Abstract
Magnetotactic bacteria (MTB) in the phylum Nitrospirae synthesize up to hundreds of intracellular bullet-shaped magnetite magnetosomes. In the present study, a watermelon-shaped magnetotactic bacterium (designated MWB-1) from Lake Beihai in Beijing, China, was characterized. This uncultivated microbe was identified as a member of the phylum Nitrospirae and represents a novel phylogenetic lineage with ≥6% 16S rRNA gene sequence divergence from all currently described MTB. MWB-1 contained 200 to 300 intracellular bullet-shaped magnetite magnetosomes and showed a helical swimming trajectory under homogeneous magnetic fields; its magnetotactic velocity decreased with increasing field strength, and vice versa. A robust phylogenetic framework for MWB-1 and all currently known MTB in the phylum Nitrospirae was constructed utilizing maximum-likelihood and Bayesian algorithms, which yielded strong evidence that the Nitrospirae MTB could be divided into four well-supported groups. Considering its population densities in sediment and its high numbers of magnetosomes, MWB-1 was estimated to account for more than 10% of the natural remanent magnetization of the surface sediment. Taken together, the results of this study suggest that MTB in the phylum Nitrospirae are more diverse than previously realized and can make important contributions to the sedimentary magnetization in particular environments.
INTRODUCTION
Biomineralization of magnetic minerals has been discovered in a broad range of organisms, including birds, fishes, mollusks, insects, and microorganisms (23, 53, 55). A typical example of biomineralization is found in magnetotactic bacteria (MTB), a morphologically and phylogenetically diverse group of microorganisms that form special intracellular organelles, called magnetosomes (3, 5). Magnetosomes are membrane-enveloped, nano-sized, high-purity crystals of iron oxide magnetite and/or iron sulfide greigite, usually arranged into one or more linear chains (21). These specific organelles help MTB to sense and swim along the earth's magnetic field, a behavior known as magnetotaxis (8). In conjunction with aerotaxis and chemotaxis, magnetotaxis facilitates the location of MTB to their favorable positions in vertical chemical gradients in the oxic-anoxic transition zone (OATZ) (11, 40). The ubiquity and abundance of MTB near the OATZ suggest their potentially important roles in the geochemical cycling of iron and sulfur in nature (48). Molecular approaches based on 16S rRNA gene sequencing have provided evidence for the phylogenetic heterogeneity of MTB. Most discovered MTB are affiliated with the Alphaproteobacteria, but MTB belonging to the Gammaproteobacteria, the Deltaproteobacteria, and Nitrospirae have also been described (2, 28).
One of the most intriguing examples of MTB is “Candidatus Magnetobacterium bavaricum,” a magnetite-producing MTB within the deep-branching bacterial phylum Nitrospirae that was first discovered in Lake Chiemsee in Upper Bavaria, Germany (49, 54). “Ca. Magnetobacterium bavaricum” is a large, rod-shaped bacterium ranging from 8 to 10 μm in length (49). The most distinguishing characteristic of “Ca. Magnetobacterium bavaricum” is its ability to form as many as 1,000 bullet-shaped magnetite magnetosomes arranged into two to six rosette-like bundles of chains, which are parallel to the long axis of the cell (22). Due to its abundance in the microenvironment, “Ca. Magnetobacterium bavaricum” is thought to play important roles in microbial ecology and natural remanent magnetization (NRM) in certain sediment layers (20, 42, 43, 49).
Another well-characterized Nitrospirae MTB is LO-1, recently discovered in southern Nevada (27). LO-1 cells are ovoid, with an average size of 3.5 μm by 2.7 μm. The phylogenetic divergence between LO-1 and “Ca. Magnetobacterium bavaricum” is as high as 9%, based on 16S rRNA gene analysis. Unlike its giant counterpart, LO-1 has only 100 to 200 bullet-shaped magnetite magnetosomes arranged into three braid-like bundles of chains. High-resolution transmission electron microscopy (HRTEM) analysis has revealed that LO-1 cells have a three-layered membrane profile (27).
Besides “Ca. Magnetobacterium bavaricum” and LO-1, several different types of MTB affiliated with the phylum Nitrospirae have also been discovered in various sediments in the Wallersee, Germany (MHB-1) (10), Lake Miyun, China (groups 1 and 2) (29–31, 35), and Great Boiling Springs, Nevada (HSMV-1) (25). In addition, bacteria that are morphologically similar to “Ca. Magnetobacterium bavaricum” and other Nitrospirae MTB have also been found in sediments worldwide, including those in Brazil (36), France (18), Japan (52), and the United States (37), but their phylogenetic affiliations have not been identified yet. These results suggest that MTB of the phylum Nitrospirae are likely globally distributed. However, since attempts to isolate these MTB in pure culture have not been successful, our present knowledge of the diversity and phylogenetic breadth of the Nitrospirae MTB is still limited.
