Abstract
On the basis of pIP501, a green fluorescent protein (GFP)-tagged monitoring tool was constructed for quantifying plasmid mobilization among Gram-positive bacteria and between Gram-positive Enterococcus faecalis and Gram-negative Escherichia coli. Furthermore, retromobilization of the GFP-tagged monitoring tool was shown from E. faecalis OG1X into the clinical isolate E. faecalis T9.
TEXT
The mechanisms of conjugative transfer in Gram-negative bacteria are fairly well understood (e.g., see references 2, 9, 18, 33, 36, and 41), whereas conjugation in Gram-positive bacteria has been studied in greater detail for only the last decade leading to the first model of a type IV secretion-like system in Gram-positive bacteria (1, 2, 16, 17, 41). The antibiotic resistance plasmid pIP501 from Streptococcus agalactiae has a very broad host range for conjugative plasmid transfer and mobilization. Its host range includes virtually all tested Gram-positive bacteria, including the multicellular filamentous streptomycetes and Gram-negative Escherichia coli (17, 24).
For Gram-positive systems, molecular tools for in situ detection of horizontal gene transfer by conjugation are still very limited in contrast to Gram-negative systems (4, 7, 8, 29, 37, 38). Nieto and Espinosa (30) have constructed a green fluorescent protein (GFP)-tagged derivative of plasmid pMV158 from Streptococcus pneumoniae, pMV158GFP, that was shown to be mobilizable to different low-GC Gram-positive bacteria like Enterococcus faecalis and Lactococcus lactis. Lorenzo-Díaz and Espinosa applied pMV158GFP to intra- and interspecies mobilization between different Gram-positive bacteria in large-scale filter mating assays (26).
Recently, Babic and coworkers (3) demonstrated conjugative transfer of the integrative and conjugative element ICEBs1 from Bacillus subtilis donor cells to B. subtilis recipient cells in real time using a lacO or LacI-GFP system for visualization of transfer events.
Here, we report the construction and mobilization of a GFP-tagged mobilizable plasmid based on the pIP501 tra region to monitor horizontal gene transfer between Gram-positive bacteria and between Gram-positive and Gram-negative bacteria by the formation of a green fluorescent phenotype in transconjugants. The mobilizable plasmid is based on a nisin-inducible expression system (NICE) and replicates in both Gram-positive and Gram-negative bacteria.
Plasmid construction.
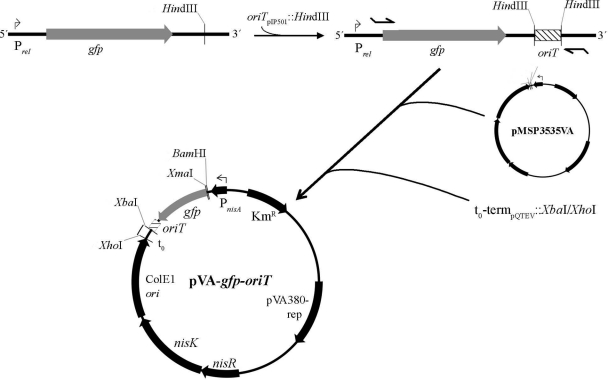
The oriT region from the broad-host-range plasmid pIP501 (oriTpIP50) was subcloned with primer pair oriT-HindIII-fw (fw stands for forward) and oriT-HindIII-re (re stands for reverse) (see Table S1 in supplemental material) via HindIII into plasmid pJPrelGFP encoding a gfp gene improved for expression in prokaryotes (28, 31). All bacterial strains and plasmids used in this work are described in Table 1. The gfp-oriTpIP501 cassette was inserted into the E. coli shuttle plasmid pMSP3535VA (6) via XmaI/XbaI with primer pair Prel-gfp-XmaI-fw and oriT-XbaI-re under the control of a nisin-inducible nisA promoter. Then, the λ phage t0 terminator was cloned downstream of the oriT region via XbaI and XhoI sites with primer pair t0-term-XbaI-fw and t0-term-XbaI-re using the expression vector pQTEV (35) as the template, thus generating plasmid pVA-gfp-oriT (Fig. 1).
