Abstract
Vibrio cholerae continues to pose a health threat in many developing nations and regions of the world struck by natural disasters. It is a pathogen that rapidly adapts to aquatic environments and the human small intestine. Small regulatory RNAs (sRNAs) may contribute to this adaptability. Specifically, the mannitol operon sRNA (MtlS sRNA; previously designated the IGR7 sRNA) is transcribed antisense to the 5′ untranslated region of the mtl operon, encoding the mannitol-specific phosphotransferase system. Mannitol is a six-carbon sugar alcohol that accumulates in the human small intestine, the primary site of V. cholerae colonization. To better understand the V. cholerae mtl operon at a molecular level, we investigated mtlA expression in the presence of various carbon sources and the role of the MtlS sRNA. We observed that MtlA protein is present only in cells grown on mannitol sugar, whereas MtlS sRNA is expressed during growth on all sugars other than mannitol. In contrast, mtlA mRNA is expressed in similar amounts regardless of the carbon source used for bacterial growth. These observations suggest that the regulation of MtlA protein expression is a posttranscriptional event. We further demonstrate that MtlS sRNA overexpression repressed MtlA synthesis without affecting the stability of the messenger and that this process is largely independent of Hfq. We propose a model in which, when carbon sources other than mannitol are present, MtlS sRNA is transcribed, base pairs with the 5′ untranslated region of the mtlA mRNA, occluding the ribosome binding site, and inhibits the synthesis of the mannitol-specific phosphotransferase system.
INTRODUCTION
Cholera continues to contribute toward a large proportion of diarrhea-related mortality (40). Vibrio cholerae, the causative agent of cholera, is a natural member of aquatic ecosystems, and pathogenic strains are found in both freshwater and estuaries in areas where cholera is endemic (5, 42). Cholera is caused by ingestion of this Gram-negative bacterium in contaminated food or water; upon colonization of the human small intestine, secretion of cholera toxin into the intestinal lumen causes the profuse watery diarrhea characteristic of cholera.
Aquatic reservoirs and the human small intestine represent two distinct environments, containing different nutrients (44, 51). Within both environments, fluxes in the carbon source may occur frequently (15, 47, 48). Genomewide transcriptional changes take place as the bacteria transition between the aquatic reservoir and the host, suggesting that different sets of gene products allow V. cholerae to persist and thrive in these two disparate environments (32, 36, 44, 52). This characteristic is likely key to the fitness of V. cholerae as a facultative pathogen.
Previous studies with V. cholerae suggest that during both colonization of the host small intestine and survival in their natural aquatic habitats, the bacteria are actively scavenging carbohydrates for energy metabolism (17, 18, 32, 36, 52). Upon infection in the rabbit ileal loop model of cholera, the V. cholerae genes encoding enzyme I (EI; VC0965) and histidine protein (HPr; VC0966) are upregulated (52); two separate studies also identified EI as necessary for the colonization of the mouse small intestine (16, 32). V. cholerae EI is also a regulator of biofilm-associated growth (18); these bacteria are known to form biofilms on the surfaces of plants, insects, and plankton found in their aquatic reservoirs. EI and HPr are highly conserved cytoplasmic components of the phosphoenolpyruvate (PEP)-carbohydrate phosphotransferase system (PTS), which catalyzes the uptake and concomitant phosphorylation of numerous carbohydrates in both Gram-negative and Gram-positive bacteria (6). Carbohydrate specificity resides in enzyme II (EII), which consists of various combinations of hydrophilic domains (domains A and B) and hydrophobic integral membrane domains (domains C and D). In the PTS multienzyme cascade, a phosphate group is transferred sequentially from PEP to EI, to HPr, to the specific EII, and finally to the carbohydrate as it is transported across the membrane.
Small regulatory RNAs (sRNAs) are often employed in bacterial mechanisms of stress adaptation, and there are precedents in V. cholerae and related bacteria for regulation by sRNAs of the expression of genes involved in carbon metabolism (11, 26, 29, 43, 50). For example, in Escherichia coli, a trans-acting sRNA regulates the synthesis of PtsG, the glucose-specific EIIBC of the PTS. Following sugar uptake by the PTS, metabolic bottlenecks may cause the accumulation of intracellular sugar phosphates, which can be toxic for a cell (37). The SgrS sRNA is induced upon such phosphosugar stress and inhibits the translation of ptsG mRNA by base pairing with the transcript through partial complementarity and the aid of the RNA chaperone Hfq (50). RNase E, a major endoribonuclease, forms a multiprotein complex called the RNA degradosome; RNase E can also bind to Hfq (39). These observations collectively suggest that upon pairing with ptsG mRNA, the SgrS sRNA and Hfq ultimately target the message for rapid degradation by RNase E, decreasing glucose transport until the phosphosugar stress is alleviated.
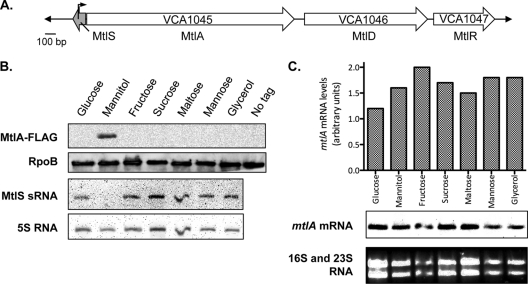
Our lab recently identified the IGR7 sRNA in V. cholerae through direct cloning and massively parallel sequencing (25). This 120-nucleotide sRNA is transcribed antisense to the 5′ untranslated region (UTR) of the mtl operon, encoding the mannitol-specific PTS (16, 22) (Fig. 1A). The 3.9-kb operon comprises three genes, organized as mtlADR, encoding a mannitol-specific enzyme IIABC (EIIMtl) component (MtlA; VCA1045), a mannitol-1-phosphate dehydrogenase (MtlD; VCA1046), and a mannitol operon transcriptional regulator (MtlR; VCA1047) (22). We observed that IGR7 sRNA expression was largely dependent on the carbon source in the growth medium and that the sequence of this sRNA is highly conserved in all Vibrionaceae; in all these species, the sRNA gene is present upstream of and opposite the mtlA homolog (25). Thus, we have renamed this sRNA the MtlS sRNA, for mannitol operon sRNA. Our preliminary results suggest that, in contrast to the SgrS sRNA, which regulates the glucose-specific PTS in trans, the MtlS sRNA represses mtlA expression in cis (25).
Fig 1.
