Abstract
DECIPHER is a new method for finding 16S rRNA chimeric sequences by the use of a search-based approach. The method is based upon detecting short fragments that are uncommon in the phylogenetic group where a query sequence is classified but frequently found in another phylogenetic group. The algorithm was calibrated for full sequences (fs_DECIPHER) and short sequences (ss_DECIPHER) and benchmarked against WigeoN (Pintail), ChimeraSlayer, and Uchime using artificially generated chimeras. Overall, ss_DECIPHER and Uchime provided the highest chimera detection for sequences 100 to 600 nucleotides long (79% and 81%, respectively), but Uchime's performance deteriorated for longer sequences, while ss_DECIPHER maintained a high detection rate (89%). Both methods had low false-positive rates (1.3% and 1.6%). The more conservative fs_DECIPHER, benchmarked only for sequences longer than 600 nucleotides, had an overall detection rate lower than that of ss_DECIPHER (75%) but higher than those of the other programs. In addition, fs_DECIPHER had the lowest false-positive rate among all the benchmarked programs (<0.20%). DECIPHER was outperformed only by ChimeraSlayer and Uchime when chimeras were formed from closely related parents (less than 10% divergence). Given the differences in the programs, it was possible to detect over 89% of all chimeras with just the combination of ss_DECIPHER and Uchime. Using fs_DECIPHER, we detected between 1% and 2% additional chimeras in the RDP, SILVA, and Greengenes databases from which chimeras had already been removed with Pintail or Bellerophon. DECIPHER was implemented in the R programming language and is directly accessible through a webpage or by downloading the program as an R package (http://DECIPHER.cee.wisc.edu).
INTRODUCTION
The small subunit (SSU) rRNA molecule has been used extensively as a phylogenetic marker since the late 1980s (18), and nowadays, 16S rRNA sequences are essential for microbial identification (2, 19). As the number of publicly available SSU rRNA sequences has increased, several repositories that curate and align the sequences have emerged. These databases also provide useful tools for data analysis and interpretation. For instance, the Ribosomal Database Project (RDP) (4) is a major repository of bacterial and archaeal 16S rRNA sequences and offers tools for browsing, classification, probe checking, and sequence matching, among others. The SILVA rRNA database (12) contains small subunit (SSU) and large subunit (LSU) rRNA sequences of bacteria, archaea, and eukarya, while Greengenes (6) offers a variety of sequence analysis tools.
One of the challenges in maintaining the ever-expanding rRNA databases is in the implementation of strategies to ensure that they are populated only with good-quality sequences. The most common type of sequence anomaly is the chimera, which is composed of two or more distinct sequences concatenated into a single one. The presence of chimeras in a database artificially increases measurements of diversity (10, 13, 16), and when chimeras are comprised of sequences from different lineages, they could be misinterpreted as representing novel lines of descent. In addition, the presence of chimeras in database repositories can lead to erroneous interpretations of specificity when using the database for the purpose of primer or probe design.
In 2005, Ashelford et al. (3) created Pintail, a program for detection of sequence abnormalities (mainly chimeras) and used it to estimate that about 5% of 16S rRNA sequences held in public repositories had substantial anomalies. Subsequently, RDP and SILVA implemented quality control filters based on Pintail to prevent populating their databases with unchecked anomalous sequences. Greengenes uses a chimera check based on Bellerophon, another algorithm created for this purpose (9). More recently, ChimeraSlayer (8) was introduced as a program with improved chimera detection, especially for chimeras created from two closely related parent sequences, and Uchime (7) has been described as having a higher detection rate than ChimeraSlayer. Eliminating chimeric sequences is now understood by the research community as an important step to take before submitting sequences to public databases or before assembling sequences from short fragments produced by next-generation sequencing approaches (8, 10, 13). It is now common practice to use one or more programs to remove chimeras before sequence submission. In addition, sequence repositories such as RDP (5), SILVA (12), and Greengenes (6) provide a second line of defense by flagging possible chimeras after submission.
Nevertheless, when routinely using RDP's “good quality” database for probe design, we still encounter chimeric sequences that affect data interpretation. Therefore, we developed a novel approach for chimera detection in the 16S databases. Chimeric regions within a query sequence are identified by detecting short sequence fragments that are uncommon within a reference phylogenetic group where the sequence is classified but much more common in another phylogenetic group.
Using this approach, we have confirmed that a number of chimeric sequences have evaded existing sequence anomaly detection methods and are presently populating the 16S databases. Here we describe the method and the results of chimera detection in the main 16S rRNA repositories and benchmark the new method against Pintail, ChimeraSlayer, and Uchime. In addition, to help in the detection of chimeras before sequence submission, we introduce DECIPHER (http://DECIPHER.cee.wisc.edu), a publicly available web-based tool specific for detection of chimeric 16S rRNA sequences by the use of the novel search-based approach. For standalone implementations, the DECIPHER R package, source code, and associated documentation are available for download under the terms of the GNU General Public License.
MATERIALS AND METHODS
16S reference phylogenetic groups.
The data set of “good quality” unaligned sequences available from RDP was used for creating a higher-quality reference data set free of detectable chimeras. The downloaded set of sequences (RDP release 10, update 22) contained 1,251,070 bacterial and 62,055 archaeal sequences. A total of 280 reference phylogenetic groups (see Table S1 in the supplemental material) were created from this sequence set by combining sequences from similar hierarchical levels. The goal of this step was to establish reference groups with a sufficiently high number of related sequences (i.e., greater than 500) so that the search for common and uncommon sequence fragments was meaningful as a chimera detection approach.