This study presents a detailed analysis of the morphology, phylogenetic position, and swimming behavior of a population of MTB (named MWB-1), affiliated with the phylum Nitrospirae, detected in Lake Beihai in Beijing, China. We additionally utilize maximum-likelihood (ML) and Bayesian methods to perform a robust phylogenetic analysis of all currently known MTB in the phylum Nitrospirae. Furthermore, we make a rough estimate of the potential contribution of MWB-1 magnetite magnetosomes to the NRM of Lake Beihai surface sediments. Our results will provide new insights into the phylogenetic diversity of Nitrospirae MTB and their contributions to surface sedimentary magnetization.
MATERIALS AND METHODS
Sediment collection and MTB enrichment.
Surface sediment (depth, 5 to 10 cm) was collected from Lake Beihai, located in central Beijing, China (39°55′N, 116°23′E), in August 2009. The lake is 1 to 3 m deep and has a surface area of about 0.39 km2. The temperature and pH during the sampling period were 27 to 29°C and 7.44 to 7.49, respectively; the concentration of dissolved oxygen in surface sediment (depth, 5 to 10 cm) ranged from 0.12 to 0.14 mg/liter as determined by using an HQ40d oxygen meter (Hach Company, CO). Collected sediment was quickly transferred to 600-ml plastic flasks, covered with approximately 100 ml of lake water, transported to the laboratory, and incubated at room temperature without disturbance. MTB in the sediment were magnetically enriched using the “MTB trap” method described by us previously (19, 32, 33). The collected MTB cells were washed twice and were then resuspended in sterile distilled water.
Light microscopy and TEM analyses.
A drop of sediment (about 30 μl) was placed on a glass coverslip to check the presence of MTB by using the hanging-drop method (13) under an Olympus light microscope (Research Microscope BX51) at magnifications of ×400 and ×1,000. Because of their unique morphology (see Fig. 1A), the number of MWB-1 cells could be counted directly by the method described by Flies et al. (9). For transmission electron microscopy (TEM) observation, energy-dispersive X-ray (EDX) analysis, and high-resolution TEM (HRTEM) analysis, a 20-μl drop of a magnetic enrichment reagent was deposited on a Formvar-carbon-coated grid and was allowed to air dry. The grid was rinsed with sterile distilled water and was then observed under a JEM-2100 transmission electron microscope with an accelerating voltage of 200 kV. Digital Micrograph software (Gatan, Pleasanton, CA) was used for image processing (filtering of HRTEM images) and for obtaining fast Fourier transform (FFT) patterns. For the observation of flagella, the air-dried cells were washed with sterile distilled water, stained with 1% aqueous uranyl acetate for 1 min, and examined under the JEM-2100 TEM.
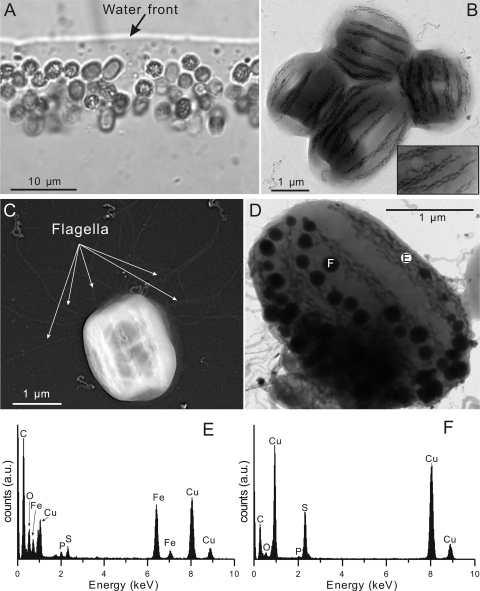
Fig 1.
(A to D) Morphology of MWB-1 cells from Lake Beihai as revealed by light microscopy (A) and transmission electron microscopy (B to D). The image in panel C is inverted. (E and F) Energy-dispersive X-ray analysis of a bullet-shaped magnetosome crystal (E) and an electron-dense granule (F) in the MWB-1 cell shown in panel D. The carbon, phosphor, and copper in the spectra are from the cellular background and the copper mesh grid, which is covered with carbon film.
PCR, cloning, and DNA sequencing.