MtlS sRNA and MtlA protein are inversely related. (A) Genomic organization of genes encoding the V. cholerae mannitol-specific PTS components and the MtlS sRNA (shaded arrow). The transcription start site for mtlA is indicated by the arrow. (B) MtlA-FLAG protein and MtlS sRNA levels in V. cholerae mtlA-FLAG. Cell lysates and total RNA were prepared from a V. cholerae mtlA-FLAG strain grown to mid-exponential phase at 37°C in minimal medium supplemented with the indicated carbon source (0.4%). Cell lysates from equal numbers of cells were analyzed by an immunoblot assay using HRP-conjugated anti-FLAG and anti-RpoB antibodies. A cell lysate from wild-type V. cholerae (untagged mtlA) grown in mannitol medium was loaded in the “No tag” lane as a control. RNA samples (2 μg) were analyzed by Northern hybridization using DNA probes specific to MtlS sRNA and 5S RNA. The data are representative of at least three independent experiments. (C) mtlA-FLAG RNA levels in V. cholerae mtlA-FLAG. The same RNA samples used in the experiment for which results are shown in panel B (2 μg) were analyzed by Northern hybridization using a DNA probe specific to mtlA. 16S rRNA and 23S rRNA, stained with ethidium bromide, are shown as loading controls. The histogram above the blot shows the mean intensities of the mtlA bands normalized to the intensity of 16S rRNA; each bar corresponds to the band located directly below. The data are representative of at least three independent experiments.
A naturally occurring sugar alcohol found in many plants, mannitol is poorly absorbed by humans and may accumulate in the small intestine, the primary site of V. cholerae colonization (51). Little is known about the mtlA operon in V. cholerae (22). When the mtlA gene is knocked out or knocked down, V. cholerae is unable to grow on a medium with mannitol as the only carbon source (16, 25). In addition, mtlA and mtlD are downregulated 2.5- and 6.25-fold, respectively, upon biofilm formation, and a 2-fold increase in the expression of these genes has been observed during colonization in the rabbit ileal loop (36, 52). These studies suggest that differential regulation of the PTS, and specifically the mtl operon, may be important to the life cycle of V. cholerae.
To better understand the regulation of the V. cholerae mtl operon at a molecular level, we investigated mtlA expression in the presence of various carbon sources and the role of the MtlS sRNA. Here we show that the MtlS sRNA regulates mtlA expression posttranscriptionally. Because the MtlS sRNA does not appear to affect the stability of mtlA mRNA, we propose a model whereby, in the presence of nonmannitol carbon sources, mtlS expression is activated and the sRNA anneals to the 5′ UTR of mtlA, preventing the translation of the PTS protein.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Table 1. The strains used in this work were El Tor strain N16961 ΔtcpA and derivatives (Table 1). The tcpA mutant is highly attenuated for virulence (14) and was used for safety purposes. Importantly, this mutant strain produced phenotypes identical to those of the wild-type strain N16961 (3) with regard to the experiments discussed in this report; hereafter, “wild type” refers to the N16961 ΔtcpA strain. Plasmids with oriR6K were propagated in Escherichia coli DH5αλpir; all other plasmids were propagated in E. coli DH5α. Plasmids for generating mutations in V. cholerae were constructed in the allelic exchange vector pCVD442 (8, 49). All mutations were constructed by splicing by overlap extension (SOE)-PCR using the primers listed in Table 2 (45). Briefly, DNA fragments of approximately 600 bp upstream and downstream of each mutation were amplified by PCR from V. cholerae N16961 genomic DNA, annealed together by complementary sequences in the R1 and F2 primers, and then PCR amplified with the F1 and R2 primers. The final PCR product was ligated into pCVD442 using the SacI and SphI restriction sites. Plasmids were conjugated into V. cholerae N16961 or V. cholerae N16961 ΔtcpA from E. coli SM10λpir as described previously (23). After one passage in LB broth with streptomycin, sucrose-resistant colonies were selected and were subsequently screened for the desired mutation by PCR with the F0 and R0 primers. All strains were grown at 37°C in aerated LB broth or M9 minimal medium supplemented with 0.1% trace metals (5% MgSO4, 0.5% MnCl2 · 4H2O, 0.5% FeCl3, 0.4% nitrilotriacetic acid) and 0.4% carbon source. When necessary, cultures were supplemented with 0.02% arabinose (Gold Biotechnology) to induce the expression of genes inserted into pJML01. Antibiotics were used at the following concentrations: streptomycin, 100 μg/ml; ampicillin, 100 μg/ml.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| N16961 | El Tor, Inaba; Smr | 3 |
| JL1 | N16961 ΔtcpA; Smr | This study |
| JL2 | N16961 ΔtcpA mtlA-FLAG; Smr | This study |
| JL54 | N16961 ΔtcpA mtlA-FLAG; Δhfq Smr | This study |
| JL55 | N16961 ΔtcpA mtlA-FLAG; ΔmtlR Smr | This study |
| JL24 | N16961 ΔtcpA ΔmtlR; Smr | This study |
| JL17 | N16961 ΔtcpA mtlA-FLAG; pJML01 Smr Apr | This study |
| JL18 | N16961 ΔtcpA mtlA-FLAG; pMtlS Smr Apr | This study |
| JL19 | N16961 ΔtcpA mtlA-FLAG; pAS-MtlS Smr Apr | This study |
| JL78 | N16961 ΔtcpA mtlA-FLAG; pJBA111 Smr Apr | This study |
| E. coli | ||
| DH5α | F− Δ(lacZYA-argF)U169 recA1 end A1 hsdR17 supE44 thi-1 gyrA96 relA1 | Laboratory strain |
| DH5αλpir | F− Δ(lacZYA-argF)U169 recA1 end A1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | Laboratory strain |
| SM10λpir | thi recA thr leu tonA lacY supE RP4-2-Tc::Mu λ::pir | Laboratory strain |
| Plasmids | ||
| pCVD442 | oriR6K mobRP4 sacB; Apr | 8 |
| pHT3 | pCVD442 ΔtcpA10; Apr | 49 |
| pAC3340 | pCVD442 mtlA::FLAG | This study |
| pJL36 | pCVD442 Δhfq; Apr | This study |
| pAC3336 | pCVD442 ΔmtlR; Apr | This study |
| pJML01 | pBAD24 derivative with +1 start of transcription after NheI site; Apr | 25 |
| pMtlS | pJML01::mtlS; Apr | 25 |
| pAS-MtlS | pJML01::antisense-mtlS; Apr | 25 |
| pJBA111 | plac-GFP(LVA) | 2 |
Table 2.
