stress response exhibited by the double mutant was specific in nature.
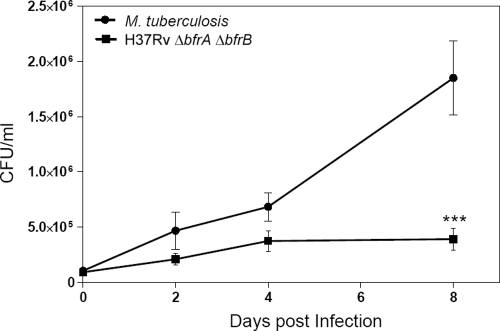
H37Rv ΔbfrA ΔbfrB exhibits attenuated growth in human macrophages.
The intracellular growth of H37Rv ΔbfrA ΔbfrB as well as the parental strain was analyzed in human macrophage THP-1 cells. The uptake of the wild type and the H37Rv ΔbfrA ΔbfrB mutant in the macrophages was found to be comparable (19 and 16%, respectively). Initially, no significant difference was observed in the growth of H37Rv ΔbfrA ΔbfrB and the parental strain; however, thereafter the wild-type strain continued to grow normally but H37Rv ΔbfrA ΔbfrB exhibited an almost complete attenuation of its growth. At 8 days postinfection, we observed a 5-fold difference in the CFU between H37Rv ΔbfrA ΔbfrB and the parental strain (Fig. 3). These results substantiate the importance of these iron storage genes in the growth and survival of the pathogen in the host macrophages.
Fig 3.
Influence of deletion of bfrA and bfrB genes on the growth of M. tuberculosis in THP-1 cells. THP-1 cells were infected with M. tuberculosis or the H37Rv ΔbfrA ΔbfrB mutant at an MOI of 1:5 (bacterium to macrophage), and the number of intracellular viable bacteria was determined for 8 days. Cells were lysed, and appropriate dilutions of mycobacteria were plated onto MB7H11 agar to determine the CFU. The H37Rv ΔbfrA ΔbfrB mutant exhibited a marked attenuation in its growth compared to that of M. tuberculosis H37Rv at 8 days postinfection. The values are represented as the means (± standard errors) of three independent infections, and the experiment was repeated three times. ***, P < 0.001 (two-way ANOVA).
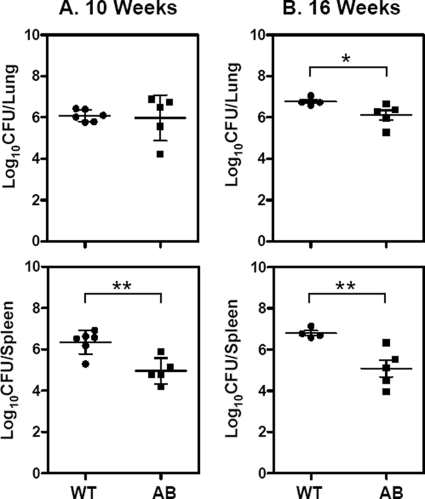
Loss of BfrA and BfrB results in a markedly reduced dissemination of M. tuberculosis to spleen.
We evaluated the influence of the deletion of bfrA and bfrB on the ability of the pathogen to grow and cause disease in the host by using a guinea pig model of experimental tuberculosis. The guinea pig model of low-dose, airborne tuberculosis infection with virulent M. tuberculosis has been used most commonly to elucidate the events in the pathogenesis of pulmonary tuberculosis. When guinea pigs are infected with fewer than 10 CFU of virulent M. tuberculosis, it has been observed that the pathogen disseminates from lungs to the pulmonary lymph nodes via hematogenous spread within 10 to 12 days postinfection and appears in spleens ∼3 weeks postinfection (18, 19). Bacilli start reseeding in the lung by ∼4 weeks to form secondary granulomas.
