Abstract
The genus Bartonella was detected by PCR in 5.7% (12/212) of wild carnivores from Northern Spain. Based on hybridization and sequence analyses, Bartonella henselae was identified in a wildcat (Felis silvestris), Bartonella rochalimae in a red fox (Vulpes vulpes) and in a wolf (Canis lupus), and Bartonella sp. in badgers (Meles meles).
TEXT
The zoonotic vector-borne pathogens present complex cycles in nature, which include reservoir hosts and hematophagous arthropods, which play a role as vectors. Among them, the genus Bartonella includes more than 30 species or subspecies that can affect mammals worldwide, and at least 14 are considered to be pathogenic to humans (26). In Spain, six different Bartonella species have been described so far (B. henselae, B. quintana, B. clarridgeiae, B. taylorii, B. alsatica, and B. vinsonii subsp. berkhoffii), affecting humans, cats, dogs, rabbits, and small mammals (10, 19, 21, 23–25). More recently, the use of new molecular tools has allowed us to detect a large repertoire of Bartonella species and genotypes circulating among the small mammals in the Basque Country, Northern Spain (12).
In Europe, as well as in the Basque Country, there is little information regarding the prevalence of vector-borne diseases in carnivores, and given their intense exposure to arthropod vectors, they could represent a good marker for the circulation of these pathogens. Herein we present our results on the presence of Bartonella isolates in carnivores collected in the Basque Country.
Between 2001 and 2006, 212 wild carnivores, belonging to 10 different species (Table 1), that were either found dead or hunted (the majority of the red foxes) were collected in the Basque Country. Carcasses were transported to the laboratory for a complete necropsy, and tissue samples were collected and stored individually. Whenever possible, animals were subjected to a detailed external examination for ectoparasite collection and identification (13, 18).
Table 1.
Presence of Bartonella species in wild carnivores from the Basque Country
| Host species | Scientific name | No. of animals | No. Bartonella positive (%) | Species |
|---|---|---|---|---|
| American mink | Mustela vison | 3 | 0 | |
| Beech marten | Martes foina | 26 | 0 | |
| Eurasian badger | Meles meles | 75 | 9 (12) | Bartonella sp. |
| Pine marten | Martes martes | 14 | 0 | |
| Polecat | Mustela putorius | 5 | 0 | |
| Red fox | Vulpes vulpes | 62 | 1 (1.6) | Bartonella rochalimae |
| Small-spotted genet | Genetta genetta | 13 | 0 | |
| Weasel | Mustela nivalis | 5 | 0 | |
| Wildcat | Felis silvestris | 6 | 1 (16.7) | Bartonella henselae |
| Wolf | Canis lupus | 3 | 1 (33.3) | Bartonella rochalimae |
| Total | 212 | 12 (5.7) |
The presence of Bartonella spp. was analyzed with a seminested PCR targeting the citrate synthase gene (gltA), using primers CS 140f (6) and BhCS1137n (20) in the first run and BhCS781p and BhCS1137n (20) in the second, following the author's protocols with minor modifications. DNA extraction was performed with a QIAamp DNA blood minikit (Qiagen, Hilden, Germany) from a homogenate of liver and spleen previously digested with proteinase K (Invitrogen, Carlsbad, CA). DNA samples were also analyzed by following three different PCR-reverse line blot hybridization (PCR-RLB) protocols previously described to detect Coxiella burnetii, Anaplasma phagocytophilum, spotted fever group rickettsiae, Borrelia spp., and Francisella spp. (2, 17).
Samples positive for Bartonella spp. were confirmed by a PCR-RLB specific for Bartonella, targeting the 16S rRNA and the 16S-23S rRNA intergenic transcribed spacer (ITS), and by sequencing (10, 12). Sequences were compared with those available in the GenBank database by nucleotide sequence homology using BLAST (1) to identify the closest relative. Furthermore, the gltA sequences were aligned with reference sequences from GenBank with the ClustalX software (15).