Primers and probes used in this study
| Function and primer or probe | Sequencea |
|---|---|
| Northern blotting | |
| b-MtlS | 5′-Biotin-CCGAACCGTTACTACGATTAAATTCAAACGGAACATCC-3′ |
| b-5S | 5′-Biotin-CTGTTTCGTTTCACTTCTGAGTTCGGGATGGAA-3′ |
| IR800-5S | 5′-IRD800-CTGTTTCGTTTCACTTCTGAGTTCGGGATGGAA-3′ |
| b-mtlA | 5′-Biotin-GCAGTAATAAAGCCCCACGCAATAAAAGCGCCAATATTC-3′ |
| T7 mtlSfor | 5′-GGATCCTAATACGACTCACTATAGGGAAAAACCCGTTGGTGATTCCATTCG-3′ |
| mtlSrev | 5′-TCCCCCGTTGGATGTTCCG-3′ |
| T7mtlAfor | 5′-GGATCCTAATACGACTCACTATAGGGTCACGCCCATCGTGGTGATG-3′ |
| mtlArev | 5′-GGTTATGAAGAATATTGGCGC-3′ |
| T7 rpsLfor | 5′-GGATCCTAATACGACTCACTATAGGGTTGTAGGTTGTGACCTTCACCACC-3′ |
| rpsLrev | 5′-GGTTCGTAAGCCACGTGCTAAGC-3′ |
| Construction of V. cholerae mtlA-FLAG insertion | |
| F1 | 5′-GGCGCATGCGTAACGAATATGGCCATCAACTCACTCC-3′ |
| R1 | 5′-GCAAAAACGTTTACTTGTCATCGTCGTCCTTGTAGTCTGCCGCTTGGCTGGTGGCC-3′ |
| F2 | 5′-AAGCGGCAGACTACAAGGACGACGATGACAAGTAAACGTTTTTGCTCCTGAGGCAAAACG-3′ |
| R2 | 5′-GGCGAGCTCTTGCAACGGTGGCACAATGCGATC-3′ |
| F0 | 5′-CATTACCAATGCCCTCGATGAC-3′ |
| R0 | 5′-GTTTCTCTCACCGCCTGAGG-3′ |
| Construction of V. choleraehfq deletion | |
| F1 | 5′-GGCGCATGCGCAGGCGTTACACGATCAGTTAC-3′ |
| R1 | 5′-CGATCCAGAAATGGGTCTTGTAGAGATTGCC-3′ |
| F2 | 5′-CCCATTTCTGGATCGTCCAGCAGAGAAGTCT-3′ |
| R2 | 5′-GCGAGCTCTTACGCAAAGTAGGATCGAG-3′ |
| F0 | 5′-CCAACGAAATAAGCTGTGTATAATGTG-3′ |
| R0 | 5′-CTGCCTGTTACCACATGCAGTG-3′ |
| Construction of V. choleraemtlR deletion | |
| F1 | 5′-GGCGCATGCAAAGGTCACCGTACCATCCGTG-3′ |
| R1 | 5′-CACCTAAACGTTACATTTTAAGACTACCGATAACCGCATTTTTTC-3′ |
| F2 | 5′-GTAGTCTTAAAATGTAACGTTTAGGTGCACGCCGATTTC-3′ |
| R2 | 5′-GGCGAGCTCGTTGAGCCAGCTCGGACAAATG-3′ |
| F0 | 5′-AACCGATAACCTGATGGCGTTTGTC-3′ |
| R0 | 5′-ACACAAAGGTGCGAATATCGCCG-3′ |
Restriction sites are underlined; the FLAG tag sequence is italicized.
Western blotting.
Total-cell lysates (∼107 cells) were mixed with 5× sample buffer (250 mM Tris-Cl [pH 6.8], 10% sodium dodecyl sulfate [SDS], 50% glycerol, 10% β-mercaptoethanol, 0.5% bromophenol blue or orange G), heated to 95°C for 10 min, and separated on a 10% SDS-containing polyacrylamide gel. Proteins were subsequently transferred to either a nylon (Amersham) or a nitrocellulose (Licor) membrane at 4°C using a wet-transfer apparatus (Bio-Rad) and glycine buffer (25 mM Tris, 192 mM glycine, 20% methanol). For chemiluminescent detection, a horseradish peroxidase (HRP)-conjugated anti-FLAG primary antibody (Abcam) was utilized at a dilution of 1:5,000, and an anti-RpoB primary antibody (Abcam) was utilized at a dilution of 1:10,000. An HRP-conjugated rabbit anti-mouse secondary antibody (Abcam) was applied at a dilution of 1:5,000. Immunodetection was achieved via use of chemiluminescent detection reagents (Thermo Scientific). The densitometry of bands was calculated using ImageJ (NIH). For infrared (IR) fluorescence detection, anti-FLAG and anti-green fluorescent protein (GFP) primary antibodies (Abcam) were utilized at a dilution of 1:5,000, and the anti-RpoB primary antibody was used as described above. IR680-conjugated goat anti-rabbit and IR800-conjugated goat anti-mouse secondary antibodies (Licor) were each applied at a dilution of 1:10,000. IR fluorescence was detected using an Odyssey Imager (Licor). The densitometry of bands was calculated using Odyssey Application software (Licor). All results were graphed using Prism (GraphPad).
Northern blot analysis.
In all cases, total RNA was isolated by acid-phenol extraction, as described previously (30). For sRNA analysis, total RNA in Loading Buffer II (Ambion) was run on a 10% denaturing polyacrylamide gel and was transferred to a nylon membrane (Amersham) in 1× Tris-borate-EDTA (TBE) by using a wet-transfer apparatus (Bio-Rad) according to the manufacturer's instructions. For mRNA analysis, total RNA in 6× loading buffer (0.4% bromophenol blue, 0.4% xylene cyanol, 50% glycerol) was separated on a 0.8% agarose gel containing ethidium bromide (0.25 μg/ml). The RNA was transferred to a nylon membrane (Amersham) by capillary action (Ambion NorthernMax kit). For all blotting, RNA was subjected to UV cross-linking after transfer and was prehybridized in ULTRAhyb-Oligo (Ambion) according to the manufacturer's instructions. In some cases, hybridizations were performed using DNA probes (Table 2) end labeled with either biotin or an IR dye (Integrated DNA Technologies). Otherwise, Northern blotting was performed with RNA probes transcribed from PCR-derived templates (Table 2) with T7 promoters by using biotin-16-UTP and T7 RNA polymerase (Promega) according to the manufacturer's instructions. All blots were washed and imaged as described in the BrightStar BioDetect kit (Ambion) (for DNA probe detection with streptavidin-conjugated alkaline phosphatase) or the Odyssey Northern blot analysis protocol (Licor) (for RNA probe detection with IR680-labeled streptavidin).
RESULTS
The MtlS sRNA and the MtlA protein are inversely related.