Guinea pigs were infected with H37Rv ΔbfrA ΔbfrB as well as the parental strain by using the aerosol route of infection. During the early stages of infection (up to 5 weeks), no significant differences were observed in the bacillary load in the lung and spleen of animals infected with H37Rv ΔbfrA ΔbfrB and the parental strain (data not shown). However, when the animals were euthanized at 10 weeks postinfection, a significant reduction in the spleen bacillary load was observed in the case of the animals infected with H37Rv ΔbfrA ΔbfrB compared to that of the animals infected with the parental strain (1.40 log10 fewer bacilli; P < 0.01) (Fig. 4A). At this time point, we did not find any difference in the bacillary load in lungs of animals belonging to either group. However, when the postchallenge period was extended to 16 weeks, a significant reduction in the lung bacillary load was observed in the case of animals infected with H37Rv ΔbfrA ΔbfrB compared to that of the animals infected with the parental strain (0.7 log10 fewer bacilli; P < 0.05) (Fig. 4B). At this time point, the H37Rv ΔbfrA ΔbfrB-infected guinea pigs exhibited a further substantial reduction in the spleen bacillary load (52-fold fewer bacilli) compared to that of the animals infected with the parental M. tuberculosis strain (Fig. 4B). There was no significant difference in the bacillary load in lung and spleen of guinea pigs infected with either H37Rv ΔbfrA or H37Rv ΔbfrB or their respective complemented strains at various time points during the study, indicating that the iron storage function in the absence of either BfrA or BfrB is compensated for by the other partner (data not shown). However, the marked reduction of spleen and lung bacillary load in the guinea pigs infected with H37Rv ΔbfrA ΔbfrB suggests that both BfrA and BfrB together play a crucial role in hematogenous spread and pathogenesis in the host.
Fig 4.
Influence of disruption of bfrA and bfrB genes of M. tuberculosis on the growth of the pathogen in guinea pigs. The figure depicts the bacillary load in the lungs and spleen of guinea pigs (n = 6) infected with M. tuberculosis (WT) and the H37Rv ΔbfrA ΔbfrB mutant (AB) at 10 (A) and 16 (B) weeks postinfection. Guinea pigs infected with H37Rv ΔbfrA ΔbfrB (AB) exhibited a significantly reduced bacillary load in spleen compared to animals infected with M. tuberculosis (WT). Each data point represents the log10 CFU value for an individual animal, and the bars depict means (± standard errors) for each group. Missing data points represent the animals that succumbed to disease before the time of euthanasia. *, P < 0.05; **, P < 0.01 (Student's t test).
BfrA and BfrB disruption results in a markedly reduced pathology.
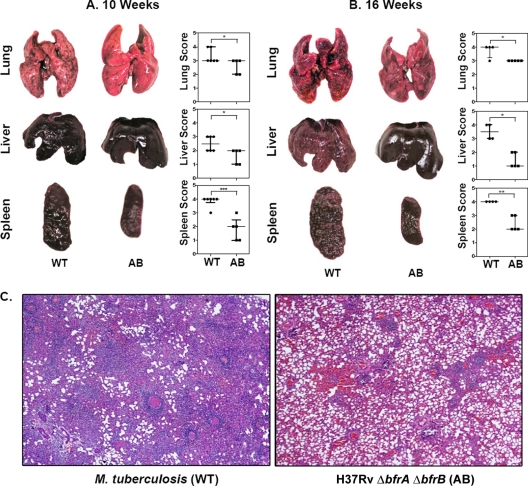
Commensurately to the bacillary burden at 5 weeks postinfection, we observed no significant differences in the pathological damage in lung, liver, and spleen of guinea pigs infected with the mutants or the parental strain (data not shown). At 10 weeks postinfection, guinea pigs infected with the wild-type strain exhibited the extensive involvement of lung and spleen with the presence of numerous large tubercles. In addition, numerous small-sized tubercles were observed in the liver. Noticeably, guinea pigs infected with the H37Rv ΔbfrA ΔbfrB strain exhibited a marked reduction in splenic pathology, with the presence of only a few small tubercles (P < 0.001). Moreover, lungs and liver also exhibited a moderately reduced pathology compared to that of guinea pigs infected with the parental strain (Fig. 5A). The gross pathological observations were further substantiated by histopathology. At 5 weeks postinfection, guinea pigs infected with M. tuberculosis and H37Rv ΔbfrA ΔbfrB exhibited scattered areas of granulomatous inflammation encompassing ∼30% of the lung section, with no significant difference observed between these two groups. In the liver, only a mild granulomatous inflammation was observed in these groups (5 to 10%) (data not shown). Further, at 10 weeks postinfection, animals infected with the wild-type strain exhibited numerous coalescing granulomas, covering ∼40 to 50% of the area of the lung sections. However, by this time point, a significant reduction in pulmonary pathology was observed in the case of animals infected with H37Rv ΔbfrA ΔbfrB compared to the level for animals infected with the parental strain. Strikingly, lungs of H37Rv ΔbfrA ΔbfrB-infected animals exhibited only a mild inflammation with a substantial restoration of normal lung architecture (Fig. 5C). However, in liver, animals infected with H37Rv ΔbfrA ΔbfrB as well as the parental strain exhibited only a mild hepatitis with granulomas covering ∼10 to 15% of liver sections.