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli XL10 | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10(Tetr) Amy Cmr] | Stratagene |
| Enterococcus faecalis | ||
| JH2-2 | Rifr Fusr | 23 |
| OG1RF | Rifr Fusr | 10 |
| OG1X | Smr | 22 |
| T9 | Tetr | 21 |
| Bacillus subtilis subsp. natto DSM 4451 | Smr | DSMZb |
| Plasmids | ||
| pIP501 | Tra Cmr MLSr | 12 |
| pMSP3535VA | pVA380-1 and ColE1 replicons nisRK PnisA Kmr | 6 |
| pJPrelGFP | Prel gfp | 31 |
| pQTEV | Pt4lacIq His7 Ampr | 35 |
| pJPrelGFP-oriT | pJPrelGFP oriTpIP501 | This work |
| pVA-gfp-oriT | pMSP3535VA gfp oriTpIP501 t0-termpQTEV | This work |
MLSr, macrolide-lincosamide-streptogramin B resistance; Hte, high transformation efficiency; Amy, amylase.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen (German Collection of Microorganisms and Cell Cultures).
Fig 1.
The pIP501 oriT region was subcloned into plasmid pJPrelGFP via HindIII. The gfp-oriT cassette was then cloned into plasmid pMSP3535VA via XmaI and XbaI sites, followed by insertion of the t0 termination sequence via XbaI and XhoI, generating pVA-gfp-oriT.
Assessment of green fluorescence by FACS.
The fluorescence of E. faecalis OG1X(pVA-gfp-oriT) was quantified by fluorescence-activated cell sorting (FACS) (FACScan flow cytometer; BD Biosciences, Heidelberg, Germany) after induction of the nisA promoter with 100 ng · ml−1 nisin on brain heart infusion (BHI) (Condalab, Madrid, Spain) agar plates supplemented with streptomycin (1,000 μg · ml−1) and kanamycin (2,000 μg · ml−1). More than 95% of the analyzed cells were shifted to a green fluorescent phenotype, indicating efficient induction of the nisA promoter and expression of GFP with the plasmid construct (data not shown).
pIP501-mediated mobilization of the GFP-tagged plasmid to E. faecalis and B. subtilis.
pVA-gfp-oriT was tested for its ability to be mobilized by pIP501 in filter matings. The donor E. faecalis OG1X harboring pIP501 and pVA-gfp-oriT and the recipient E. faecalis JH2-2 were grown to an optical density at 600 nm (OD600) of 0.5, mixed in a 1:10 ratio, and passed through a sterile nitrocellulose membrane filter (0.45 μm) (Millipore, Schwalbach, Germany). After overnight incubation on BHI agar at 37°C, the cells were recovered in 1 ml phosphate-buffered saline (PBS), and serial dilutions were plated on BHI agar with kanamycin (400 μg · ml−1) and/or fusidic acid (50 μg · ml−1) to enumerate transconjugants and recipients, respectively. The mean mobilization rate from three independent experiments was 1.42 · 10−5 ± 3.11 · 10−6 transconjugants per recipient cell. Mobilization rates were approximately three times lower than pIP501 transfer rates in experiments under the same conditions, suggesting a possible cis-acting preference of the Orf1 relaxase in agreement with observations made for the TraA relaxase of plasmid pRetCF2d and relaxase Orf28 of the conjugative transposon Tn1549 (32, 40).
Transconjugants were verified by green fluorescent phenotype after induction of the nisA promoter on BHI agar supplemented with kanamycin (400 μg · ml−1), fusidic acid (50 μg · ml−1), and nisin (100 ng · ml−1) and amplification of the gfp gene by PCR with primer pair Prel-gfp-XmaI-fw and oriT-XbaI-re (data not shown).
Since pIP501 can be transferred and stably maintained in various Gram-positive bacteria, we tried to mobilize pVA-gfp-oriT to B. subtilis in a triparental mating with the help of pIP501: E. faecalis JH2-2(pVA-gfp-oriT), E. faecalis OG1RF(pIP501), and B. subtilis subsp. natto DSM 4451 were grown to an OD600 of 0.5 and mated overnight at 30°C in a ratio of 1:1:10. Serial dilutions were plated on Luria-Bertani agar supplemented with kanamycin (20 μg · ml−1) and/or streptomycin (1,000 μg · ml−1) to enumerate transconjugants and recipients, respectively. The mean mobilization rate obtained from three independent experiments was 4.10 · 10−6 ± 4.74 · 10−7 transconjugants per recipient. Thus, the mobilization rates are similar to those obtained for mobilization of pMV158 from E. faecalis OG1X to B. subtilis MB46 SL601 by pIP501 (24). Mobilization of pVA-gfp-oriT was verified by a green fluorescent phenotype for B. subtilis subsp. natto DSM 4451(pVA-gfp-oriT) (not shown) after induction of the nisA promoter on LB agar supplemented with kanamycin (20 μg · ml−1), streptomycin (1,000 μg · ml−1), and nisin (100 μg · ml−1) and amplification of the gfp gene by PCR (data not shown).