Nearly complete 16S rRNA genes were amplified directly from magnetically enriched MTB cells in which MWB-1 was dominant by using the universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) as described previously (31, 34). PCR was performed using a T-Gradient thermocycler (Whatman Biometra, Göttingen, Germany). The PCR amplification program consisted of 5 min at 95°C; 30 cycles of 1.5 min at 92°C, 1 min at 50°C, and 2 min at 72°C; and a final extension at 72°C for 10 min. The PCR product was purified by 0.8% (wt/vol) agarose gel electrophoresis.
The purified PCR product was cloned into the pMD19-T vector and chemically competent DH5α cells (both from TaKaRa, Dalian, China) by following the manufacturer's instructions. The transformed cells were incubated overnight on Luria-Bertani agar plates with ampicillin. Clones were randomly selected and were sequenced using an ABI 3730 genetic analyzer (Beijing Genomics Institute, Beijing, China).
Phylogenetic analysis.
The resulting sequences were checked for chimera formation with the Bellerophon server (17) before being compared with the GenBank database (4) by using the BLAST program to search for related sequences with high similarities. The sequences retrieved in this study, together with all 13 currently published 16S rRNA gene sequences of MTB in the phylum Nitrospirae, 8 Alphaproteobacteria MTB 16S rRNA gene sequences, and the 16S rRNA gene sequence of 1 non-MTB bacterium from the phylum Nitrospirae, were aligned using MUSCLE (7). Gblocks was then used to eliminate poorly aligned and noisy portions of the alignment (6). The cured alignment consisted of 1,239 positions. Phylogenetic trees were constructed utilizing maximum-likelihood (ML) and Bayesian methods. For model-based phylogenetic analyses, evolutionary models of substitution were evaluated by MODELTEST (46), and the general time-reversible (GTR) model incorporating a proportion of invariant sites and a gamma distribution (GTR+I+G) substitution matrix was chosen as the best substitution model. ML phylogenetic analysis was performed by PhyML, version 3.0 (14), and was bootstrapped using 100 replicates. The Bayesian tree was constructed using MrBayes, version 3.1.1 (47), with two MrBayes runs using 1,000,000 Markov chain Monte Carlo samples every 100 generations, and the first 2,500 trees were discarded as burn-in.
FISH.
Fluorescence in situ hybridization (FISH) analysis was carried out according to the work of Pernthaler et al. (44) with some modifications. Briefly, 20 μl of MTB enrichment fluid was placed directly on an agarose-coated (0.02%) welled slide and was air dried. The sample was fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 30 min and was then dehydrated for 3 min each in 50%, 80%, and 100% ethanol. A Cy3-labeled probe specific for most Nitrospirae MTB (5′-GCCATCCCCTCGCTTACT-3′; named BaP in this study) (30, 49) and the 6-carboxyfluorescein (FAM)-labeled universal bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) (1) were used for hybridization, which was performed at 46°C for 3 h. After washing for 20 min at 48°C, the sample was counterstained with 10 μl of a 1-μg/ml 4′,6-diamidino-2-phenylindole (DAPI) solution for 3 min, rinsed with distilled water, air dried, and observed with a BX51 fluorescence microscope (Olympus Optical, Tokyo, Japan).
Analysis of swimming behavior.
The swimming behavior of MWB-1 cells in rotating magnetic fields was observed in the Bacteriodrome (Petersen Instruments, Munich, Germany), a setup of two pairs of computer-controlled Helmholtz coils oriented perpendicularly to each other to generate a homogeneous magnetic field rotating in the horizontal plane at an adjustable intensity (45). The Bacteriodrome permitted quick and accurate calculation of the swimming speed and behavior of a single MTB cell (40, 41, 45). Rotating homogeneous magnetic fields (from 0.2 to 1.4 mT) were applied, and the swimming trajectories of MWB-1 cells were subsequently recorded by a charge-coupled device camera (frame rate, 24 frames per second [fps]). The magnetotactic velocity (VM) of bacteria was calculated as πD/T, where D is the diameter of the circular swimming path and T is the period of one cycle (40, 45).
NRM measurement.
Surface sediment (depth, 5 to 10 cm) from Lake Beihai was air dried, and its NRM values were measured by using a superconducting rock magnetometer, model 760 (2G Enterprises, Mountain View, CA). The average NRM value was determined from three replicates.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in GenBank under accession no. JN630580.
RESULTS
Cell morphology and magnetosomes.