Fig 5.
Influence of disruption of bfrA and bfrB genes of M. tuberculosis on gross pathological lesions and histopathological damage in organs of infected guinea pigs. The figure depicts representative photographs of gross pathological lesions in lung, liver, and spleen of guinea pigs (n = 6) infected with M. tuberculosis (WT) and H37Rv ΔbfrA ΔbfrB (AB) euthanized at 10 (A) and 16 (B) weeks postinfection. Guinea pigs infected with H37Rv ΔbfrAΔbfrB resulted in fewer and smaller lung, liver, and spleen lesions compared to animals infected with M. tuberculosis. Each data point represents the score of an individual animal, and the bars depict medians (± interquartile ranges) for each group. Missing data points represent the animals that succumbed to disease before the time of euthanasia. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test). (C) Influence of the disruption of bfrA and bfrB genes of M. tuberculosis on the histopathological damage to the organs of infected guinea pigs. The lung tissues were fixed in 10% buffered formalin and were embedded in paraffin. Subsequently, 5-μm-thick sections were cut and stained with hematoxylin and eosin (H&E) for histopathological examination. The figure depicts a representative photograph of the extent of pathological damage to animals (n = 6) infected with either M. tuberculosis (A) or H37Rv ΔbfrA ΔbfrB (B) at ×40 magnification at 10 weeks postinfection. H37Rv ΔbfrA ΔbfrB-infected animals showed reduced granulomatous infiltration, with only a few small and discrete granulomas, compared to that of M. tuberculosis-infected animals.
At 16 weeks postinfection, lung, liver, and spleen of guinea pigs infected with the wild-type strain exhibited heavy involvement with the presence of numerous large tubercles. In contrast, animals infected with H37Rv ΔbfrA ΔbfrB exhibited only the moderate involvement of lungs and liver (P < 0.05). However, the most striking differences were observed in the spleen of guinea pigs. The animals infected with H37Rv ΔbfrA ΔbfrB exhibited minimal involvement with only a few small visible tubercles, compared to splenomegaly in the case of animals infected with the parental strain (P < 0.01) (Fig. 5B).
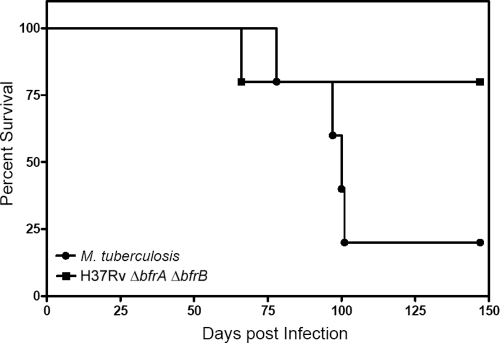
In addition to differences observed in the bacillary load and pathological damage, a marked difference was observed in the survival of animals. We observed that 5 out of 6 guinea pigs died in the case of infection with the wild-type strain, whereas only 1 out of 6 guinea pigs died in the case of infection with the double mutant up to 21 weeks postinfection, after which the experiment was terminated (Fig. 6).