Reference groups were limited to a maximum of 10,000 sequences to facilitate computational optimization of the search algorithm, except for genera or unclassified groups that already had more than 10,000 sequences (e.g., Staphylococcus genus or the “unclassified_Bacteria” group; see Table S1 in the supplemental material). Some of the reference groups had to be defined as having less than 500 sequences, because their combination with other groups at the same hierarchical level was not logical. For instance, the phyla Korarchaeota and Nanoarchaeota within the domain Archaea are both represented in the database by single genera with less than 500 sequences each. A potential combination of these phyla with other groups at the same hierarchical level would require inclusion of the Crenarchaeota and Euryarchaeota phyla. Such a diverse group would prevent detection of chimeras within the entire Archaea domain by the use of the search-based algorithm. Thus, Korarchaeota and Nanoarchaeota appear as individual groups in the reference group set. The resulting 280 reference phylogenetic groups are presented in Table S1 in the supplemental material, along with a list of the genera or unclassified groups contained in each reference group.
Chimera detection method.
The evaluation of whether a query sequence is a chimera takes the following steps.
First, the query sequence is classified with 51% confidence using the RDP Classifier software (17) and then assigned to one of the 280 reference phylogenetic groups (see Table S1 in the supplemental material) based upon this classification.
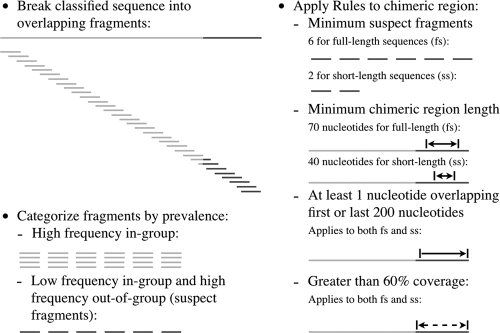
Then, a set of 30-nucleotide-long overlapping fragments is formed from the sequence, beginning every fifth nucleotide and continuing for the length of the sequence (Fig. 1). For instance, a sequence of 1,400 nucleotides would result in 275 fragments 30 nucleotides long that begin at nucleotide positions 1, 6, 11, etc. Fragments containing wild-card characters (i.e., N) are excluded.
Fig 1.
Steps used by DECIPHER to determine whether a sequence is a chimera. The differently shaded lines represent the different pieces of a chimeric sequence, with the darker color representing the chimeric region. Sequence fragments exhibiting low in-group frequency and high out-of-group frequency are marked as suspect fragments. These suspect fragments must meet the additional rules shown in the figure in order for the sequence to be deemed a chimera.
The presence or absence of these fragments within other sequences in the classified reference group (i.e., in-group search) and outside the classified group (i.e., out-of-group search) forms the basis for detection of chimeras in this algorithm. That is, if the query sequence is a chimera, then some fragments are likely to have very few matches within their own reference phylogenetic group but a large number of matches to another reference group.
Thus, a search for each fragment is conducted in the set of all sequences within the classified reference group. The number of sequences containing the 30-mer fragment, allowing a maximum of one mismatch, is recorded as the number of hits in-group. Fragments with 5 or more hits in-group are excluded from further analysis, as such large numbers of hits in-group are not indicative of a potential chimeric fragment. A second search through the remaining fragments is done while allowing two mismatches, which is a more lenient evaluation. Those fragments with more than 9 lenient hits in-group are also excluded, leaving a set of 30-mer fragments that are rare within their own phylogenetic reference group. This is the initial set of fragments suspected to correspond to a chimeric region (Fig. 1).
To determine whether these uncommon fragments within the classified group are more common in another phylogenetically different group, a search for the presence of these fragments (with a maximum of one mismatch allowed) is then conducted in each of the other phylogenetic groups in the reference set. If the number of hits found in another group exceeds 20 times the number of hits detected in-group for a fragment, then it is considered suspect. If a fragment has no hits in-group (i.e., a similar fragment was not found in the reference group) and it has more than 20 hits out-of-group, then it is also considered suspect.
When the RDP classifier tool places the query sequence in an unclassified group, other groups sharing the same line of descent are skipped in the out-of-group search, because unclassified groups may not be phylogenetically coherent. For instance, if a query sequence is classified as unclassified_Actinobacteria, then the search for out-of-group hits excludes all reference groups within Actinobacteria but does not exclude classified or unclassified reference groups in other lineages. Likewise, if the reference group where a query sequence is classified has a reference group of unclassified organisms at the same hierarchical level, then this group is also skipped in the out-of-group search. An important consequence of the former restriction is that sequences assigned to the unclassified_Bacteria or unclassified_Archaea group cannot be evaluated by DECIPHER. Using RDP's classifier tool with a 51% confidence level ensures that this is not a common problem. In the data sets analyzed in this study, less than 5% of sequences fell into this category.
The set of suspect fragments resulting from the steps described above must meet several more criteria in order for a query sequence to be identified as a chimera (Fig. 1). Two different sets of criteria were defined, depending on whether DECIPHER is used to evaluate assembled, nearly complete 16S sequences or short sequences. For full sequences (fs_DECIPHER), there must be six or more suspect fragments belonging to a sequence. In addition, the identified chimeric region, defined as the distance between the start of the first fragment and end of the last fragment, must be at least 70 nucleotides long, and the chimeric region must have at least one nucleotide overlapping the first or last 200 nucleotides of the sequence. Finally, the combined ranges of all suspect fragments belonging to another reference group must cover more than 60% of the total chimeric region.