A light microscope showed the presence of a peculiar population of large, watermelon-shaped MTB (about 2.1 × 103 cells/ml) and small magnetotactic cocci (about 5.6 × 102 cells/ml) in the sediments from Lake Beihai, Beijing, China (Fig. 1A). The watermelon-shaped bacteria had an average length of 2.8 μm and an average width of 2.4 μm (Fig. 1B). The cell had multiple flagella originating from a single pole on the bacterial surface, as shown by TEM observation (Fig. 1C). MWB-1 formed 4 to 7 bundles of magnetosome chains that were parallel to the long axis of the cell. Interestingly, the magnetosome chains were slightly curved along the cell envelope (Fig. 1B), indicating their close proximity to the inner cytoplasmic envelope, a pattern similar to the magnetosome architecture in “Ca. Magnetobacterium bavaricum” (22). Each chain was composed of 2 to 5 twisted strands of bullet-shaped magnetosomes, the tips of which were not always consistently oriented and were sometimes pointed in the opposite direction (Fig. 1B, inset). There were 200 to 300 magnetosomes per cell, and their average length and width were about 116 and 40 nm, respectively. EDX analysis of magnetosomes revealed that they were composed of iron and oxygen (Fig. 1E), which formed iron oxide. Besides magnetosomes, about 37% of identified MWB-1 cells contained more than 30 electron-dense granules with diameters between 100 and 250 nm (Fig. 1D). EDX analysis indicated that these granules were mainly composed of sulfur (Fig. 1F).
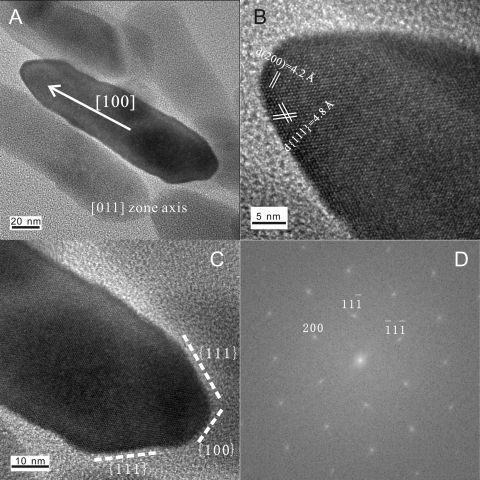
Further observation by HRTEM revealed that the mineral phase of MWB-1 magnetosomes was magnetite. As shown in Fig. 2, the measured d spacings of magnetosomes were 4.8, 4.2, 2.9, and 2.5 Å, corresponding to planes 111, 200, 220, and 311 of face-centered cubic magnetite. Low-temperature magnetic remanence measurement has indicated that the Verwey transition temperature of MWB-1 was 112 K, which was similar to that of other bacterial magnetite (usually between 86 and 117 K) and further confirmed the magnetite composition of MWB-1 magnetosomes (data not shown). In addition, mature MWB-1 magnetosome crystals were highly elongated, and their final elongation direction was along the [100] crystal direction (Fig. 2), similar to the morphology of the magnetosomes in LO-1 and another “Ca. Magnetobacterium bavaricum”-like bacterium (27, 29).
Fig 2.
Typical high-resolution transmission electron microscopy images (A to C) and the fast Fourier transform pattern (D) of the same magnetosome crystal recorded from the [011] zone axis of magnetite.
FISH and phylogenetic analysis.
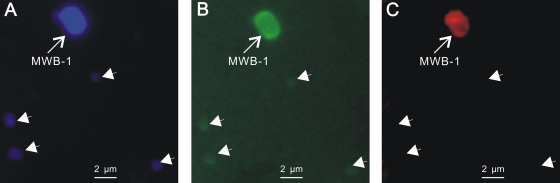
The phylogenetic affiliation of MWB-1 was determined after amplification and sequencing of the 16S rRNA gene sequence. A BLAST search of a public database indicated that the sequence retrieved in this study was affiliated with the phylum Nitrospirae and was only 94% similar to the uncultivated Nitrospirae MTB LO-1 (GenBank accession no. GU979422) (27). FISH analysis was then performed to confirm that the newly identified organism MWB-1 was associated with the phylum Nitrospirae. An MTB enrichment culture containing MWB-1 was stained with DAPI and was hybridized with the universal bacterial probe EUB338 (1) and the specific probe BaP (30, 49). As shown in Fig. 3, only MWB-1 could be hybridized with the “Ca. Magnetobacterium bavaricum”-specific probe.
Fig 3.
Specific detection of MWB-1 associated within the phylum Nitrospirae through FISH. The same microscopic field is shown after staining with DAPI (A), after hybridization with the FAM-labeled universal bacterial probe EUB338 (B), and after hybridization with the Cy3-labeled specific probe BaP (C). Only MWB-1 (indicated by arrows) hybridized with the specific probe; the other cells (indicated by arrowheads) were not stained.