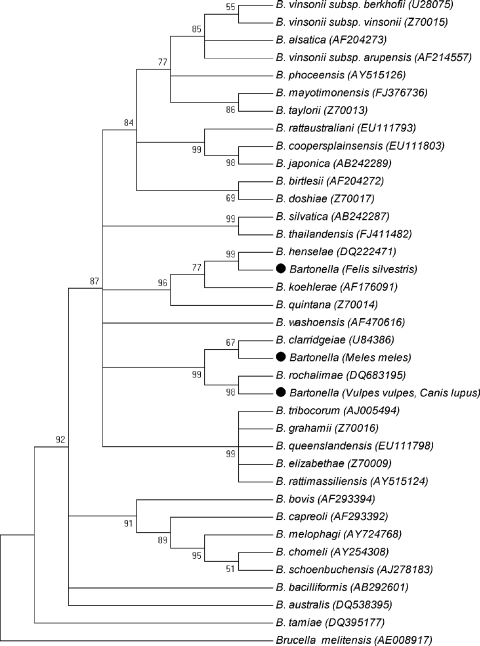
Pairwise distance matrices were determined with the Kimura two-parameter method with MEGA3.1 software (16), and phylogenetic trees were constructed by applying the neighbor-joining algorithm with the internal-branch test for evaluation of their topology, using 1,000 replicates. The dendrogram was collapsed by using a cutoff bootstrap value of 50 (Fig. 1).
Fig 1.
Neighbor-joining tree based on gltA gene sequences, built with reference sequences from GenBank and those obtained in this study (black dots). GenBank accession numbers are indicated within brackets.
Fisher's exact test was applied to determine statistical differences between some variables in the prevalence of Bartonella (age and sex of carnivores, annual season in which they were collected, and ectoparasites present on them). The overall level of statistical significance was the standard 5% (P < 0.05).
The presence of Bartonella spp. was determined in 12 samples, giving an overall prevalence of 5.7% (12/212). Four carnivore species were found to be infected with three different Bartonella genotypes (Table 1). The distribution of the positive animals, grouped by age, sex, location, and season, is shown in Table 2.
Table 2.
Carnivores infected with Bartonella spp., location and season of collection, and ectoparasites found on them
| Host species | Age | Sex | Location | Season | Ectoparasite(s) found |
||
|---|---|---|---|---|---|---|---|
| Licea | Fleasb | Ticksc | |||||
| Eurasian badger | Young | Male | South | Winter | T. melis | I. hexagonus | |
| Eurasian badger | Young | Male | South | Winter | T. melis | I. ricinus | |
| Eurasian badger | Adult | Male | South | Winter | T. melis | P. melis | I. hexagonus, I. ricinus |
| Eurasian badger | Young | Male | South | Spring | T. melis | I. hexagonus, I. ricinus | |
| Eurasian badger | Adult | Female | South | Winter | T. melis | C. trichosa | I. canisuga |
| Eurasian badger | Adult | Male | North | Winter | T. melis | C. trichosa | Ixodes sp. |
| Eurasian badger | Adult | Male | North | Winter | C. trichosa | D. reticulatus | |
| Eurasian badger | Adult | Male | South | Spring | R. pusillus | ||
| Eurasian badger | Adult | Female | South | Autumn | T. melis | C. trichosa | R. pusillus |
| Red fox | Young | Female | South | Spring | P. melis | Ixodes sp. | |
| Wildcat | Young | Male | South | Spring | |||
| Wolf | Young | Male | South | Summer | T. canis | I. ricinus | |
T. melis, Trichodectes melis.
P. melis, Paraceras melis; C. trichosa, Chaetopsylla trichosa.
I. hexagonus, Ixodes hexagonus; I. ricinus, Ixodes ricinus; I. canisuga, Ixodes canisuga; D. reticulatus, Dermacentor reticulatus; R. pusillus, Rhipicephalus pusillus.
Regarding age, 11.3% (6/53) of the young animals showed positive results, compared to 4.7% (6/129) of the adults (P = 0.1), which is in agreement with several studies developed in cats and dogs (8, 22), probably due to an immune system that is not fully developed in young specimens or to the acquisition of immunoresistance in the adults after successive contacts with these pathogens.