Our previous results suggested that mtlS and mtlA expression exhibited an inverse relationship in the three carbon sources tested: glucose, mannitol, and glycerol (25). To expand on our initial observations, we examined MtlS sRNA and MtlA levels in V. cholerae grown with either PTS sugars (glucose, mannitol, fructose, sucrose, or mannose) or non-PTS sugars (maltose and glycerol) as the sole carbon source. Northern and Western blot analyses indicated that MtlA is present only in cells grown in mannitol sugar, whereas the MtlS sRNA is expressed during growth in all sugars other than mannitol (Fig. 1B). These results confirm that an inverse relationship exists between MtlS sRNA and MtlA protein expression.
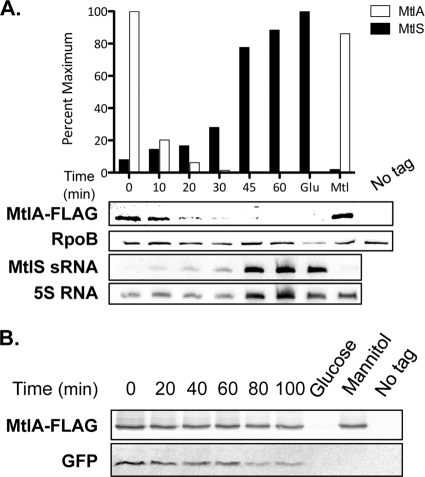
To further explore this inverse relationship, we analyzed MtlS sRNA and MtlA levels as bacteria were shifted from a mannitol to a glucose medium. V. cholerae was grown in mannitol medium, a condition under which MtlA protein levels are high and MtlS sRNA expression is low. Once the cells reached mid-exponential phase, they were collected, resuspended in fresh glucose medium, and allowed to continue growing for an additional 60 min. RNA and protein samples were harvested from cells at 0, 10, 20, 30, 45, and 60 min after the switch of the carbon source. Total RNA was analyzed by Northern hybridization using a probe specific for MtlS sRNA, and total protein was analyzed by immunoblotting for FLAG epitope-tagged MtlA.
Prior to the shift in the carbon source (0 min), MtlS sRNA was undetectable while MtlA protein was abundant (Fig. 2A). Within 10 min after switching of the carbon source, MtlS sRNA was detected and the amount of MtlA protein was decreased by ∼80%. This trend continued over the course of the experiment; the transporter levels were rapidly depleted, and MtlA was not detectable after ∼45 min. These results further support an inverse relationship between the sRNA and the transporter protein and also suggest that the expression of the sRNA may have a direct effect on MtlA protein expression. We noted, however, that compared to the disappearance of MtlA, the increase in MtlS sRNA levels was more gradual over the course of the experiment, reaching half-maximal levels only after ∼40 min had elapsed.
Fig 2.
MtlA is rapidly depleted in the absence of mannitol. (A) MtlS sRNA induction and MtlA repression in glucose medium. A V. cholerae mtlA-FLAG strain was grown to mid-exponential phase in mannitol medium at 37°C, at which time cells were harvested, moved to glucose medium, and grown at 37°C. Cell lysates and total RNA were isolated from the culture at 0, 10, 20, 30, 45, and 60 min after the switch of the carbon source. Cell lysates from equal numbers of cells were analyzed by immunoblot assays using HRP-conjugated anti-FLAG and anti-RpoB antibodies. RNA samples (2 μg) were analyzed by Northern hybridization using DNA probes specific to MtlS sRNA and 5S RNA. The histogram above the blot shows the mean intensities of the MtlA-FLAG and MtlS sRNA bands, normalized to RpoB and 5S RNA intensities, respectively. Each bar corresponds to the band located directly below. MtlA levels are expressed relative to those at 0 min. MtlS sRNA levels are expressed relative to those in the glucose control sample. (B) MtlA is a stable protein. V. cholerae mtlA-FLAG GFP-LVA strains were grown to mid-exponential phase at 37°C in mannitol medium. Cell lysates were collected 0, 20, 40, 60, 80, and 100 min after treatment with 100 μg/ml chloramphenicol and were analyzed by immunoblot assays using anti-FLAG and anti-GFP antibodies. Cell lysates and total RNA from V. cholerae mtlA-FLAG grown to mid-exponential phase in glucose medium or mannitol medium were used as controls, as was a cell lysate from wild-type V. cholerae (untagged mtlA) grown in mannitol medium. The data are representative of at least three independent experiments.
We were intrigued by the rapid disappearance of the MtlA protein and questioned whether the transporter had a short half-life or whether the removal of mannitol from the growth medium promoted active MtlA proteolysis. To distinguish between these two possibilities, we grew V. cholerae to mid-exponential phase and treated the cells with chloramphenicol to inhibit protein synthesis (51). We observed that under these conditions, MtlA was largely stable over the course of the experiment, for as long as 100 min after the addition of the translational inhibitor; we also analyzed levels of a GFP variant expressed in the same strain and observed that the levels of the control protein did decrease over time, as expected (Fig. 2B) (2). These results suggest that the shift in the carbon source from a mannitol to a glucose medium not only activates MtlS sRNA expression but also triggers active degradation of existing MtlA protein.
MtlS sRNA expressed in trans represses MtlA synthesis.
We observed previously that the ability of V. cholerae to grow in mannitol medium is diminished when the MtlS sRNA is overexpressed (25) (see Fig. S1 in the supplemental material). To directly test whether this growth impairment is due to effects on MtlA protein expression, we analyzed MtlA levels in cells overexpressing MtlS sRNA. We placed mtlS expression under the control of an arabinose-inducible promoter in pMtlS and followed steady-state levels of MtlA upon MtlS sRNA induction (13). V. cholerae carrying this plasmid was grown in mannitol medium, a condition under which MtlA levels are high, to mid-exponential phase before the addition of arabinose to induce mtlS expression. Total protein was collected from cells at 0, 30, and 60 min after the addition of the inducer. MtlA protein levels were then analyzed by immunoblotting.
Prior to induction, MtlA levels were high, as expected. Within 30 min after the induction of MtlS sRNA expression, MtlA levels were significantly reduced (Fig. 3A). By 60 min after induction, no MtlA protein was detected. Cells carrying the cloning vector alone maintained constant levels of MtlA protein during the same period (Fig. 3A). These results support a direct role for the MtlS sRNA in repressing MtlA protein expression.
Fig 3.