Fig 6.
Influence of disruption of bfrA and bfrB genes of M. tuberculosis on the survival of guinea pigs postinfection. Guinea pigs infected with M. tuberculosis H37Rv/pJV53 or H37Rv ΔbfrA ΔbfrB through the aerosol route were monitored for survival (n = 6). The experiment was started with a bacillary load of 5 to 10 CFU in lungs at 1 day postinfection. Bacillary burden in lungs and spleen was monitored at 5, 10, and 16 weeks postinfection. Remaining guinea pigs (n = 6) were monitored for survival up to 21 weeks postinfection.
DISCUSSION
Iron availability during M. tuberculosis infection is regarded as a critical factor that influences the progression and magnitude of disease. However, to develop strategies to interfere with the iron metabolism of this pathogen, a better understanding of the importance of iron in the physiology of M. tuberculosis and in the growth of the pathogen in the host is required at the genetic level. In this study, we have generated mutants of M. tuberculosis lacking bfrA (Rv1876) and bfrB (Rv3841) encoding the iron storage proteins. We show that the mutant of M. tuberculosis, H37Rv ΔbfrA ΔbfrB, which lacks the function of both bfrA and bfrB, has significantly reduced growth under iron-deprived conditions, is markedly vulnerable to oxidative stress, and exhibits the attenuation of growth in human macrophages. Moreover, reduced bacillary load in lung and spleen of H37Rv ΔbfrA ΔbfrB-infected guinea pigs, resulting in a significant reduction in pathology, clearly implies that these proteins play a crucial role in the pathogenesis of M. tuberculosis.
Mycobacteria are continuously exposed to oxidative stress generated by the activated macrophages that they inhabit (4). When we evaluated the ability of M. tuberculosis mutants lacking the function of bfrA and bfrB to resist oxidative stress, we observed that simultaneous mutations in bfrA and bfrB in M. tuberculosis (H37Rv ΔbfrA ΔbfrB) tremendously reduced its ability to withstand oxidative stress, implying the role of these iron storage proteins in restricting oxidative damage. BfrA and BfrB are iron storage proteins that reduce the freely available ferrous form, thereby limiting the production of oxygen radicals by Fenton reaction and protecting the bacteria from the harmful oxidative damage. When the M. tuberculosis mutants lacking the function of a single Bfr protein (BfrA or BfrB) were evaluated for their ability to withstand oxidative stress, it was observed that these mutants also exhibited a moderate ability to withstand the oxidative damage; however, the magnitude of influence was less than that of the double mutant. Our observations thus clearly demonstrate the importance of these iron storage proteins in the mycobacterial response to oxidative stress and are in agreement with previous studies which have shown the increased sensitivity of the ideR mutant of M. tuberculosis to both H2O2 and plumbagin. The expression of bfrA and bfrB is known to be regulated by the binding of Fe2+-activated IdeR to operator sites upstream of the bfrA and bfrB transcriptional start point (TSP). The deletion of these IdeR binding sites or the inactivation of ideR has been shown to result in the enhanced sensitivity of these mutants to oxidative agents such as H2O2 and plumbagin (10, 22). Similar studies in the case of Pseudomonas aeruginosa and Brucella abortus have shown that BfrA protects the organism against the onslaught of hydrogen peroxide by sequestering the excess free iron from the system and thus preventing iron-induced oxidative damage (2, 16). Bfr- and Ftn-deficient mutants of Salmonella enterica serovar Typhimurium and Campylobacter jejuni, respectively, have been shown to exhibit a significantly higher sensitivity to H2O2 and paraquat than the parental strain (26, 28). These findings demonstrate that bacterioferritins and ferritins make a significant contribution to iron storage as well as to protection from intracellular iron overload, which is responsible for iron-mediated oxidative stress. Interestingly, the ferritin mutants of Helicobacter pylori and Escherichia coli exhibited no change in their ability to withstand oxidative stress mediated by paraquat and H2O2, respectively (1, 29). These studies demonstrate that bacterioferritins and ferritins may have diverse roles in different bacterial species.