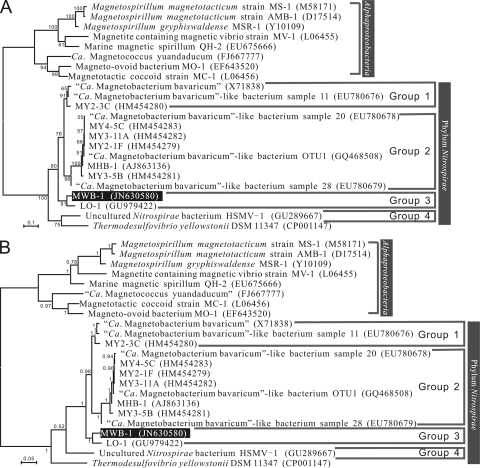
A robust phylogenetic analysis of MWB-1 and all currently known Nitrospirae MTB was performed using both the ML algorithm and a Bayesian approach. The ML and Bayesian trees were similar, and both revealed that the MWB-1 sequence formed a monophyletic group together with LO-1 (Fig. 4). In addition, four well-supported groups of Nitrospirae MTB were consistently formed in the ML and Bayesian trees, the reliability of which is indicated by the high bootstrap support and high Bayesian posterior probability values (Fig. 4A and B).
Fig 4.
Maximum-likelihood phylogenetic tree (A) and Bayesian phylogenetic tree (B) of 16S rRNA gene sequences showing the relationship between MWB-1 and closely related magnetotactic bacteria. The GTR+I+G evolutionary model of substitution was used. Note that the topology of the ML analysis was congruent with that of the Bayesian tree. Numbers at nodes are ML bootstrap proportions (A) or posterior probability values (B). GenBank accession numbers are given in parentheses.
Magnetotactic swimming behavior.
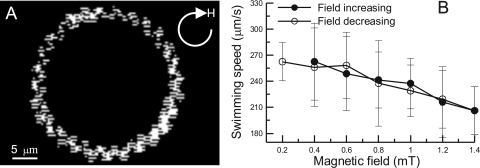
In contrast to other bacteria, MTB swim along magnetic fields due to their intracellular magnetosome chains. Therefore, magnetotactic swimming behavior has been used to characterize MTB (13, 15, 40, 41, 45, 51). MWB-1 swam along the direction of an applied magnetic field and displayed a north-seeking magnetotactic behavior. Bacteriodrome analysis of MWB-1 revealed a circular and helical trajectory under a homogeneous rotating field (Fig. 5A; see also movie S1 in the supplemental material). On the basis of the work of Pan et al. (40), we have calculated the magnetotactic swimming velocity (VM; parallel to the magnetic field line) of MWB-1 cells. Interestingly, the VM of MWB-1 was not constant but decreased as the strength of the magnetic field increased, and this decrease in VM was nearly reversible after the strength of the magnetic field was decreased (Fig. 5B).
Fig 5.
(A) Typical observed swimming trajectory of MWB-1 in a homogeneous rotating magnetic field. (B) Magnetotactic swimming velocities of MWB-1 versus magnetic-field strength.
DISCUSSION
This study describes a novel population of uncultivated MTB, MWB-1, which has 200 to 300 bullet-shaped magnetite magnetosomes arranged in 4 to 7 bundles of chains. Its phylogenetic position in the phylum Nitrospirae was confirmed by 16S rRNA gene amplification and FISH analysis.
The cells of MWB-1 were morphologically similar to those of Nitrospirae MTB LO-1, discovered in the United States (27). Close inspection, however, reveals several differences. For example, the average size of MWB-1 cells is smaller than that of LO-1 cells (2.8 by 2.4 μm for MWB-1 versus 3.5 by 2.7 μm for LO-1), but the average number of magnetosomes and the number of chain bundles per MWB-1 cell are much higher than those in LO-1 cells (200 to 300 magnetosomes arranged in 4 to 7 bundles of chains for MWB-1 versus 100 to 200 magnetosomes usually arranged in 3 bundles of chains for LO-1). In addition, the magnetotactic swimming speeds of MWB-1 cells (as high as 260 μm/s) are much faster than those of LO-1 cells (average, 116 μm/s). Moreover, the phylogenetic divergence between MWB-1 and LO-1 is as high as 6%, greater than the cutoff point (3%) for species definition (50), suggesting that MWB-1 likely represents a novel species in the phylum Nitrospirae.