By sex, the percentages of positive males (7.1%; 9/127) and females (3.7%; 3/81) were similar (P = 0.2). Seasonally, similar percentages of specimens collected during winter (6.9%; 6/87) and spring (7.7%; 4/52) had positive results, while those specimens found during summer (1/34) and autumn (1/39) showed a prevalence rate below 3% (in all cases, the P value was >0.3). Geographically, a higher number of positive animals corresponded to individuals collected in the South of the Basque Country (10.2%; 10/98) than in the North (1.8%; 2/114) (P = 0.01). This lower Bartonella abundance might represent less adequate environmental conditions and, therefore, a higher risk of stress for the microorganism's survival. More detailed studies should be carried out to determine the factors present in each area that favor the transmission of these bacteria.
The gltA sequence obtained from the wildcat showed a 100% similarity with B. henselae (GenBank accession no. DQ222471). In the case of the samples from the fox and the wolf, the gltA sequences showed 100% similarity with B. rochalimae (GenBank accession no. DQ683195). It is noteworthy that a high percentage of badgers (12%) was infected with Bartonella sp. The nine gltA sequences obtained from badgers were identical among them (GenBank accession no. GU570947) and did not correspond to any of the sequences deposited in GenBank. Regarding the percentage of similarity, the closest relative was B. clarridgeiae, with 96.4% similarity (GenBank accession no. U84386). This result was also supported by the dendrogram, where the badger's sequence was placed in the same clade as B. clarridgeiae (Fig. 1). Moreover, the sequences of 16S rRNA and ITS (GenBank accession no. EU098127 and EU98132, respectively) from the 9 badgers were also identical among them and were also closely related to B. clarridgeiae (10). The future isolation in culture of this organism and a further characterization of additional genes will clarify the taxonomic position of this bacterium.
The results shown here indicate that wild carnivores in the Basque Country are infected with at least three different Bartonella species. B. henselae and B. rochalimae are zoonotic agents (7, 9, 27), and their description in three species of wild carnivores may suggest that these animals may be important in the maintenance of this pathogen in wild ecosystems in the Basque Country and that transmission to humans may be possible through the bite of arthropod vectors that have previously fed on them. On the other hand, the identification of B. rochalimae in a wolf (33.3%) and a fox (1.6%) constitute the first description of this Bartonella species in Spain, although it has previously been described in raccoons, coyotes, and foxes in the United States, France, and Hungary (14). Although the competent reservoir host for B. rochalimae is still unknown, these data and our results point out that carnivore species could play this role. Finally, the third species is strongly associated with badgers, and its risk for human health remains unknown.
Ticks, fleas, and lice were collected from the carnivores, and the percentages of infestation are summarized in Table 3, with badgers and foxes being the most parasitized species. Among carnivores infected with Bartonella spp., the majority were infested by ticks (11/12), and in 88.9% of cases by Ixodes sp. (Table 2), the more frequent tick genus in the area (3). Also, most of them were infested by lice (8/12), mainly by Trichodectes melis (77.8%), and fleas (6/12), mainly by Chaetopsyla trichosa (55.6%). Animals with positive results showed the highest prevalence of infestation by ectoparasites (91.7%; P = 0.03). More specifically, animals with Bartonella spp. showed the highest prevalence of infestation by ticks (83.3%; P = 0.006) and lice (66.7%; P = 0.005) but not by fleas (50%; P = 0.1).
Table 3.