For short sequences (ss_DECIPHER), the criteria were relaxed such that a chimeric region of at least 40 nucleotides that included at least two suspect fragments was required. The rules of overlapping the first or last 200 nucleotides and the 60% coverage are also kept in ss_DECIPHER, but these rules become less important as the sequence length decreases. Although both DECIPHER options can handle sequences of any length, fs_DECIPHER is a conservative option with a very low rate of false positives and moderately lower chimera detection capabilities, while ss_DECIPHER detects more chimeras, albeit with a higher level of false-positive detections. Based on the rates of false-positive and false-negative detections (see Fig. S1 in the supplemental material), ss_DECIPHER is recommended for sequences of any length, while fs_DECIPHER is recommended only for sequences longer than 600 nucleotides.
The specific values of the different parameters used by the two versions of DECIPHER were determined by comparing results obtained with hundreds of manually checked sequences originating from the RDP database, as well as sets of artificial chimeras. To manually check each query sequence, we use the Probe Match tool of RDP to find sequences that matched 40- to 60-nucleotide-long fragments taken from different locations within the query sequence. If the sequences that matched the different fragments belonged to different genera, then we aligned the query sequence with the matched sequences by the use of BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and visually inspected the alignments to identify potential breakpoints within the query sequence. If breakpoints were evident by visual inspection, then the sequence was determined to be a chimera. The calibration parameters were primarily aimed at providing a very low rate of false-positive detections with a high efficiency of chimera detection. For instance, the length of the overlapping fragments was set to 30 nucleotides to maximize detection of very short chimeric regions within a sequence, and the selections of the thresholds for in-group and out-of-group hits were calibrated to minimize false-positive identifications. Other parameters, such as the length of the identifying fragment and its distance to the end of the sequence as they affect the rates of false-positive and false-negative identifications, are described in more detail in supplementary documentation using a set of artificially generated chimeras (see Fig. S2 and S3 in the supplemental material).
Implementation.
The method described above was implemented in the R programming language (14). The slow search speed was one of the main challenges in applying this method, since each suspect fragment is compared to a reference database that contains more than one million sequences. To speed up the search, we made use of the Aho-Corasick dictionary-matching algorithm (1) implemented as part of Biostrings (11). In this algorithm, a trusted band of known nucleotides is defined for each fragment to be searched. We defined the trusted band as the first five nucleotides of each fragment when performing the in-group search that allowed 1 mismatch, and then the trusted band was shifted by five nucleotides for the search allowing 2 mismatches. For the out-of-group search, the trusted band was returned to the first 5 nucleotides of each fragment. Additionally, the trusted band cannot contain ambiguity characters (e.g., S for C or G), so fragments with a trusted band containing these characters were excluded from the search. Ambiguity characters outside the trusted band were determined to be perfect matches or mismatches according to conventional IUPAC notation.
RESULTS AND DISCUSSION
Preparation of a chimera-free reference sequence set.
We used the more conservative version of DECIPHER (fs_DECIPHER) to determine how many chimeras could be found in the RDP database of “good quality” sequences (i.e., the RDP database already screened with Pintail) while minimizing the erroneous flagging of nonchimeric sequences. The goal was to produce a higher-quality data set that could be used as a reference for the detection of chimeras. As a starting point, the downloaded sequences were used as the reference database and every sequence in the database was tested. A total of 12,470 sequences were determined to be chimeras. These chimeras were then removed from the reference database, and the search process was repeated. Since the search results changed between runs, it was possible to continue finding additional chimeras by successively updating the reference database and searching again for potential chimeras not detected in earlier runs. The number of chimeras found in each subsequent run was much smaller than the number in the previous run, until the process approached zero newly detected chimeras. Overall, 12 runs were made, yielding a total of 18,484 chimeras, corresponding to 1.41% of the “good quality” RDP database. With a false-positive detection rate of 0.15% (see below), we estimate that about 2,000 of these chimeras could potentially represent false-positive detections. The resulting database, free of chimeras detectable with fs_DECIPHER, became the reference database for the rest of the study.
A list of all the sequences identified as chimeras can be found in the supplementary documentation (see Table S2 in the supplemental material). Of all the newly identified chimeras from RDP, 54.9% were formed between two sequences in the same phylum, 40.4% corresponded to chimeras formed between two sequences in different phyla, and 0.18% were formed between sequences belonging to the domains Bacteria and Archaea. The low percentage of chimeras formed between sequences in different domains reflects the relatively low number of studies that have used universal primer sets simultaneously targeting both domains. The higher number of chimeras formed between sequences in the same phylum compared to those formed from sequences of different phyla is more difficult to explain because of the multitude of factors that may influence these observations. For instance, sequencing experiments targeting a specific group of organisms produce chimeras only from sequences within the same phylum, thus contributing to the greater percentage of in-phylum chimeras. It is also possible that chimeras are more likely to be formed between closely related parents (8), which would also contribute to a higher number of in-phylum chimeras. However, it is also important that the analysis was done with the RDP database that has already been screened by Pintail, so Pintail's chimera detection was also influencing the percentages observed. Regardless, the high percentage of chimeras identified as being formed from two different phyla (i.e., distantly related parents) is an important indicator that these types of chimeras are also likely to be formed and should be targeted by chimera detection programs.
Comparing databases.
In addition to evaluating the good-quality data set downloaded from RDP, fs_DECIPHER was also used to investigate the presence of chimeras in the SILVA and Greengenes databases. SILVA had 1,096,710 bacterial and archaeal sequences with a Pintail score of 100 (perfect score) in their database in October 2010. In these sequences, we detected 12,231 chimeras (1.1%). In the set of Greengenes sequences that Bellerophon had declared to be “not chimeric” (377,150 sequences), we found 7,136 chimeras (1.9%). Thus, all three 16S repositories contain relatively similar percentages of chimeras that were undetected by Pintail (RDP and SILVA) or Bellerophon (Greengenes).