MTB in the phylum Nitrospirae were originally described as comprising only “Ca. Magnetobacterium bavaricum” (49) but have recently been recognized to contain bacterial species showing a broad range of morphological properties and with high phylogenetic diversity (10, 25, 27, 30, 31). However, no thorough phylogenetic analysis of the Nitrospirae MTB considering all known sequences has been conducted yet. Here we have used ML and Bayesian algorithms to construct phylogenetic trees, which are more rigorous than distance-based methods (i.e., neighbor joining), in order to perform an accurate phylogenetic reconstruction of all known Nitrospirae MTB. Both ML and Bayesian analyses consistently reveal that all currently known MTB sequences in the phylum Nitrospirae can be divided into four major evolutionary groups (Fig. 4). “Ca. Magnetobacterium bavaricum” (49) and two clones from Lake Miyun sediment in China (30, 31) are found to constitute group 1, with giant rod-shaped cells. The second group (group 2) includes MHB-1 from Germany (10) and seven sequences from magnetic collections from China (30, 31). As revealed by FISH, group 2 includes rod-shaped and coccoid-to-ovoid bacteria that are smaller than the bacteria in group 1. The newly identified bacterium MWB-1 forms the third phylogenetic group (group 3) together with LO-1 (27). Moreover, a moderately thermophilic bacterium from the United States, HSMV-1, which synthesizes only a single magnetosome chain, forms the fourth group owing to its low identity to other MTB (<90%) (25). This robust phylogeny has several important implications for the diversity and evolution of the Nitrospirae MTB: (i) these bacteria, especially those in groups 1, 2, and 3, are globally distributed; (ii) the morphotypes of MTB in distinct lineages are apparently different; and (iii) MTB within the phylum Nitrospirae show considerable phylogenetic diversity.
The swimming velocity of MWB-1 can reach 260 μm/s under a magnetic field with a strength of 0.2 mT (Fig. 5B), making them faster than most described MTB (12). High velocity is believed to help bacteria locate the nutrient resources in a microenvironment quickly (38). MWB-1 exhibits a helical trajectory similar to those of Alphaproteobacteria magnetotactic cocci harboring a single magnetosome chain (26, 39, 40) and LO-1 in the Nitrospirae (27). The helical trajectories of magnetotactic cocci are due to the inclination between the magnetosome chain and the flagellar propulsion axis (40). We thus hypothesize that for MWB-1, the same thing is happening; that is, although MWB-1 harbors several magnetosome chains, the total magnetic moment of a single MWB-1 cell is not colinear with the flagellar propulsion axis. This lack of collineation additionally causes the inverse correlation between the VM of MWB-1 and the strength of applied homogeneous magnetic fields (Fig. 5B), as suggested by Pan et al. (40). Our result provides additional evidence to support the notion that, for particular MTB, magnetotactic efficiency may be evolutionarily optimized under the earth's magnetic field and will be reduced under higher magnetic fields (40). In addition, this property is phylogeny independent and is probably widely spread in different phylogenetic groups of MTB.
Stable single-domain magnetite crystals of MTB are thought to play important roles in sedimentary magnetization in various environments, including lacustrine, microbial mat, hemipelagic, pelagic, and carbonate platforms (24). Due to its relatively high population densities and its high numbers of intracellular magnetosomes, MWB-1 may make potentially important contributions to the magnetization of surface sediments. The cell magnetization of MWB-1, calculated from the volume of magnetite magnetosomes (116 nm by 40 nm by 40 nm, for an estimated 200 to 300 magnetosomes) and the saturation magnetization value (480 electromagnetic units [emu]/cm3 for magnetite), is approximately 1.8 × 10−11 to 2.7 × 10−11 emu/cell. Given the average number of about 2.1 × 103 cells/ml for living MWB-1 as revealed by the hanging-drop method, we can estimate that the total magnetic contribution of MWB-1 to sediments is on the order of 10−8 emu/ml. The average NRM of surface sediment was determined to be 2.2 × 10−7 emu/ml. Therefore, bacterial magnetite from living MWB-1 alone accounts for more than 10% of the NRM of this sediment layer. If we further consider the cumulative succession of bacterial populations and the fossil magnetosomes preserved in sediments, the contribution calculated above is likely to be an underestimate. Therefore, taking our findings together with those of other studies (15, 16, 42), it is reasonable to suggest that, besides abiogenic magnetic minerals, magnetite magnetosomes formed by MTB, such as MWB-1, are important carriers of NRM in freshwater surface sediments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yinzhao Wang and Wenfang Wu for assistance in the laboratory. We also acknowledge Bi Li and Haitao Chen for help with field sampling. We are very grateful to three anonymous reviewers for their valuable comments.