Parasitization of animals by ticks, fleas, and lice
| Host species | No. of animals parasitized/total no. examined (%) |
|||
|---|---|---|---|---|
| Any ectoparasite | Ticks | Fleas | Lice | |
| American mink | 3/3 (100) | 3/3 (100) | 1/3 (33.33) | 0/3 (0) |
| Beech marten | 9/26 (34.62) | 6/26 (23.08) | 4/26 (15.38) | 6/26 (23.08) |
| Eurasian badger | 59/75 (78.67) | 40/75 (53.33) | 26/75 (34.67) | 47/67 (70.15) |
| Pine marten | 3/14 (21.4) | 2/14 (14.29) | 2/14 (14.29) | 0/14 (0) |
| Polecat | 0/5 (0) | 0/5 (0) | 0/5 (0) | 0/5 (0) |
| Red fox | 39/62 (62.9) | 33/62 (53.23) | 22/62 (35.48) | 0/61 (0) |
| Small-spotted genet | 5/12 (41.67) | 5/12 (41.67) | 1/12 (8.33) | 0/12 (0) |
| Weasel | 2/5 (40) | 1/5 (20) | 1/5 (20) | 0/5 (0) |
| Wildcat | 1/6 (16.67) | 1/6 (16.67) | 0/6 (0) | 0/5 (0) |
| Wolf | 1/2 (50) | 1/2 (50) | 0/2 (0) | 1/2 (50) |
The presence of Bartonella spp. was also analyzed by gltA PCR in 68 adult ticks collected from 12 badgers (36 ticks) and 9 foxes (32 ticks), with all the samples being negative. Lice and fleas could not be further analyzed because the identification methods employed and storage conditions left this material unable to be analyzed by PCR.
The carnivores with higher levels of infestation by ectoparasites (wolf, badger, and fox) are those with a higher rate of infection by Bartonella spp., except in the case of the wildcat, which did not show any ectoparasites. However, except in the case of hunted foxes, which were collected at the time of their death, the rest of the animals spent various times between death and the collection of carcasses, which may have caused the release of ectoparasites and, therefore, an underestimation of the prevalence of parasitism. Also, the host specificity of some ectoparasites (i.e., T. melis and C. trichosa in badgers) could be related to the Bartonella-host associations observed in this study. Bartonella bacteria are widely distributed among badgers in our study area, and the fact that most of the badgers were infested with lice, fleas, and ticks indicates a need to determine the vectors involved in its transmission.
All samples were negative for the presence of other vector-borne bacteria, although all of them, except Francisella tularensis, are well documented in our study area in ticks (2, 4, 5) and small mammals (2, 11). Therefore, the absence of these pathogens in the specimens studied suggests that these carnivore species may not be involved in the epidemiologic cycle of these pathogens in the Basque Country.
In summary, the results shown here indicate that wild carnivores in the Basque Country are infected with at least three different Bartonella species, two of them zoonotic and a third strongly associated with badgers, whose risk for human health is unknown.
Nucleotide sequence accession numbers.
The gltA sequence of the Bartonella sp. found in badgers has been deposited in GenBank under accession no. GU570947.
ACKNOWLEDGMENTS
This work was supported by fellowships from the Department of Agriculture, Fisheries and Food and from the Department of Commerce, Industry and Tourism of the Basque Government and by grants PI10/00051 and MPY 025/09, funded by FIS Instituto de Salud Carlos III, Ministry of Science and Innovation.
We thank the Grupo Alavés para la Defensa y Estudio de la Naturaleza (GADEN), Asociación de Cotos de Caza de Álava (ACCA), and Diputaciones Forales de Álava, Bizkaia y Gipuzkoa for providing animal samples used in this study. We also thank A. L. García and J. Barandika for laboratory assistance and J. C. Beaucournu (Faculty of Medicine of Rennes, France) and M. P. Martín (National Museum of Natural Sciences, CSIC, Spain) for identification of fleas and lice.