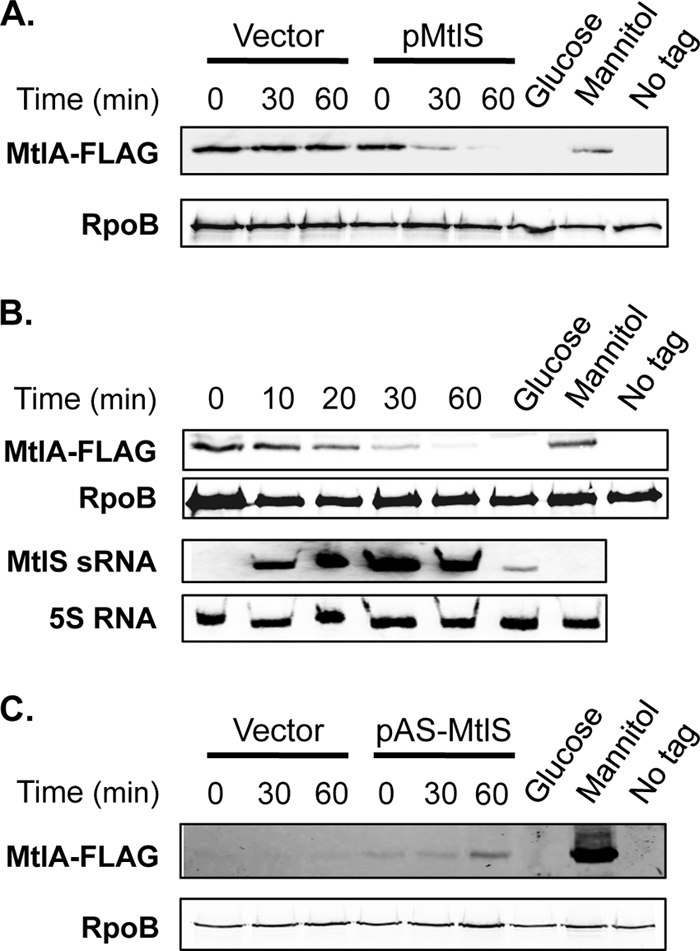
MtlS sRNA induction reduces MtlA protein levels. Strains of V. cholerae mtlA-FLAG containing an arabinose-inducible copy of MtlS sRNA (A and B), an RNA complementary to MtlS sRNA (AS-MtlS) (C), or an empty-vector control (A and C) were grown to mid-exponential phase in mannitol medium (A and B) or glycerol medium (C) and were stimulated with arabinose. Cell lysates were collected at the indicated time points (from the start of induction), and equal amounts of cells were subjected to immunoblot analysis using anti-FLAG and anti-RpoB antibodies. For panel B, total-RNA samples (2 μg) were collected at the indicated time points and were analyzed by Northern hybridization using an RNA probe specific to MtlS sRNA and a 5S RNA-specific DNA probe. For all panels, cell lysates and total RNA from V. cholerae mtlA-FLAG grown to mid-exponential phase in glucose medium (Glucose) or mannitol medium (Mannitol) were used as controls, as were cell lysates from wild-type V. cholerae (untagged mtlA) grown in mannitol medium (No tag).
The decrease in MtlA levels in this experiment, however, appeared to follow kinetics different from those we observed when the bacteria were shifted from a mannitol to a glucose medium (Fig. 2A). We therefore repeated the MtlS sRNA overexpression experiment using more time points in order to better define the half-life of MtlA when MtlS sRNA expression was induced in cells grown in mannitol (Fig. 3B). In agreement with our previous experiments, prior to induction, MtlS sRNA was undetectable while MtlA levels were abundant. Within 10 min, MtlS sRNA was expressed at high levels and MtlA amounts were reduced, disappearing completely by 60 min post-MtlS sRNA induction. We noted, however, that the half-life of MtlA under these conditions was approximately 15 min, at least 3 times longer than the transporter half-life observed when V. cholerae was shifted from a mannitol to a glucose medium.
We also constructed a plasmid, pAS-MtlS, that allows for arabinose-inducible expression of an RNA that is complementary to MtlS sRNA (AS-MtlS RNA) and thus was predicted to base pair with MtlS sRNA and effectively “knock down” its activity (25). V. cholerae harboring this plasmid was grown in glycerol medium, a condition under which MtlA levels are normally low (Fig. 1B). Because the pBAD-based expression vector used to overexpress AS-MtlS RNA is repressed in the presence of glucose (13), the bacteria were grown in glycerol medium for these experiments; this strain grows similarly to the wild type in glycerol medium (see Fig. S1 in the supplemental material). When AS-MtlS RNA expression was induced, MtlA protein was observed even in the absence of mannitol (Fig. 3C). From these results, we conclude that AS-MtlS RNA prevented the base pairing of MtlS sRNA with the mtlA mRNA, thereby relieving the MtlS sRNA-mediated repression of MtlA synthesis. We note, however, that MtlA protein levels were lower than those in wild-type V. cholerae grown in mannitol medium (Fig. 3C). We postulate that this was due to incomplete inhibition of MtlS sRNA activity, because the AS-MtlS RNA must compete with other MtlS sRNA-binding partners (see below), and that this plasmid-expressed antisense transcript may not be as efficient as other RNAs at finding and binding to MtlS sRNA.
Expression of MtlA protein is regulated at the posttranscriptional level.
To investigate the mechanism of MtlS sRNA-mediated repression of MtlA expression, we analyzed mtlA mRNA expression in V. cholerae grown in both PTS and non-PTS sugars. In contrast to our observations with MtlS sRNA or MtlA, we observed that mtlA mRNA is expressed in similar amounts regardless of the carbon source used for bacterial growth (Fig. 1C). It is worth noting that the results we obtained through Northern blot analysis differ from those previously reported in which mtlA mRNA levels, as measured by quantitative reverse transcription-PCR (qRT-PCR), did change with differing carbon sources (25). A possible explanation for this discrepancy is that the primers used for the qRT-PCR experiments hybridized to the 5′ end of the mtlA transcript, which may undergo processing or alterations in the absence of mannitol, whereas the Northern blot analyses indicate that mtlA transcripts as a whole remain largely unaffected by changes in the carbon source. Additionally, the mRNA levels shown in Fig. 1C do suggest that there are some minor differences in mtlA mRNA levels, to which qRT-PCR may be highly sensitive. Nevertheless, given the sum total of the data presented in Fig. 1B and C, we believe that the changes in mtlA mRNA levels are relatively minor compared to the large difference in MtlA protein levels observed when the bacteria are grown in different carbon sources. Therefore, we believe these observations indicate that the regulation of MtlA protein is a posttranscriptional event. We also performed 5′ RACE (rapid amplification of 5′ cDNA ends) analysis on the mtlA transcript and identified the start of transcription, which is 74 nucleotides before the start codon; the sRNA and the mRNA therefore share 70 nucleotides of perfect complementarity (see Fig. S2 in the supplemental material).