Our studies of human macrophage cells further substantiate the role of BfrA and BfrB in the survival of M. tuberculosis against host-induced stress. The growth kinetics of H37Rv ΔbfrA ΔbfrB up to 8 days postinfection revealed severe attenuation of the apparent growth of the mutant compared to that of the parental strain. Earlier it was reported that the downregulation of transferrin receptors in the activated macrophages limits the availability of iron inside the macrophages, leading to the impairment of M. tuberculosis growth in these iron-restrictive environments (5). This triggers the release of iron-sequestering molecules by the pathogen into the macrophage cytoplasm for acquiring iron along with a consequent increase in the expression of iron storage proteins. Since BfrA and BfrB are the only two iron storage proteins in M. tuberculosis, their absence perhaps results in the insufficient supply of iron to the cell, leading to the concomitant attenuation of growth. The attenuation of M. tuberculosis growth in macrophages in response to the deficiency of iron in host macrophages has been reported earlier in several studies (9, 11).
The most substantial evidence for the role of bacterioferritins in M. tuberculosis pathogenesis emerges from our guinea pig studies, wherein at 10 weeks postinfection a marked reduction was observed in the CFU of H37Rv ΔbfrA ΔbfrB in the spleen of guinea pigs compared to that of the parental strain (25-fold reduction). The bacillary load of H37Rv ΔbfrA ΔbfrB compared to that of the parental strain was further reduced when the disease was allowed to progress up to 16 weeks of infection. At this time point, a 52-fold lower bacillary load was observed in the spleen along with a 5-fold reduction in the lung of guinea pigs infected with the H37Rv ΔbfrA ΔbfrB strain compared to that of infection with the parental strain. Thus, we show that BfrA and BfrB together are required for the survival and pathogenesis of M. tuberculosis in the guinea pig model, as measured by bacillary load in lung and spleen and the pathological insult to the organs.
In conclusion, this study demonstrates that BfrA and BfrB proteins play a crucial role in protecting the pathogen against oxidative stress encountered during infection. In addition, BfrA and BfrB proteins are important for the survival and hematogenous spread of the pathogen. We have reported earlier the crystal structures of BfrA and BfrB of M. tuberculosis. M. tuberculosis BfrA and BfrB, unlike most other ferritins, especially those of humans, have extended C-terminal regions, and BfrA contains an altered heme pocket unique to M. tuberculosis (12). Our crystallographic data as well as biochemical studies implicate this extended C-terminal region in the iron entry from the 3-fold channels to the ferroxidase center and in making iron more readily accessible for the oxidation (14). The unique structures of these iron storage proteins, which are not found in ferritins, and the essential requirement of BfrA and BfrB proteins in the survival of the pathogen inside the host, as shown in this study, clearly establish these proteins as attractive drug targets for the development of new therapeutic molecules against mycobacterial infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Graham F. Hatfull and Julia C van Kessel, University of Pittsburgh, PA, for providing the reagents for mycobacterial recombineering method; Bappaditya Dey and Ruchi Jain for their help in designing the guinea pig experiments; Ruchi Jain and Priyanka Chauhan for critical proofreading of the manuscript; and Rakesh Gupta, Vibha Gupta, and Vikram Saini for useful discussions. Technical assistance provided by Bahadur Singh, Sandeep Kumar, and Priti Singh is highly acknowledged.
P.V.R. is thankful to DBT, India, for a research fellowship. This work was supported by a research grant from the Department of Biotechnology, Government of India.