Footnotes
Published ahead of print 2 December 2011
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Barandika JF, et al. 2007. Tick-borne zoonotic bacteria in wild and domestic small mammals in northern Spain. Appl. Environ. Microbiol. 73:6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barandika JF, et al. 2011. Differences in questing tick species distribution between Atlantic and continental climate regions in Spain. J. Med. Entomol. 48:13–19 [DOI] [PubMed] [Google Scholar]
- 4. Barral M. 1998. Estudio de la infección por Borrelia burgdorferi, grupo Ehrlichia phagocytophila y virus de la encefalitis ovina en las poblaciones de ixódidos de la Comunidad Autónoma Vasca Ph. D. thesis. University of Zaragoza, Zaragosa, Spain [Google Scholar]
- 5. Barral M, et al. 2002. Distribution of Borrelia burgdorferi sensu lato in Ixodes ricinus (Acari: Ixodidae) ticks from the Basque Country, Spain. J. Med. Entomol. 39:177–184 [DOI] [PubMed] [Google Scholar]
- 6. Birtles RJ, Raoult D. 1996. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int. J. Syst. Bacteriol. 46:891–897 [DOI] [PubMed] [Google Scholar]
- 7. Breitschwerdt EB, Kordick DL. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chang CC, et al. 2000. Coyotes (Canis latrans) as the reservoir for a human pathogenic Bartonella sp.: molecular epidemiology of Bartonella vinsonii subsp. berkhoffii infection in coyotes from central coastal California. J. Clin. Microbiol. 38:4193–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eremeeva ME, et al. 2007. Bacteremia, fever, and splenomegaly caused by a newly recognized bartonella species. N. Engl. J. Med. 356:2381–2387 [DOI] [PubMed] [Google Scholar]
- 10. García-Esteban C, et al. 2008. Molecular method for Bartonella species identification in clinical and environmental samples. J. Clin. Microbiol. 46:776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gil H, Barral M, Escudero R, García-Pérez AL, Anda P. 2005. Identification of a new Borrelia species among small mammals in areas of northern Spain where Lyme disease is endemic. Appl. Environ. Microbiol. 71:1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gil H, et al. 2010. Variability of Bartonella genotypes among small mammals in Spain. Appl. Environ. Microbiol. 76:8062–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gil-Collado J, Guillén-Llera J, Zapatero-Ramos LM. 1979. Claves para la identificación de los Ixodoidea españoles (adultos). Rev. Iber. Parasitol. 39:107–111 [Google Scholar]
- 14. Henn JB, et al. 2009. Bartonella rochalimae in raccoons, coyotes, and red foxes. Emerg. Infect. Dis. 15:1984–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins DG, Sharp PM. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237–244 [DOI] [PubMed] [Google Scholar]
- 16. Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 17. Long GW, et al. 1993. Detection of Francisella tularensis in blood by polymerase chain reaction. J. Clin. Microbiol. 31:152–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Manilla G. 1998. Acari, Ixodida. Fauna d'Italia. ; Calderini, Bologna, Italy [Google Scholar]
- 19. Márquez FJ. 2010. Molecular detection of Bartonella alsatica in European wild rabbits (Oryctolagus cuniculus) in Andalusia (Spain). Vector Borne Zoonotic Dis. 10:731–734 [DOI] [PubMed] [Google Scholar]
- 20. Norman AF, Regnery R, Jameson P, Greene C, Krause DC. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pons I, et al. 2008. Seroprevalence of Bartonella spp. infection in HIV patients in Catalonia, Spain. BMC Infect. Dis. 8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pons I, et al. 2005. Prevalence of Bartonella henselae in cats in Catalonia, Spain. Am. J. Trop. Med. Hyg. 72:453–457 [PubMed] [Google Scholar]
- 23. Roura X, Breitschwerdt E, Lloret A, Ferrer L, Hegarty B. 2005. Serological evidence of exposure to Rickettsia, Bartonella, and Ehrlichia species in healthy or Leishmania infantum-infected dogs from Barcelona, Spain. Int. J. Appl. Res. Vet. Med. 3:129–137 [Google Scholar]
- 24. Solano-Gallego L, Hegarty B, Espada Y, Llull J, Breitschwerdt E. 2006. Serological and molecular evidence of exposure to arthropod-borne organisms in cats from northeastern Spain. Vet. Microbiol. 118:274–277 [DOI] [PubMed] [Google Scholar]
- 25. Solano-Gallego L, Llull J, Osso M, Hegarty B, Breitschwerdt E. 2006. A serological study of exposure to arthropod-borne pathogens in dogs from northeastern Spain. Vet. Res. 37:231–244 [DOI] [PubMed] [Google Scholar]
- 26. Vayssier-Taussat M, Le Rhun D, Bonnet S, Cotte V. 2009. Insights in Bartonella host specificity. Ann. N. Y. Acad. Sci. 1166:127–132 [DOI] [PubMed] [Google Scholar]
- 27. Wormser GP. 2007. Discovery of new infectious diseases—Bartonella species. N. Engl. J. Med. 356:2346–2347 [DOI] [PubMed] [Google Scholar]