There are several possible mechanisms by which MtlS sRNA could affect MtlA protein synthesis at the posttranscriptional level. MtlS sRNA could alter MtlA synthesis either by (i) affecting the stability of mtlA mRNA by binding the messenger and recruiting RNases, (ii) repressing translation initiation through occlusion of the ribosome binding site, or (iii) a combination of the two mechanisms (10). Northern blotting was used to determine the effect of MtlS sRNA on mtlA transcript stability. Rifampin was added to wild-type cultures to terminate transcription, after which the mtlA transcripts were monitored over time. The analysis was performed for V. cholerae strains grown in both glucose and mannitol media. Figure 4A shows that, under both growth conditions tested, the mtlA mRNA is a highly stable transcript. The control shows that the added rifampin has the expected effect on the rpsL transcript (24). Moreover, we observed no difference in mtlA mRNA levels between V. cholerae strains grown in glucose versus mannitol medium over the course of the experiment (60 min) (Fig. 4A). Because MtlS sRNA levels are higher in glucose than in mannitol medium, if the sRNA did reduce the stability of mtlA mRNA, we would expect to find mtlA mRNA to be less stable in bacteria grown in glucose medium than in those grown in mannitol medium. Instead, our data suggest that the presence of MtlS sRNA does not affect the stability of the mtlA mRNA. Using the same rifampin-treated cells, we also determined that MtlS sRNA has a half-life of approximately 20 min, which is in line with those of other known sRNAs (Fig. 4B) (1, 27, 28, 34). Thus, in the first 30 min of the experiment for which results are shown in Fig. 4A, MtlS sRNA is still present and likely is available to base pair with mtlA mRNA. These results further indicate that MtlS sRNA does not affect mtlA transcript stability and suggest that MtlS sRNA negatively impacts MtlA synthesis by blocking the translation of the transporter protein.
Fig 4.
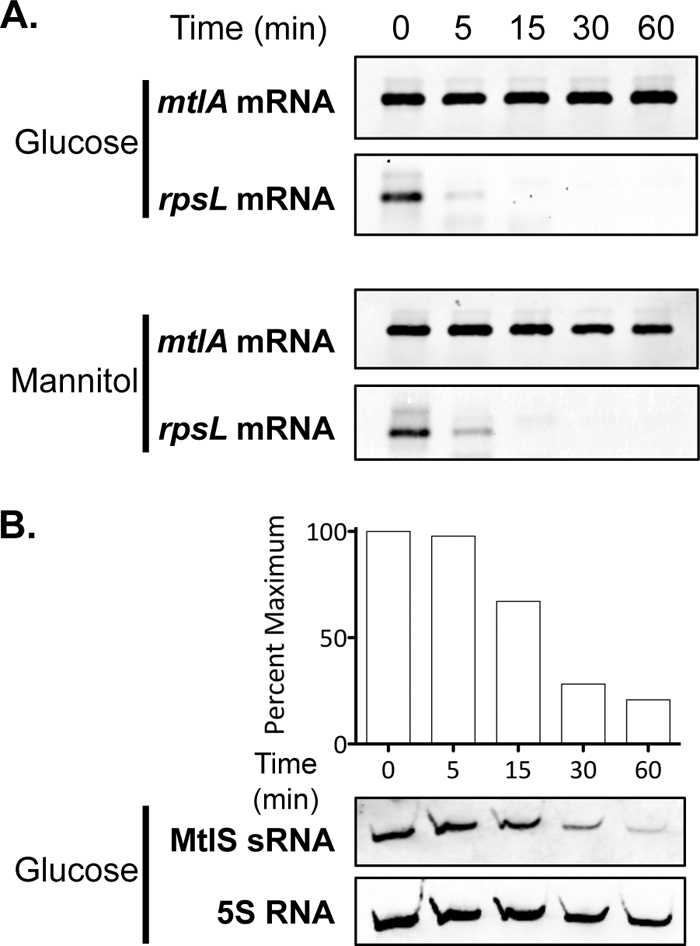
mtlA mRNA is a stable transcript. (A) V. cholerae mtlA-FLAG strains were grown to mid-exponential phase at 37°C in glucose or mannitol medium. Total-RNA samples (1 μg) prepared at 0, 5, 15, 30, and 60 min after treatment with 100 μg/ml rifampin were analyzed by Northern hybridization using RNA probes specific for mtlA and rpsL. (B) The same total-RNA samples (2 μg) used for the experiment for which results are shown in panel A were analyzed by Northern hybridization using an RNA probe specific to MtlS sRNA and a 5S RNA-specific DNA probe. The histogram above the MtlS sRNA and 5S RNA blots shows the mean intensities of the MtlS sRNA bands, normalized to the intensity of 5S RNA. Each bar corresponds to the band located directly below. MtlS sRNA levels are expressed relative to those at 0 min. The data are representative of at least three independent experiments.
Hfq is not necessary for the regulation of MtlA synthesis by MtlS sRNA.
Many of the best-characterized sRNAs require Hfq for activity (24, 35, 46). Originally identified as a host factor for the replication of the RNA phage Qβ in E. coli (9), Hfq is an RNA binding protein that has been studied extensively for its role in sRNA-mediated gene regulation. In several cases, Hfq has been shown to aid sRNA activity by enhancing the rate of duplex formation between sRNA and target RNA (19, 41). To test whether MtlS sRNA requires Hfq for its activity, we analyzed MtlA protein levels in wild-type and Δhfq strains. The Δhfq strain features a markerless deletion of the Hfq open reading frame. Consistent with previous observations, the hfq mutant does not exhibit any major growth defects under the experimental conditions used here (7) (see Fig. S3 in the supplemental material). We observed that MtlA synthesis in the mutant strain remained unchanged from that in the wild type, indicating that Hfq is not needed to repress MtlA expression in glucose medium (Fig. 5A). Northern blot analysis further revealed that mtlA mRNA levels in the wild-type and hfq strains are also similar (Fig. 5B).
Fig 5.