Evaluation of DECIPHER with artificially generated chimeras.
We compared ss_DECIPHER and fs_DECIPHER to Pintail, ChimeraSlayer, and Uchime. Bellerophon was excluded from analysis due to a high rate of false positives detected in preliminary tests (13%), which agrees with the observations of Haas et al. (8). To evaluate Pintail, we used the WigeoN reimplementation (8), and for Uchime, we used its implementation in mothur (15). For ChimeraSlayer and Uchime runs, we used the gold data set described by Haas et al. (8) as the reference set of sequences.
Simple two-parent and more complex three- and four-parent chimeras were formed by combining segments from different parent sequences joined at random breakpoints. In the generation of all the artificial chimeras, each parent sequence was required to be represented by a minimum of 30 nucleotides. In addition, the length of the artificial sequences was randomly adjusted from 80 nucleotides to full length, with some data sets restricted to sequences shorter than either 600 or 300 nucleotides. Most chimeras were formed from aligned sequences randomly chosen from the 7,451 type-strain bacterial sequences available from RDP (release 10, update 22), while one data set was formed from the 284 aligned type-strain archaeal sequences found in RDP. Each set of artificial chimeras contained 1,000 sequences.
When evaluated with the data set of simple two-parent chimeras, ss_DECIPHER and fs_DECIPHER detected 88% and 75% of the chimeras, while Uchime, ChimeraSlayer, and WigeoN detected 73%, 56%, and 47%, respectively. The 2,000 parent sequences in the data set were also evaluated to estimate the rate of false-positive detections (assuming that all type-strain sequences are not chimeric), with fs_DECIPHER having the lowest false-positive rate (0.15%), followed by ChimeraSlayer (0.70%), WigeoN (0.85%), Uchime (1.5%), and ss_DECIPHER (1.6%). Since all the parent sequences were part of the reference data set, a more conservative calculation of false positives for fs_DECIPHER and ss_DECIPHER was performed after removing all type-strain sequences from the reference data set. This had no effect on the false positives seen with fs_DECIPHER but increased the ss_DECIPHER false-positive rate to 2.1%.
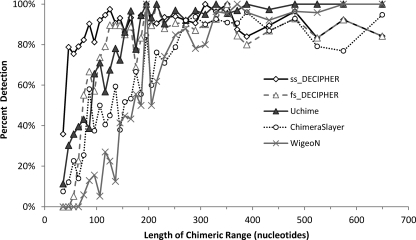
The most significant parameter influencing the rate of detection was the chimeric range, defined in the simple artificial chimeras as the shorter of the two segments contributed by the parents to the chimera. Figure 2 shows a comparison of chimera detection results as a function of the chimeric range and identifies ss_DECIPHER as the only method capable of a high rate of detection of chimeras (85%) with chimeric ranges between 40 and 125 nucleotides. Chimeras with smaller chimeric ranges, between 30 and 40 nucleotides long, were more difficult to detect, with ss_DECIPHER detecting the most (36%), followed by Uchime (11%) and ChimeraSlayer (8%), while fs_DECIPHER and WigeoN did not detect any chimeras. Uchime and fs_DECIPHER followed ss_DECIPHER in overall performance. They had poor detection of chimeras with chimeric fragments shorter than 100 nucleotides but a relatively high detection rate when the chimeric range was between 100 and 250 nucleotides. ChimeraSlayer and WigeoN were not suitable for detection of chimeras with chimeric ranges below 200 nucleotides. For chimeric ranges of more than 400 nucleotides, Uchime and WigeoN had the highest detection efficiencies (99% and 96%, respectively), followed by ss_DECIPHER, fs_DECIPHER, and ChimeraSlayer, with 89%, 88%, and 87%, respectively.
Fig 2.
Chimera detection by ss_DECIPHER, fs_DECIPHER, ChimeraSlayer, WigeoN (Pintail), and Uchime as a function of chimeric range length. The artificial chimera set (see the Two_Part-1 dataset in the supplemental materials) contained a total of 1,000 simple chimeras formed by combining two parent sequences. The chimeras in this data set were binned according to the length of the chimeric range, and the average of the chimeric range in each bin is shown in this figure.
The high rate of detection of short chimeric ranges by ss_DECIPHER is possible because the fundamental unit of detection in DECIPHER is 30 nucleotides and because ss_DECIPHER requires only an identifying chimeric range of at least 40 nucleotides (Fig. 1). Importantly, this basic unit is independent of the overall length of the sequence. The conservative fs_DECIPHER method requires the presence of an identifying chimeric range of at least 70 nucleotides in length; hence its poorer performance with shorter chimeric ranges. The low efficiency of chimera detection in ChimeraSlayer when the chimeric range is short can be explained by the fact that this algorithm uses 30% of the query sequences at each end of the sequence as the basic fragments to perform the searches in the reference database (8). Since these basic fragments are of various sizes, ChimeraSlayer's detection efficiency depends on the overall length of the sequence. Thus, if a short chimeric range (e.g., 40 nucleotides long) is present in a nearly full sequence (e.g., 1,500 nucleotides long), ChimeraSlayer is likely to miss it, since the majority of the 30% end (460 nucleotides) that contains the very small chimeric range would correspond to the same parent represented by the other end of the sequence. This problem is not as significant for shorter sequences, in which the small chimeric range has a stronger influence in the evaluation of potential parent sequences. For example, out of 33 artificial chimeras with chimeric ranges between 41 and 50 nucleotides long, ChimeraSlayer missed the detection of all chimeras with overall sequences more than 300 nucleotides long (i.e., 27 chimeras). On the other hand, ChimeraSlayer detected 4 out of 6 chimeras with overall sequence lengths between 164 and 281 nucleotides, for which 30% of the sequence closely matched the size of the chimeric fragment. Uchime seems to have the same problem as ChimeraSlayer, although it is not as severe. For the same subset of sequences with short chimeric ranges, Uchime failed to detect all chimeras in sequences longer than 523 nucleotides (i.e., 18 chimeras) but detected 10 out of the 15 chimeras in the shorter sequences. The improvement of Uchime compared to ChimeraSlayer could have resulted from the fact that Uchime uses the entire query sequence, split into four nonoverlapping sections, for searches in the reference database (7) rather than using only the sequence ends as ChimeraSlayer does. Nevertheless, the lengths of Uchime's searchable sections are also dependent on the length of the query sequence, and therefore, Uchime has the same difficulty as ChimeraSlayer in detecting the short chimeric ranges within long sequences.