Interestingly, even after induction of the nisA promoter with different nisin concentrations (10, 100, and 1,000 ng · ml−1), only 5% of the Bacillus transconjugants expressed a green fluorescent phenotype. It has been previously demonstrated that the NICE system can be used for efficient inducible gene expression in Gram-positive bacteria like Lactococcus, Bacillus, and Enterococcus (6, 11, 27). However, Hirt and coworkers (20) demonstrated weak expression of the pCF10 encoded surface protein PrgB with plasmid pMSP3535 in B. subtilis. This finding together with our results leads to the hypothesis that the NICE vectors pMSP3535 and pMSP3535VA might not be suitable for efficient gene expression in Bacillus species. Thus, GFP expression levels in our case might not have reached a certain threshold in most B. subtilis cells that is necessary for developing a green fluorescent phenotype (15).
pIP501-mediated mobilization from the Gram-positive E. faecalis to the Gram-negative E. coli.
pVA-gfp-oriT was shown to be suited to monitor the transfer of plasmid from the Gram-positive E. faecalis to the Gram-negative E. coli by formation of a green fluorescent phenotype in an E. coli recipient. The donor E. faecalis OG1X(pVA-gfp-oriT), the helper E. faecalis OG1RF(pIP501), and the recipient E. coli XL10 were grown to an OD600 of 0.5 and mated overnight at 37°C in a ratio of 1:1:10. Serial dilutions were plated on Luria-Bertani agar supplemented with kanamycin (50 μg · ml−1) and/or tetracycline (10 μg · ml−1) to enumerate transconjugants and recipients, respectively. pIP501-mediated mobilization of pVA-gfp-oriT from E. faecalis OG1X to E. coli XL10 occurred with a rather low frequency of 2.30 · 10−8 ± 2.07 · 10−8 transconjugants per recipient cell. However, the nisA promoter in E. coli was functional, leading to a green fluorescent phenotype in approximately 95% of the induced E. coli cells (data not shown). Trieu-Cuot and coworkers (39) showed mobilization of shuttle vectors containing the RK2 oriT region from Gram-negative E. coli to Gram-positive E. faecalis with mobilization frequencies in the same range.
Retromobilization of pVA-gfp-oriT into the clinical strain E. faecalis T9.
To investigate whether mobilization of pVA-gfp-oriT can also occur into pathogenic enterococcal strains, we tested the clinical E. faecalis T9 isolate (21). Molecular characterization of E. faecalis T9 revealed the presence of pSK41-like nes and traE, traG, and traK genes confirmed by dot blotting (Fig. 2; see Table S1 in the supplemental material for the primers used in dot blots), thus indicating the presence of a putative conjugative element in E. faecalis T9. However, plasmid isolation by the method of Woodford et al. (43) with the modifications of Werner et al. (42) and plasmid profiling by S1 nuclease macrorestriction (5, 13, 25) of E. faecalis T9 did not confirm the presence of plasmids (not shown).
Fig 2.
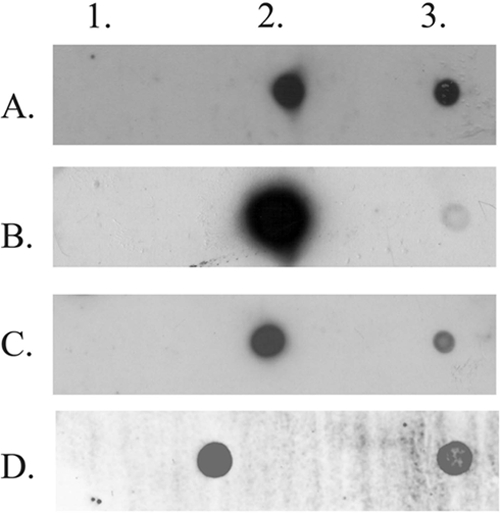
Dot blot hybridization of nes (A), traE (B), traG (C), and traK (D). PCR products were generated with digoxigenin-labeled pSK41-derived probes. Spot 1, negative control (no DNA template was applied to the PCR); spot 2, PCR product of S. aureus(pSK41) as a positive control, spot 3, PCR product of E. faecalis T9.