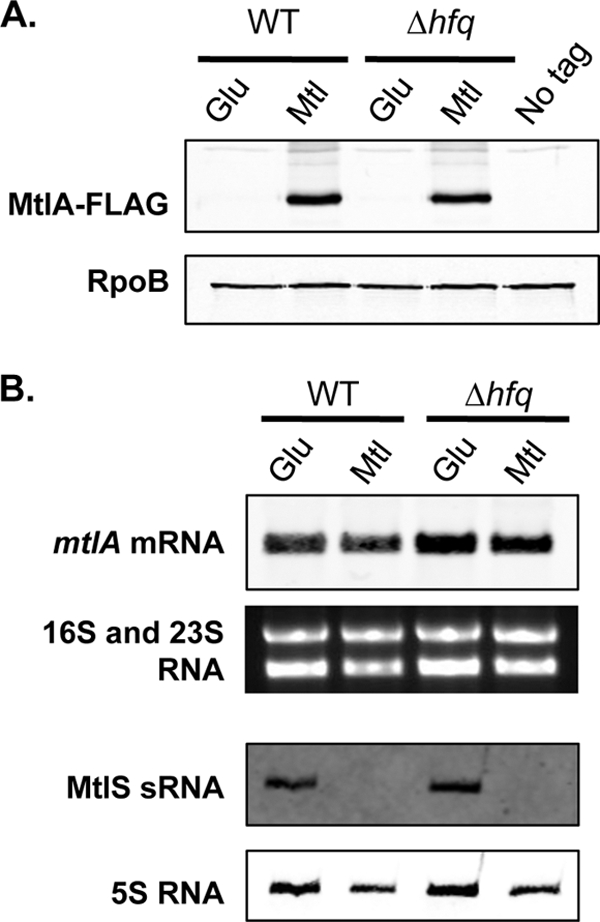
The regulation of MtlA synthesis is independent of Hfq. The V. cholerae mtlA-FLAG strain and its hfq mutant derivative were grown to the mid-exponential phase in glucose (Glu) or mannitol (Mtl) medium at 37°C. Cell lysates and total-RNA (2 μg) samples were used for immunoblot analysis with anti-FLAG and anti-RpoB antibodies (A) and for Northern hybridization using RNA probes specific to mtlA and MtlS sRNA (B). 5S RNA was analyzed using a 5S RNA-specific DNA probe. 16S rRNA and 23S rRNA were stained with ethidium bromide as loading controls. The same total-RNA samples were used in both the mtlA and MtlS sRNA assays. The data are representative of at least three independent experiments.
It is well established that Hfq-interacting small RNAs are markedly less stable in the absence of Hfq (20, 30, 34, 38, 43, 46). To explore whether Hfq plays a role in MtlS sRNA stability, we assessed MtlS sRNA levels by Northern blot analysis of RNA from wild-type and hfq mutant V. cholerae strains. In agreement with our observation that Hfq is not involved in regulating MtlA synthesis, the levels of MtlS sRNA in the wild type and the hfq mutant were equal in both glucose and mannitol media (Fig. 5B).
MtlR is not an effector of mtlS expression.
The mtl operon in V. cholerae includes a putative transcriptional regulator, MtlR. We observed that strains with a markerless deletion of the MtlR open reading frame exhibited a lag phase slightly shorter than that of the wild-type strain after dilution from rich LB into minimal mannitol medium (see Fig. S4 in the supplemental material). We reasoned that the observed change in the growth rate might be the result of the mutant synthesizing MtlA protein under conditions when transporter expression is normally repressed. We therefore hypothesized that MtlR could be an activator of mtlS expression. To test our hypothesis, we compared MtlS sRNA expression in the wild type and the mtlR mutant by Northern blotting. Our results indicate that there is no difference in MtlS sRNA expression between the wild type and the mtlR mutant (Fig. 6). It is therefore unlikely that MtlR regulates the expression of the MtlS sRNA.
Fig 6.
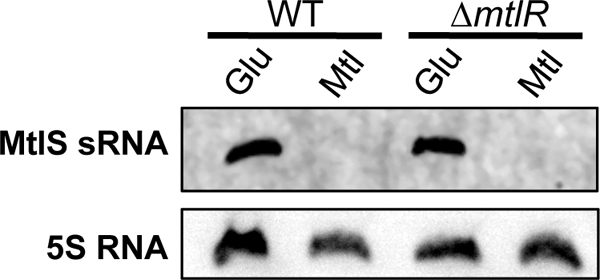
MtlS sRNA is not activated by MtlR. The wild-type V. cholerae strain and a mtlR mutant derivative were grown to mid-exponential phase in glucose (Glu) or mannitol (Mtl) medium at 37°C. Total-RNA samples (1 μg) were used for Northern hybridization with an RNA probe specific to MtlS sRNA and a DNA probe specific to 5S RNA. The data are representative of at least three independent experiments.
DISCUSSION
MtlS sRNA contributes to a growing list of sRNAs involved in carbon source transport or regulation (11). One of the first regulatory sRNAs identified, the Spot42 sRNA, is a trans-acting sRNA that negatively regulates the synthesis of galactose kinase (GalK) in E. coli by preventing the initiation of translation of the galK mRNA (29). Spot42 sRNA synthesis itself is repressed 3- to 5-fold in the absence of glucose, resulting in a concomitant increase in GalK synthesis and the ability of the cells to use galactose for energy. Also in E. coli, the SgrS sRNA is induced upon phosphosugar stress and downregulates the glucose-specific PTS transporter PtsG (50). Intriguingly, in V. cholerae, the TarA sRNA negatively regulates PtsG synthesis, but in a manner independent of phosphosugar stress (43). All three of these sRNAs, moreover, are trans-acting sRNAs and base pair with target mRNAs in an Hfq-dependent fashion.
Here we sought to define the role of the MtlS sRNA, a cis-encoded sRNA in V. cholerae, and found that this antisense RNA controls the synthesis of the mannitol-specific PTS transporter MtlA through an Hfq-independent mechanism. MtlS sRNA shares 70 nucleotides of perfect complementarity with the mtlA 5′ UTR, and we observed that the sRNA and the MtlA protein are expressed in an inverse relationship, while the mtlA mRNA is constitutively transcribed. Surprisingly, the mtlA mRNA is a very stable transcript regardless of the intracellular levels of the sRNA (Fig. 4A). Consistent with these observations, we propose a model where in the presence of sugars other than mannitol, the MtlS sRNA is synthesized and binds the mtlA messenger, occluding the ribosome binding site and impairing translation initiation. Initial collisions of two divergently elongating RNA polymerases may explain the slow kinetics of MtlS sRNA accumulation observed when V. cholerae was shifted from a mannitol to a glucose medium (Fig. 2A).
The mannitol permease is highly conserved with respect to the mannitol transport system in other organisms (16, 22). Sequence analysis shows, however, that MtlS sRNA is present only in Vibrio species (25). Northern blot analysis also failed to identify an sRNA in the 5′ UTR of E. coli mtlA (25). We speculate that the MtlS sRNA could easily have come into existence via a small number of point mutations that yielded an active promoter within the mtlA coding region in V. cholerae. Preliminary data from our lab suggest that in addition to MtlS sRNA, the MtlR protein may also regulate MtlA expression (data not shown). Thus, it would seem that two distinct regulators, MtlR and MtlS sRNA, act on mtlA to control the synthesis of the transporter protein. The cis-acting MtlS sRNA may provide a particularly rapid response to changes in the carbon source due to its proximity to its target. As soon as MtlS sRNA is expressed, it should be able to base pair with an mtlA transcript that is transcribed on the opposite strand. We hypothesize that this pairing is rapid and irreversible. This model is further supported by our observations that an AS-MtlS RNA transcript expressed from a plasmid cannot fully inhibit MtlS sRNA activity, most likely because the AS-MtlS RNA is outcompeted by the mtlA mRNA for MtlS sRNA binding sites (Fig. 3C).