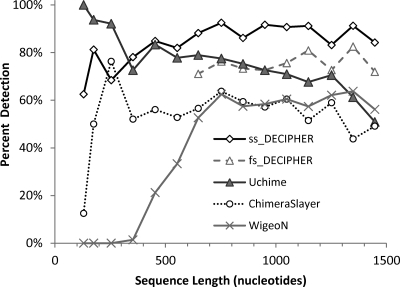
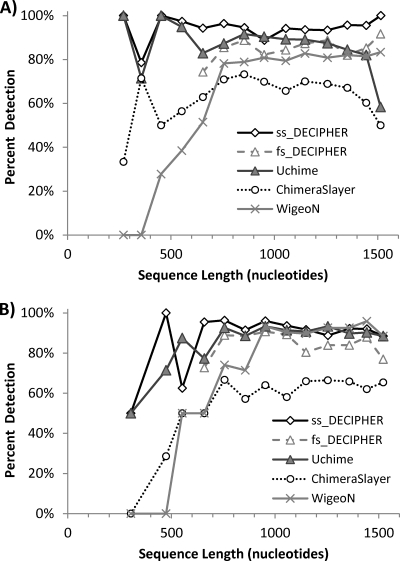
Other variables such as overall sequence length and the divergence between the parents of the chimera were also analyzed. Figure 3 shows the comparison as a function of sequence length for the same set of artificial chimeras used in Fig. 2 (Two_Part-1 data set). Except for sequences between 100 and 300 nucleotides long, ss_DECIPHER outperformed the other methods. Uchime was the best method for sequences less than 300 nucleotides long (95% detection), followed by ss_DECIPHER (80% detection). Interestingly, Uchime's performance deteriorated as the total sequence length increased, reaching a detection rate as low as 51% for sequences longer than 1,400 nucleotides. In contrast, ss_DECIPHER maintained a high level of detection for long sequences (82 to 93%). ChimeraSlayer and WigeoN had the lowest performance, averaging 55% and 60% detection rates, respectively, for sequences longer than 800 nucleotides (Fig. 3). For shorter sequences, ChimeraSlayer maintained a similar level of detection, except for sequences shorter than 200 nucleotides, whereas WigeoN was unable to detect chimeras in sequences shorter than 400 nucleotides. The more conservative fs_DECIPHER option, evaluated only for sequences longer than 600 nucleotides, also outperformed ChimeraSlayer, WigeoN, and Uchime in the level of detection of full-length chimeric sequences.
Fig 3.
Comparison of chimera detection by ss_DECIPHER, fs_DECIPHER, ChimeraSlayer, WigeoN (Pintail), and Uchime as a function of sequence length for simple chimeras formed by combining two parent sequences at a random breakpoint. The artificial chimera set (Two_Part-1) contained a total of 1,000 chimeras of random length, which were binned according to sequence length for this figure. All sequences were analyzed with ss_DECIPHER, while only sequences > 600 nucleotides long were analyzed with fs_DECIPHER.
The general trend of these results was confirmed with an independent set of 1,000 artificial chimeras (Two_Part-2 data set) formed from two parent sequences (see Fig. S4 in the supplemental material). The overall detection rates in this second set were 88% for ss_DECIPHER, 75% for fs_DECIPHER, 71% for Uchime, 55% for ChimeraSlayer, and 49% for WigeoN, in agreement with the results from the first set of artificial chimeras (Fig. 3). The false positives in this second set, estimated from the evaluation of the 2,000 parent sequences, were similar to the data from the first set, with 0.2% for fs_DECIPHER, 0.6% for ChimeraSlayer, 1.1% for WigeoN, 1.4% for ss_DECIPHER, and 1.6% for Uchime. The more conservative calculation using a reference data set that did not contain any type-strain sequences had no effect on false positives with fs_DECIPHER but increased ss_DECIPHER's rate to 2.2%.
Effect of parent divergence.