This work was supported by the CAS/SAFEA International Partnership Program for Creative Research Teams (KZCX2-YW-T10), the CAS project, and NSFC grant 40821091.
Footnotes
Published ahead of print 23 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Amann R, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann R, Peplies J, Schüler D. 2007. Diversity and taxonomy of magnetotactic bacteria, p 25–36 In Schüler D. (ed), Magnetoreception and magnetosomes in bacteria. Springer, Berlin, Germany [Google Scholar]
- 3. Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217–230 [DOI] [PubMed] [Google Scholar]
- 4. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2005. Genbank. Nucleic Acids Res. 33:D34–D38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blakemore RP. 1975. Magnetotactic bacteria. Science 190:377–379 [DOI] [PubMed] [Google Scholar]
- 6. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 7. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875–4898 [DOI] [PubMed] [Google Scholar]
- 9. Flies CB, et al. 2005. Diversity and vertical distribution of magnetotactic bacteria along chemical gradients in freshwater microcosms. FEMS Microbiol. Ecol. 52:185–195 [DOI] [PubMed] [Google Scholar]
- 10. Flies CB, Peplies J, Schüler D. 2005. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71:2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frankel RB, Williams TJ, Bazylinski DA. 2007. Magneto-aerotaxis, p 1–24 In Schüler D. Magnetoreception and magnetosomes in bacteria. Springer, Berlin, Germany [Google Scholar]
- 13. Greenberg M, Canter K, Mahler I, Tornheim A. 2005. Observation of magnetoreceptive behavior in a multicellular magnetotactic prokaryote in higher than geomagnetic fields. Biophys. J. 88:1496–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 15. Hanzlik M, Winklhofer M, Petersen N. 2002. Pulsed-field-remanence measurements on individual magnetotactic bacteria. J. Magn. Magn. Mater. 248:258–267 [Google Scholar]
- 16. Hanzlik M, Winklhofer M, Petersen N. 1996. Spatial arrangement of chains of magnetosomes in magnetotactic bacteria. Earth Planet. Sci. Lett. 145:125–134 [Google Scholar]
- 17. Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 18. Isambert A, Menguy N, Larquet E, Guyot F, Valet JP. 2007. Transmission electron microscopy study of magnetites in a freshwater population of magnetotactic bacteria. Am. Miner. 92:621–630 [Google Scholar]
- 19. Jogler C, et al. 2009. Towards cloning the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75:3972–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jogler C, et al. 2010. Cultivation-independent characterization of “Candidatus Magnetobacterium bavaricum” via ultrastructural, geochemical, ecological and metagenomic methods. Environ. Microbiol. 12:2466–2478 [DOI] [PubMed] [Google Scholar]
- 21. Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501–521 [DOI] [PubMed] [Google Scholar]
- 22. Jogler C, et al. 2011. Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum. Proc. Natl. Acad. Sci. U. S. A. 108:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirschvink JL. 1989. Magnetite biomineralization and geomagnetic sensitivity in higher animals: an update and recommendations for future study. Bioelectromagnetics 10:239–259 [DOI] [PubMed] [Google Scholar]
- 24. Kopp RE, Kirschvink JL. 2008. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth Sci. Rev. 86:42–61 [Google Scholar]
- 25. Lefèvre CT, et al. 2010. Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl. Environ. Microbiol. 76:3740–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lefèvre CT, Bernadac A, Yu-Zhang K, Pradel N, Wu L. 2009. Isolation and characterization of a magnetotactic bacteria from the Mediterranean Sea. Environ. Microbiol. 11:1646–1657 [DOI] [PubMed] [Google Scholar]
- 27. Lefèvre CT, Frankel RB, Abreu F, Lins U, Bazylinski DA. 2011. Culture-independent characterization of a novel, uncultivated magnetotactic member of the Nitrospirae phylum. Environ. Microbiol. 13:538–549 [DOI] [PubMed] [Google Scholar]
- 28. Lefèvre CT, et al. 2011. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. doi:10.1038/ismej.2011.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li J, et al. 2010. Biomineralization, crystallography and magnetic properties of bullet-shaped magnetite magnetosomes in giant rod magnetotactic bacteria. Earth Planet. Sci. Lett. 293:368–376 [Google Scholar]