We also observed that the MtlA protein is rapidly degraded when V. cholerae is shifted from a mannitol to a glucose medium (half-life, ∼5 min). Since the protein itself appears highly stable, with a half-life of >100 min (Fig. 2B), we suggest that proteolysis may represent yet another mechanism by which MtlA levels are regulated in the cell. Intriguingly, when the presence of mannitol was kept constant but MtlS sRNA was overexpressed, MtlA levels still decreased at a rate suggestive of active proteolysis, with a half-life of ∼15 min (Fig. 3B). These observations suggest that there may be multiple triggers for MtlA proteolysis, including shifts in the extracellular carbon source and changes in intracellular MtlS sRNA levels.
It has been hypothesized that in V. cholerae the PTS plays an essential role in sensing, integrating, and responding to environmental cues in both aquatic reservoirs and the host (16, 17). For example, in aquatic environments, chitin, a PTS carbon source, is highly available and can be used by V. cholerae to promote both growth and competence (4, 31). The latter feature may allow V. cholerae to better adapt for survival in unique environments and may also contribute to increased pathogenicity. The use of multiple regulators for the mannitol-specific PTS further supports a central and important role of PTS substrates in the V. cholerae life cycle. Marine algae produce large amounts of mannitol and may represent an important carbon source for V. cholerae survival in aquatic habitats (51). Moreover, mannitol is known to accumulate in the human small intestine (51), where it may be used as an energy source for V. cholerae during colonization.
The sugar alcohol is also one of several compatible solutes that are known to help bacteria with osmoadaptation (21). Though a halophilic species, V. cholerae can nevertheless persist in sources of drinking water with very low salinities and can thereby come into contact with humans; ingestion of the pathogen would then expose the bacteria to the intestinal lumen, which has an osmolarity equivalent to 0.3 M NaCl or higher (12, 33). Thus, upon ingestion by the human host, V. cholerae may upregulate MtlA synthesis to scavenge for mannitol in order to combat the increase in osmolarity. We therefore propose that cells expressing elevated MtlA levels could be at a competitive advantage from both an energy and an osmoadaptation perspective during both the colonization of humans and survival in their natural aquatic habitats.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant R15 AI090606 from the National Institutes of Health to J.M.L. and by grants and scholarships from the Beta Beta Beta Research Foundation (to L.M.M.), the American Society for Microbiology (to S.A.), and Drew University.
We thank Andrew Camilli for critical reading of the manuscript.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43–53 [DOI] [PubMed] [Google Scholar]
- 2. Andersen JB, et al. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angelichio MJ, Spector J, Waldor MK, Camilli A. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67:3733–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blokesch M, Schoolnik GK. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colwell RR. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025–2031 [DOI] [PubMed] [Google Scholar]
- 6. Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol. Microbiol. 53:345–354 [DOI] [PubMed] [Google Scholar]
- 8. Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Franze de Fernandez MT, Eoyang L, August JT. 1968. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature 219:588–590 [DOI] [PubMed] [Google Scholar]
- 10. Georg J, Hess W. 2011. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75:286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Görke B, Vogel J. 2008. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 22:2914–2925 [DOI] [PubMed] [Google Scholar]
- 12. Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herrington DA, et al. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hood MA, Guckert JB, White DC, Deck F. 1986. Effect of nutrient deprivation on lipid, carbohydrate, DNA, RNA, and protein levels in Vibrio cholerae. Appl. Environ. Microbiol. 52:788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 78:1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 192:3055–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Houot L, Watnick PI. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawamoto H, Koide Y, Morita T, Aiba H. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61:1013–1022 [DOI] [PubMed] [Google Scholar]
- 20. Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. 2005. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 19:328–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kets EP, Galinski EA, de Wit M, de Bont JA, Heipieper HJ. 1996. Mannitol, a novel bacterial compatible solute in Pseudomonas putida S12. J. Bacteriol. 178:6665–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar S, Smith KP, Floyd JL, Varela MF. 2011. Cloning and molecular analysis of a mannitol operon of phosphoenolpyruvate-dependent phosphotransferase (PTS) type from Vibrio cholerae O395. Arch. Microbiol. 193:201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee SH, Angelichio MJ, Mekalanos JJ, Camilli A. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 180:2298–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lenz DH, et al. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82 [DOI] [PubMed] [Google Scholar]
- 25. Liu JM, et al. 2009. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 37:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu MY, et al. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272:17502–17510 [DOI] [PubMed] [Google Scholar]
- 27. Majdalani N, Chen S, Murrow J, St John K, Gottesman S. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382–1394 [DOI] [PubMed] [Google Scholar]
- 28. Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U. S. A. 95:12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Majdalani N, Vanderpool CK, Gottesman S. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 40:93–113 [DOI] [PubMed] [Google Scholar]
- 30. MassÉ E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 32. Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471–1491 [DOI] [PubMed] [Google Scholar]
- 33. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Møller T, et al. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23–30 [DOI] [PubMed] [Google Scholar]
- 35. Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16:1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608–15614 [DOI] [PubMed] [Google Scholar]
- 38. Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. 2004. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54:1063–1075 [DOI] [PubMed] [Google Scholar]
- 39. Morita T, Maki K, Aiba H. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19:2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prüss-Üstün A, Bos R, Gore F, Bartram J. 2008. Safer water, better health: costs, benefits and sustainability of interventions to protect and promote health. World Health Organization, Geneva, Switzerland [Google Scholar]
- 41. Rasmussen A, et al. 2009. A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol. Microbiol. 72:566–577 [DOI] [PubMed] [Google Scholar]
- 42. Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125–139 [DOI] [PubMed] [Google Scholar]
- 43. Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ. 2010. The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA. Mol. Microbiol. 78:1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schild S, et al. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Senanayake SD, Brian DA. 1995. Precise large deletions by PCR-based overlap extension method. Mol. Biotechnol. 4:13–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sledjeski DD, Whitman C, Zhang A. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sonnenburg JL, Chen CTL, Gordon JI. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Southgate DA. 1995. Digestion and metabolism of sugars. Am. J. Clin. Nutr. 62(1 Suppl):203S–210S [DOI] [PubMed] [Google Scholar]
- 49. Thelin KH, Taylor RK. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076–1089 [DOI] [PubMed] [Google Scholar]
- 51. Wang YM, van Eys J. 1981. Nutritional significance of fructose and sugar alcohols. Annu. Rev. Nutr. 1:437–475 [DOI] [PubMed] [Google Scholar]
- 52. Xu Q, Dziejman M, Mekalanos JJ. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U. S. A. 100:1286–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.