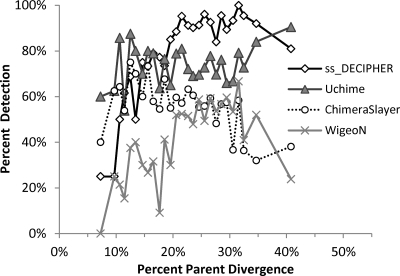
A comparison of DECIPHER with the other chimera detection methods was also made as a function of parent divergence (Fig. 4; see also Fig. S5 in the supplemental material), since this is a fundamental parameter used in the creation and benchmarking of ChimeraSlayer and Uchime (7, 8). By definition, DECIPHER does not detect chimeras formed from a combination of parents belonging to the same genus, whereas ChimeraSlayer and Uchime were designed to detect this type of chimera. In the benchmarking data set used (Two_Part-1), parent divergence ranged from 7% to 41%. For parents diverging less than 20%, Uchime outperformed the other methods, while ss_DECIPHER had the best performance when parent divergence was greater than 20% (Fig. 4). This higher detection with ss_DECIPHER was again the result of DECIPHER's superior ability to detect chimeras with short chimeric ranges as discussed previously (Fig. 2). To further evaluate the effect of parent divergence for the range of divergences described in the ChimeraSlayer benchmark study (8), an additional artificial chimera data set was created by restricting the parent divergence to less than 10%. In this case (see Fig. S6 in the supplemental material), Uchime and ChimeraSlayer outperformed DECIPHER as expected, although also with low detection rates that reflected the limitations of these programs seen with short chimeric fragments as described previously (Fig. 2). WigeoN also had low detection rates, which was in agreement with observations described in the ChimeraSlayer benchmark study (8).
Fig 4.
Comparison of chimera detection by ss_DECIPHER, ChimeraSlayer, WigeoN (Pintail), and Uchime as a function of parent divergence for simple chimeras formed by combining two parent sequences at a random breakpoint (Two_Part-1 data set). The artificial chimera set contained a total of 1,000 chimeras of random length, binned according to the divergence between parents. The comparison with fs_DECIPHER is shown in Fig. S5 in the supplemental material.
Evaluation of chimeras with random mutations.
In the benchmarking of ChimeraSlayer, Haas et al. (8) prepared a set of artificial chimeras created from parents with various degrees of divergence. The effect of mutations (i.e., substitutions, insertions, and deletions) was evaluated with sets that had 1% to 5% random mutations introduced into a set of artificially generated chimeras. Thus, we also used these data sets to evaluate the effect of random mutations on DECIPHER's ability to detect chimeras. For the subset with parent divergence greater than or equal to 20% (Table 1), a decrease in detection was observed as the percentage of mutations increased, with a small drop in detection for up to 2% mutations and with the most significant drop resulting from 4% and 5% insertion or deletion mutations. The variability was not as high with lower parent divergence, but as expected, the overall detection rate of chimeras formed from closely related parents was low (data not shown).
Table 1.
Chimera detection rate by DECIPHER as a function of random mutations, using the benchmark data set of Haas et al. (8)
| Type of evolution | % detection at indicated evolution rate (%)a |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Nucleotide substitution | 76 | 74 | 70 | 67 | 61 |
| Insertion or deletion | 73 | 69 | 54 | 45 | 36 |
| Insertion | 73 | 66 | 55 | 42 | 34 |
| Deletion | 72 | 66 | 54 | 43 | 31 |
The detection rate for 0% evolution was 76%. Only chimeras with parent divergence greater than or equal to 20% were used.
The effect of a large mutation rate on DECIPHER's ability to detect chimeras can be explained by the mutations resulting in sequences that are not properly classified by the RDP classifier tool and by mutations affecting the number of chimeric fragments identified. These problems did not occur with lower rates of artificially simulated mutations. By definition, DECIPHER allows the presence of mismatches in the identification of in-group and out-of-group fragments, likely contributing to the adequate detection of chimeras with low rates of mutations. In addition, by using taxonomic groupings made from the RDP database, the natural variability of sequences, due to mutations or sequencing errors, is inherently included in the DECIPHER analysis. Nevertheless, since the benchmark data set of Haas et al. uses mutations at random locations, which may not represent natural sequence variability present in the RDP database, the larger drop in detection as the mutation rate increased was anticipated.
Evaluation of complex chimeras.
The overall rates of detection of three-parent chimeras (Fig. 5) were 94% with ss_DECIPHER, 87% with Uchime, 85% with fs_DECIPHER, 76% with WigeoN, and 65% with ChimeraSlayer. For four-parent chimeras (Fig. 5), 92% were detected with ss_DECIPHER, 90% with Uchime, 89% with WigeoN, 85% with fs_DECIPHER, and 63% with ChimeraSlayer. Clearly, ChimeraSlayer had the lowest performance with complex chimeras, reflecting specific limitations in the design of the algorithm, which uses only the ends of the query sequence for comparisons to the reference (8). On the other hand, because DECIPHER works with the principle of identifying 30-mer fragments that do not belong to the group where the sequence is classified, it is more effective at detecting the smaller chimeric regions that would be present in the complex chimeras. Likewise, because Uchime works with the entire sequence divided into four segments, it is also efficient at detecting multiple-part chimeras (7).
Fig 5.
DECIPHER, Uchime, ChimeraSlayer, and WigeoN (Pintail) detection of chimeras formed from (A) three-parent and (B) four-parent sequences. The artificial chimera sets contained 1,000 chimeras of random length. All sequences were analyzed with ss_DECIPHER, while only sequences > 600 nucleotides long were analyzed with fs_DECIPHER (926 three-parent and 983 four-parent chimeras).
The random sets of complex chimeras used in the analysis described above resulted in a majority of the sequences (>90%) having lengths greater than 600 nucleotides. Thus, in order to further evaluate the effect of sequence length, new sets were generated, but the length was restricted to either less than 600 (see Fig. S7 in the supplemental material) or less than 300 (see Fig. S8 in the supplemental material) nucleotides. These complex chimera sets confirmed ss_DECIPHER and Uchime as having the best performance and ChimeraSlayer's and WigeoN′s inability to detect multiple-part chimeras in short sequences. For the complex chimeras (three and four parent) restricted to 600 nucleotides long, ss_DECIPHER detected 89% to 91%, Uchime detected 85% to 89%, ChimeraSlayer detected 45% to 56%, and WigeoN detected 26% to 28%. The detection rate with the complex chimeras with less than 300 nucleotides was 74% to 75% with ss_DECIPHER, 61% to 76% with Uchime, 15% to 29% with ChimeraSlayer, and 0% with WigeoN.