- 30. Lin W, Jogler C, Schüler D, Pan Y. 2011. Metagenomic analysis reveals unexpected subgenomic diversity of magnetotactic bacteria within the phylum Nitrospirae. Appl. Environ. Microbiol. 77:323–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin W, Li J, Schüler D, Jogler C, Pan Y. 2009. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Syst. Appl. Microbiol. 32:342–350 [DOI] [PubMed] [Google Scholar]
- 32. Lin W, Pan Y. 2010. Temporal variation of magnetotactic bacterial communities in two freshwater sediment microcosms. FEMS Microbiol. Lett. 302:85–92 [DOI] [PubMed] [Google Scholar]
- 33. Lin W, Pan Y. 2009. Uncultivated magnetotactic cocci from Yuandadu Park in Beijing, China. Appl. Environ. Microbiol. 75:4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin W, Tian L, Li J, Pan Y. 2008. Does capillary racetrack-based enrichment reflect the diversity of uncultivated magnetotactic cocci in environmental samples? FEMS Microbiol. Lett. 279:202–206 [DOI] [PubMed] [Google Scholar]
- 35. Lin W, Wang Y, Li B, Pan Y. 2011. A biogeographic distribution of magnetotactic bacteria influenced by salinity. ISME J. doi:10.1038/ismej.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lins U, Freitas F, Keim CN, Farina M. 2000. Electron spectroscopic imaging of magnetotactic bacteria: magnetosome morphology and diversity. Microsc. Microanal. 6:463–470 [DOI] [PubMed] [Google Scholar]
- 37. Mann S, Sparks NHC, Blakemore RP. 1987. Ultrastructure and characterization of anisotropic magnetic inclusions in magnetotactic bacteria. Proc. R. Soc. Lond. Ser. B Biol. Sci. 231:469–476 [Google Scholar]
- 38. Mitchell JG, Kogure K. 2006. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 55:3–16 [DOI] [PubMed] [Google Scholar]
- 39. Nogueira FS, Lins de Barros HGP. 1995. Study of the motion of magnetotactic bacteria. Eur. Biophys. J. 24:13–21 [Google Scholar]
- 40. Pan Y, et al. 2009. Reduced efficiency of magnetotaxis in magnetotactic coccoid bacteria in higher than geomagnetic fields. Biophys. J. 97:986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan Y, Lin W, Tian L, Zhu R, Petersen N. 2009. Combined approaches for characterization of an uncultivated magnetotactic coccus from Lake Miyun near Beijing. Geomicrobiol. J. 26:313–320 [Google Scholar]
- 42. Pan Y, et al. 2005. The detection of bacterial magnetite in recent sediments of Lake Chiemsee (southern Germany). Earth Planet. Sci. Lett. 232:109–123 [Google Scholar]
- 43. Pan Y, et al. 2005. Rock magnetic properties of uncultured magnetotactic bacteria. Earth Planet. Sci. Lett. 237:311–325 [Google Scholar]
- 44. Pernthaler J, Glockner FO, Schonhuber W, Amann R. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207–226 [Google Scholar]
- 45. Petersen N, Weiss DG, Vali H. 1989. Magnetic bacteria in lake sediments, p 231–241 In Lowes F. (ed), Geomagnetism and palaeomagnetism. NATO ASI Ser. C, no. 261. Kluwer, Dordrecht, The Netherlands [Google Scholar]
- 46. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 47. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 48. Simmons SL, Edwards KJ. 2007. Geobiology of magnetotactic bacteria, p 77–102 In Schüler D. Magnetoreception and magnetosomes in bacteria. Springer, Berlin, Germany [Google Scholar]
- 49. Spring S, et al. 1993. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Appl. Environ. Microbiol. 59:2397–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849 [Google Scholar]
- 51. Steinberger B, Petersen N, Petermann H, Weiss DG. 1994. Movement of magnetic bacteria in time-varying magnetic-fields. J. Fluid Mech. 273:189–211 [Google Scholar]
- 52. Thornhill RH, Burgess JG, Sakaguchi T, Matsunaga T. 1994. A morphological classification of bacteria containing bullet-shaped magnetic particles. FEMS Microbiol. Lett. 115:169–176 [Google Scholar]
- 53. Tian L, Lin W, Zhang S, Pan Y. 2010. Bat head contains soft magnetic particles: evidence from magnetism. Bioelectromagnetics 31:499–503 [DOI] [PubMed] [Google Scholar]
- 54. Vali H, Forster O, Amarantidis G, Petersen N. 1987. Magnetotactic bacteria and their magnetofossils in sediments. Earth Planet. Sci. Lett. 86:389–400 [Google Scholar]
- 55. Winklhofer M. 2010. Magnetoreception. J. R. Soc. Interface 7:S131–S134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.