To prove the presence of a conjugative element in E. faecalis T9, a biparental retromobilization experiment was performed: E. faecalis T9 as a helper strain and recipient and E. faecalis OG1X(pVA-gfp-oriT) as a donor were grown to an OD600 of 0.5 and mated overnight at 37°C in a ratio of 1:10 (donor/recipient [D/R]). Serial dilutions were plated on BHI agar supplemented with kanamycin (500 μg · ml−1) and/or tetracycline (10 μg · ml−1) to enumerate transconjugants and recipients, respectively. Retromobilization of pVA-gfp-oriT into E. faecalis T9 occurred with a frequency of 1.62 · 10−5 ± 1.04 · 10−5, indicating a putative pSK41-like conjugative element in E. faecalis T9. Transconjugants were verified by PCR amplification of the gfp gene, plasmid isolation, and fluorescence microscopy after induction of the nisA promoter on BHI agar supplemented with kanamycin (500 μg · ml−1), tetracycline (10 μg · ml−1), and nisin (100 ng · ml−1) (data not shown). To the best of our knowledge, retromobilization into E. faecalis has been demonstrated for the first time.
To further investigate the conjugative element present in E. faecalis T9, E. faecalis T9 harboring pVA-gfp-oriT was used as a donor to mobilize the GPF-tagged plasmid in a biparental mating to B. subtilis subsp. natto DSM 4451. The strains were grown to an OD600 of 0.5 and mated overnight at 30°C. Serial dilutions were plated on LB agar supplemented with kanamycin (20 μg · ml−1) and/or streptomycin (1,000 μg · ml−1) to enumerate transconjugants and recipients, respectively. Green fluorescent B. subtilis transconjugants were obtained with a mean mobilization rate of 3.10 · 10−7 ± 3.32 · 10−8 per recipient, indicating that mobilizable plasmids can be transferred to B. subtilis and E. faecalis by the conjugative element in E. faecalis T9. These results suggest that the pIP501 oriT region is recognized by the pSK41-like relaxase present in E. faecalis T9. In favor of this hypothesis, Garcillan-Barcia et al. reported that pIP501 and pSK41 relaxases belong to the same mobilization protein (MOB) family (14).
To date, we have no information about traits that can be transferred by the conjugative element in E. faecalis T9. Most widely spread integrative and conjugative elements (ICEs) in E. faecalis such as Tn916, Tn1545, and Tn1549 are genetically linked to genes conferring antibiotic resistance and have a broad host range (13). However, none of these transposons shows similarities to the pSK41 tra region originating from Staphylococcus aureus. Considering the clinical origin of E. faecalis T9, a pSK41-like plasmid might have been integrated into the chromosome of E. faecalis T9, thus allowing the capture of different traits from the clinical background.
It has been demonstrated that Inc18 plasmids like pIP501 might be involved in the spread of vancomycin resistance genes and the emergence of vancomycin-resistant S. aureus (VRSA) (44, 45). Recent studies indicate that the pIP501 replicon has a high prevalence in clinical Enterococcus faecium isolates and that it is often genetically linked with vanA resistance genes in enterococci (19, 34). Thus, the constructed mobilizable plasmid might be a powerful tool to screen for pIP501-like and pSK41-like conjugative elements in Enterococcus and Staphylococcus isolates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant Concordia microbial dynamics from BMWi/DLR to E.G., the European Space Agency (ESA), the French Polar Institute (IPEV), and the Italian Antarctic Programme (PNRA).
We thank Christine Bohn and Carola Fleige for their skillful technical assistance. Special thanks to Manuel Espinosa for the gift of plasmid pJPrelGFP and to Gary Dunny for the gift of plasmid pMSP3535VA.