Evaluation of archaeal sequences.
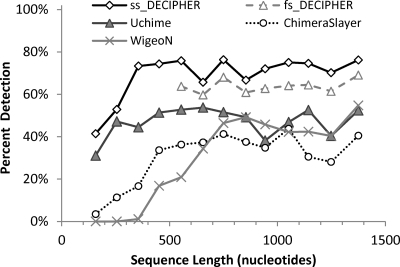
An additional evaluation was performed using a two-parent chimera data set formed exclusively with type-strain archaeal sequences. As shown in Fig. 6, DECIPHER's chimera detection rate was much higher than the rates of the other methods, achieving overall detections of 71% and 63% with ss_DECIPHER and fs_DECIPHER, respectively, while detection with the other methods were between 32% and 48%. Such a significant difference can be traced to the effect of the reference data set used for the other methods (i.e., the gold data set described by Haas et al. [8] for ChimeraSlayer and also recommended for Uchime [7]), which contains 5,181 total sequences but only 33 archaeal sequences. When we changed to a reference data set containing all 284 parent sequences used in the generation of the artificial chimeras, Uchime's detection rate increased to 76% (data not shown), illustrating that the results are dependent on the reference data set used.
Fig 6.
Comparison of chimera detection by ss_DECIPHER, fs_DECIPHER, ChimeraSlayer, WigeoN (Pintail), and Uchime as a function of sequence length for archaeal chimeras formed by combining two parent sequences at a random breakpoint. The artificial chimera set contained a total of 1,000 chimeras of random length constructed from 284 archaeal type-strain sequences. All sequences were analyzed with ss_DECIPHER, while only sequences > 600 nucleotides long were analyzed with fs_DECIPHER.
Since DECIPHER takes the comprehensive approach of using the entire 16S rRNA database as the reference set of good sequences (after removing detectable chimeras with fs_DECIPHER as discussed previously), detection is not dependent on the scope of the reference data set. Furthermore, the database can be updated as RDP grows in size or if changes are made to the sequences marked as “suspect quality” in RDP. These updates are important to ensure that novel lineages or recently defined genera, currently represented in the database with a small number of sequences, are better represented as the database is populated with additional related sequences.
Comparative abilities of the different chimera-detection methods.
Table 2 presents a qualitative summary of the chimera-finding abilities of the different programs tested. With the recent benchmarks by Haas et al. (8), Edgar et al. (7), and this study, as well as the recent release of ChimeraSlayer, Uchime, and DECIPHER, it is evident that there is currently not a single program capable of accurate detection of all possible types of chimeras. Furthermore, next-generation sequencing produces large data sets of shorter sequences compared with traditional Sanger sequencing, and therefore, the characteristics of data sets to be screened for chimeras are also changing, making earlier chimera-finding algorithms obsolete. For instance, Bellerophon has now been shown to have a rate of false positives that is too high compared to other methods, and both Bellerophon and Pintail have been demonstrated to perform poorly with short sequences (8). ChimeraSlayer was introduced as a method suitable for shorter sequences and optimized to detect chimeras formed from closely related parents, even parents within the same genus, but these advantages were quickly overshadowed by Uchime (7). Furthermore, ChimeraSlayer's development focused only on simple chimeras formed by the combination of two parent sequences, while the developers of Uchime considered the possibility of more complex chimeras, and as a result, Uchime also outperforms ChimeraSlayer in this regard. Our benchmark of DECIPHER confirms the known limitations of earlier algorithms and reveals additional limitations (Table 2). WigeoN (Pintail) is unable to detect chimeras in short sequences (<400 nucleotides) and is inefficient with the midrange sequences between 400 and 800 nucleotides in length. ChimeraSlayer does relatively well with midrange sequences but is inefficient with short sequences. ChimeraSlayer is also inefficient at detecting complex, multiple-parent chimeras, in agreement with the findings of Edgar et al. (7). DECIPHER's simple approach for chimera detection makes it less efficient at detecting chimeras from closely related parents and has limitations when sequences belong to unclassified groups according to the RDP classifier tool. However, the most salient observation in this study is the inefficiency of all the earlier methods at detecting chimeras when the chimeric range is very short (e.g., 30 to 100 nucleotides long), as shown in Fig. 2. This creates a significant limitation in Uchime, particularly when the very short chimeric ranges are in long sequences (Fig. 3). The short-sequence (ss) option of DECIPHER provides the means to detect these chimeras, although with the caveat that DECIPHER does not detect chimeras formed from sequences classified within the same genus. Nevertheless, our analysis of the sequences present in RDP's good-quality database showed that 40.4% of the detected chimeras were formed between sequences from different phyla, and therefore, chimera detection should not be limited to finding chimeras formed from closely related parents.
Table 2.
Qualitative summary of chimera-detection characteristics of the benchmarked programs
| Characteristic | Chimera detection ratea |
||||
|---|---|---|---|---|---|
| fs_DECIPHER | ss_DECIPHER | Uchime | ChimeraSlayer | Pintail (WigeoN) | |
| Detection in short sequences (100<length<400) | + | +++ | ++++ | + | + |
| Detection in midrange sequences (400<length<800) | + | ++++ | +++ | ++ | + |
| Detection in long sequences (length >800) | +++ | ++++ | +++ | ++ | ++ |
| Detection of short chimeric regions | ++ | ++++ | ++ | ++ | + |
| Detection of complex chimeras | ++++ | ++++ | ++++ | + | +++ |
| Detection of chimeras from low-divergence parents | + | + | ++++ | +++ | + |
| Independence from reference data setb | +++ | +++ | ++ | ++ | ++ |
| Low false positives | ++++ | ++ | ++ | +++ | ++ |
++++, very high rate of detection; +++, high rate of detection; ++, low rate of detection; +, very low rate of detection.