P.V.R., R.V.P., A.K., and A.K.T. conceived and designed the experiments. P.V.R., R.V.P., and A.K. conducted the experiments and analyzed the data. P.V.R. and A.K.T. wrote the manuscript. A.K.T. provided overall supervision throughout the study.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abdul-Tehrani H, et al. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almirón MA, Ugalde RA. 2010. Iron homeostasis in Brucella abortus: the role of bacterioferritin. J. Microbiol. 48:668–673 [DOI] [PubMed] [Google Scholar]
- 3. Andrews SC. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281–351 [DOI] [PubMed] [Google Scholar]
- 3a. Bardarov S, et al. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 4. Boelaert JR, Vandecasteele SJ, Appelberg R, Gordeuk VR. 2007. The effect of the host's iron status on tuberculosis. J. Infect. Dis. 195:1745–1753 [DOI] [PubMed] [Google Scholar]
- 5. Byrd TF, Horwitz MA. 1993. Regulation of transferrin receptor expression and ferritin content in human mononuclear phagocytes. Coordinate upregulation by iron transferrin and downregulation by interferon gamma. J. Clin. Investig. 91:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrondo MA. 2003. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22:1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 7a. Das Gupta SK, et al. 1993. Cloning and assessment of mycobacterial promoters by using a plasmid shuttle vector. J. Bacteriol. 175:5186–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Voss JJ, Rutter K, Schroeder BG, Barry CE., III 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Douvas GS, May MH, Crowle AJ. 1993. Transferrin, iron, and serum lipids enhance or inhibit Mycobacterium avium replication in human macrophages. J. Infect. Dis. 167:857–864 [DOI] [PubMed] [Google Scholar]
- 10. Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. 2001. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42:851–865 [DOI] [PubMed] [Google Scholar]
- 11. Gomes MS, Florido M, Pais TF, Appelberg R. 1999. Improved clearance of Mycobacterium avium upon disruption of the inducible nitric oxide synthase gene. J. Immunol. 162:6734–6739 [PubMed] [Google Scholar]
- 12. Gupta V, Gupta RK, Khare G, Salunke DM, Tyagi AK. 2009. Crystal structure of BfrA from Mycobacterium tuberculosis: incorporation of selenomethionine results in cleavage and demetallation of haem. PLoS One 4:e8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jain R, et al. 2008. Enhanced and enduring protection against tuberculosis by recombinant BCG-Ag85C and its association with modulation of cytokine profile in lung. PLoS One 3:e3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khare G, et al. 2011. Ferritin structure from Mycobacterium tuberculosis: comparative study with homologues identifies extended C-terminus involved in ferroxidase activity. PLoS One 6:e18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lounis N, Truffot-Pernot C, Grosset J, Gordeuk VR, Boelaert JR. 2001. Iron and Mycobacterium tuberculosis infection. J. Clin. Virol. 20:123–126 [DOI] [PubMed] [Google Scholar]
- 16. Ma JF, et al. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majumdar SD, et al. 2010. Co-expression of DevR and DevR(N)-Aph proteins is associated with hypoxic adaptation defect and virulence attenuation of Mycobacterium tuberculosis. PLoS One 5:e9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McMurray DN. 2001. Disease model: pulmonary tuberculosis. Trends Mol. Med. 7:135–137 [DOI] [PubMed] [Google Scholar]
- 19. McMurray DN. 1994. Guinea pig model of tuberculosis, p. 135–147. In Bloom BR. (ed.), Tuberculosis: pathogenesis, protection, and control. ASM Press, Washington, DC [Google Scholar]
- 20. Park HD, et al. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodriguez GM, Smith I. 2003. Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol. Microbiol. 47:1485–1494 [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosenkrands I, et al. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol. 184:3485–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherman DR, et al. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. U. S. A. 98:7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147–152 [DOI] [PubMed] [Google Scholar]
- 26. Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63:1495–1507 [DOI] [PubMed] [Google Scholar]
- 27. Voskuil MI, Visconti KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinburg) 84:218–227 [DOI] [PubMed] [Google Scholar]
- 28. Wai SN, Nakayama K, Umene K, Moriya T, Amako K. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 20:1127–1134 [DOI] [PubMed] [Google Scholar]
- 29. Waidner B, et al. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.