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abajy MY, et al. 2007. A type IV secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J. Bacteriol. 189:2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babic A, Berkmen MB, Lee CA, Grossman AD. 2011. Efficient gene transfer in bacterial cell chains. mBio 2(2):e00027–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. 2008. Direct visualization of horizontal gene transfer. Science 319:1533–1536 [DOI] [PubMed] [Google Scholar]
- 5. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 6. Bryan EM, Bae T, Kleerebezem M, Dunny GM. 2000. Improved vectors for nisin-controlled expression in Gram-positive bacteria. Plasmid 44:183–190 [DOI] [PubMed] [Google Scholar]
- 7. Christensen BB, et al. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dahlberg C, Bergström M, Hermansson M. 1998. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl. Environ. Microbiol. 64:2670–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34:18–40 [DOI] [PubMed] [Google Scholar]
- 10. Dunny GM, Brown BM, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eichenbaum Z, et al. 1998. Use of the lactococcal nisA promoter to regulate gene expression in Gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans RP, Macrina FL. 1983. Streptococcal R-plasmid pIP501: endonuclease site map, resistance determinant location, and construction of novel derivatives. J. Bacteriol. 154:1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freitas AR, et al. 2010. Global spread of the hylEfm colonization-virulence gene in megaplasmids of the Enterococcus faecium CC17 polyclonal subcluster. Antimicrob. Agents Chemother. 54:2660–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garcillan-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 15. Geoffroy MC, et al. 2000. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl. Environ. Microbiol. 66:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grohmann E. 2006. Mating cell-cell channels in conjugating bacteria, p 21–38 In Baluska F, Volkmann D, Barlow PW. (ed), Cell-cell channels. Landes Biosciences, Georgetown, TX [Google Scholar]
- 17. Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayes CS, Aoki SK, Low DA. 2010. Bacterial contact-dependent delivery systems. Annu. Rev. Genet. 44:71–90 [DOI] [PubMed] [Google Scholar]
- 19. Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16:541–554 [DOI] [PubMed] [Google Scholar]
- 20. Hirt H, Erlandsen SL, Dunny GM. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182:2299–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hufnagel M, Koch S, Creti R, Baldassarri L, Huebner J. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189:420–430 [DOI] [PubMed] [Google Scholar]
- 22. Ike Y, Craig RA, White BA, Yagi Y, Clewell DB. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 80:5369–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurenbach B, et al. 2003. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50:86–93 [DOI] [PubMed] [Google Scholar]
- 25. Laverde-Gomez JA, et al. 2011. A multiresistance megaplasmid bearing a hylEfm genomic island in hospital Enterococcus faecium isolates. Int. J. Med. Microbiol. 301:165–175 [DOI] [PubMed] [Google Scholar]
- 26. Lorenzo-Díaz F, Espinosa M. 2009. Large-scale filter mating assay for intra- and inter-specific conjugal transfer of the promiscuous plasmid pMV158 in Gram-positive bacteria. Plasmid 61:65–70 [DOI] [PubMed] [Google Scholar]
- 27. Mierau I, Kleerebezem M. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705–717 [DOI] [PubMed] [Google Scholar]
- 28. Miller WG, Lindow SE. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149–153 [DOI] [PubMed] [Google Scholar]
- 29. Mølbak L, Licht TR, Kvist T, Kroer N, Andersen SR. 2003. Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: characterization, direct quantification, and in situ location of transconjugant cells. Appl. Environ. Microbiol. 69:5536–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nieto C, Espinosa M. 2003. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 49:281–285 [DOI] [PubMed] [Google Scholar]
- 31. Nieto C, et al. 2006. The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol. Microbiol. 59:1280–1296 [DOI] [PubMed] [Google Scholar]
- 32. Pérez-Mendoza D, et al. 2006. The relaxase of the Rhizobium etli symbiotic plasmid shows nic site cis-acting preference. J. Bacteriol. 188:7488–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rêgo AT, Chandran V, Waksman G. 2010. Two-step and one-step secretion mechanisms in Gram-negative bacteria: contrasting the type IV secretion system and the chaperone-usher pathway of pilus biogenesis. Biochem. J. 425:475–488 [DOI] [PubMed] [Google Scholar]
- 34. Rosvoll TCS, et al. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58:254–268 [DOI] [PubMed] [Google Scholar]
- 35. Scheich C, Niesen FH, Seckler R, Bussow K. 2004. An automated in vitro protein folding screen applied to a human gynactin subunit. Prot. Sci. 13:370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74:434–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sørensen SJ, Sørensen AH, Hansen LH, Oregaard G, Veal D. 2003. Direct detection and quantification of horizontal gene transfer by using flow cytometry and gfp as a reporter gene. Curr. Microbiol. 47:129–133 [DOI] [PubMed] [Google Scholar]
- 38. Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700–710 [DOI] [PubMed] [Google Scholar]
- 39. Trieu-Cuot P, Carlier C, Martin P, Courvalin P. 1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48(1-2):289–294 [Google Scholar]
- 40. Tsvetkova K, Marvaud J-C, Lambert T. 2010. Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J. Bacteriol. 192:702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wallden K, Rivera-Calzada A, Waksman G. 2010. Type IV secretion systems: versatility and diversity in function. Cell. Microbiol. 12:1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Werner G, Klare I, Witte W. 1999. Large conjugative vanA plasmids in vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 37:2383–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Woodford N, Morrison D, Cookson B, George RC. 1993. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob. Agents Chemother. 37:681–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu W, et al. 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52:452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu W, et al. 2010. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54:4314–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.