DECIPHER depends on the RDP taxonomy, while the other methods depend on the reference data set provided by the user.
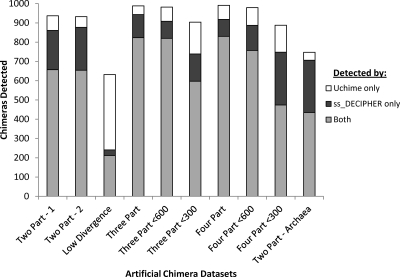
Interestingly, we find that the highest possible rate of chimera detection in our benchmark data sets is achieved simply by combining the chimera-detection advantages of ss_DECIPHER and Uchime, reaching overall detection rates of 89% to 99%, except for the data set of chimeras formed from closely related parents (Fig. 7). The chimeras uniquely detected by ss_DECIPHER mostly corresponded to those with short chimeric ranges, while the ones uniquely detected by Uchime corresponded to those formed from closely related parents and sequences that were indecipherable because they were classified by RDP's classifier tool (17) as unclassified_Bacteria or unclassified_Archaea. Nevertheless, there was surprisingly minimal sequence overlap between the false-positive detections of ss_DECIPHER and Uchime, and therefore, one may expect a higher rate of false-positive detections when using both programs to assess a single set of sequences.
Fig 7.
Comparison of chimera detection with ss_DECIPHER and Uchime for all the artificial chimera sets used in this study. All chimera sets had a total of 1,000 chimeras.
Online DECIPHER tool and standalone implementation.
It is essential not only to identify chimeras present in the public databases but also to prevent more from entering the databases. To this end, DECIPHER has been implemented as a web tool and is publicly available online (http://DECIPHER.cee.wisc.edu). On the website, a user can submit a FASTA file of unaligned or aligned 16S rRNA sequences to be checked for chimeras and select whether the analysis is done with the full-sequence or short-sequence option. DECIPHER results are returned via email. The online tool is limited to submissions of files of less than 10 Mb, which corresponds to approximately 6,500 full sequences (∼1,500 nucleotides long) or 20,000 short sequences (∼500 nucleotides long). In its current implementation on a Dell Precision Workstation T5400 with two Xeon 5405 2.00 GHz processors, 10,000 sequences of approximately 400 nucleotides were processed in 120 min (0.7 s per sequence), and 4,000 full-length sequences (>1,200 nucleotides) were processed in 113 min (1.7 s per sequence).
For users interested in processing much bigger data sets, it is possible to submit their data sets split into multiple files or to use the standalone tool that is available to run in the R programming environment. In the workstation described above, it took approximately 72 h to analyze the entire RDP database of ∼1.2 million sequences (0.25 s per sequence). The database was split into eight separate sections, each with its own processor core, which explains its speed improvement over the sequence sets. Splitting a large sequence set across multiple processes allows parallel processing, albeit without the shared memory configuration that is consistent with true parallelization.
The DECIPHER R package, source code, and supporting documentation and the 16S reference database are all available online (http://DECIPHER.cee.wisc.edu). Sequence classification requires the RDP Multiclassifier tool (17), which is available from the RDP website (http://rdp.cme.msu.edu/classifier/).
With either the online or standalone version, the results file indicates for each detected chimera the start and end positions of the identifying chimeric region along with the corresponding reference group or groups where the chimeric region is commonly found. In its current version, DECIPHER does not provide a confidence evaluation score as found in other chimera detection programs. In some cases, the detected chimeric region corresponds to only one reference group, but it is also common to find chimeric regions associated with multiple reference groups, which is an indication that the chimeric region is conserved in some branches of the phylogenetic tree.
Supplementary Material
ACKNOWLEDGMENTS
Support for this research was provided by the Water Research Foundation (WaterRF Project 4291). The University of Wisconsin—Madison gratefully acknowledges that WaterRF is the joint owner of the technical information upon which this publication is based. The University of Wisconsin—Madison thanks the Water Research Foundation for its financial, technical, and administrative assistance in funding and managing the project through which this information was discovered.
Finally, we thank Daewook Kang for his helpful evaluations of the online tool.
Footnotes
Published ahead of print 18 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aho AV, Corasick MJ. 1975. Efficient string matching—aid to bibliographic search. Commun. ACM 18:333–340 [Google Scholar]
- 2. Amann R, Fuchs BM. 2008. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6:339–348 [DOI] [PubMed] [Google Scholar]
- 3. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cole JR, et al. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 23 June 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics [Epub ahead of print.] doi:10.1093/bioinformatics/btr1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haas BJ, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 10. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pages H, Aboyoun P, Gentleman R, DebRoy S. 2010Biostrings: string objects representing biological sequences, and matching algorithms. R package version 2.16.9 R Foundation for Statistical Computing, Vienna, VA: http://www.R-project.org [Google Scholar]
- 12. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quince C, et al. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641 [DOI] [PubMed] [Google Scholar]
- 14. R Development Core Team 2010R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, VA: http://www.R-project.org [Google Scholar]
- 15. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Wintzingerode F, Gobel UB, Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229 [DOI] [PubMed] [Google Scholar]
- 17. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yarza P, et al. 2008. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.