Abstract
Bacillus subtilis and its close relatives are widely used in industry for the Sec-dependent secretory production of proteins. Like other Gram-positive bacteria, B. subtilis does not possess SecB, a dedicated targeting chaperone that posttranslationally delivers exported proteins to the SecA component of the translocase. In the present study, we have implemented a functional SecB-dependent protein-targeting pathway into B. subtilis by coexpressing SecB from Escherichia coli together with a SecA hybrid protein in which the carboxyl-terminal 32 amino acids of the B. subtilis SecA were replaced by the corresponding part of SecA from E. coli. In vitro pulldown experiments showed that, in contrast to B. subtilis SecA, the hybrid SecA protein gained the ability to efficiently bind to E. coli SecB, suggesting that the structural details of the extreme C-terminal region of SecA constitute a crucial SecB binding specificity determinant. Using a poorly exported mutant maltose binding protein (MalE11) and alkaline phosphatase (PhoA) as model proteins, we could demonstrate that the secretion of both proteins by B. subtilis was significantly enhanced in the presence of the artificial protein targeting pathway. Mutations in SecB that do not influence its chaperone activity but prevent its interaction with SecA abolished the secretion stimulation of both proteins, demonstrating that the implemented pathway in fact critically depends on the SecB targeting function. From a biotechnological view, our results open up a new strategy for the improvement of Gram-positive bacterial host systems for the secretory production of heterologous proteins.
INTRODUCTION
In most bacteria, the major route of protein transport across the cytoplasmic membrane is the general secretion (Sec) pathway (24, 40). Powered by the translocation motor SecA (32), Sec-dependent proteins are translocated across the membrane through a protein-conducting channel (SecYEG) (19). Sec substrates usually are synthesized as precursor proteins possessing an amino-terminal signal peptide. When nascent Sec substrates emerge from the ribosome, they are recognized and targeted to the Sec translocase either by the signal recognition particle (SRP)-mediated cotranslational pathway (6, 12) or, alternatively, by a posttranslational targeting mechanism that involves the SecB chaperone (1, 29, 44). SecB is found in alpha-, beta-, and gammaproteobacteria, including the Gram-negative model bacterium Escherichia coli, while it is absent from Gram-positive bacteria (36). SecB is a secretion-dedicated chaperone that interacts with newly synthesized precursor proteins and maintains them in a translocation-competent state. Besides this function as an antifolding factor, SecB also acts as a targeting factor that directly binds to the membrane-bound SecA motor protein and thereby delivers the substrate to the export sites. For the high-affinity SecB-SecA interaction, a binding contact between the negatively charged flat β-sheet surface of SecB and the zinc-containing C-terminal 22 amino acids of SecA is of crucial importance (3, 11, 43). The C-terminal region of SecA is highly conserved in bacteria, even in organisms that do not possess a SecB homologue, such as the Gram-positive model organism Bacillus subtilis (35). In fact, it has been shown previously that the C-terminal 22 amino acids of B. subtilis SecA were able to bind the E. coli SecB (37).
Due to their enormous secretion capacity, B. subtilis and some of its close relatives are widely used in industry for the secretory production of various endogenous enzymes (33). However, attempts to use these organisms for secreting heterologous proteins often have led to disappointing results (27). For example, it has been found that the SecB-dependent maltose-binding protein (MalE) from E. coli was inefficiently translocated across the B. subtilis plasma membrane (4, 5). It has been speculated that the lack of SecB contributes to the relatively poor translocation efficiency of MalE in B. subtilis. In fact, the coexpression of E. coli SecB in B. subtilis resulted in an approximately 2-fold enhancement of MalE precursor processing, an effect that has been attributed to the anti-folding activity of SecB (5).
In the present study, we have investigated the requirements for the installation of a functional SecB-SecA protein-targeting pathway in B. subtilis. In vitro interaction analysis revealed that the full-length B. subtilis SecA protein did not efficiently bind to either the E. coli SecB or the Haemophilus influenzae SecB. In contrast, hybrid SecA proteins beSecA (for B. subtilis-E. coli SecA) and bhSecA (for B. subtilis-H. influenzae SecA), in which the C-terminal 32 amino acids of the B. subtilis SecA have been replaced by the counterpart of the SecA proteins from E. coli or H. influenzae, respectively, efficiently bound SecB from E. coli or from H. influenzae, indicating that the structural details of the C tail of the SecA proteins dictate whether it can tightly bind to the SecB chaperone or not. Using a poorly exported MalE variant (MalE11) as a reporter protein, we have shown that the coexpression of beSecA and E. coli SecB in B. subtilis resulted in a significant improvement of MalE11 secretion, and that this improvement was dependent on the SecB targeting function. Interestingly, similar secretion stimulation also was observed for the SecB-independent alkaline phosphatase (PhoA) from E. coli. Taken together, our results indicate that we successfully introduced a functional SecB-SecA protein targeting pathway into B. subtilis, and that this pathway can improve the secretory production of heterologous proteins.
MATERIALS AND METHODS
Plasmids, bacterial strains, media, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. E. coli and B. subtilis strains were grown in Luria-Bertani (LB) medium consisting of 1% tryptone, 0.5% yeast extract, and 1% NaCl. If appropriate, media were supplemented with 100 μg ml−1 ampicillin, 100 μg ml−1 kanamycin, 5 μg ml−1 chloramphenicol, or 5 μg ml−1 erythromycin. secB gene expression in B. subtilis was induced by the addition of 0.5% (wt/vol) xylose unless indicated otherwise.
Table 1.
Strains and plasmids used in this study
| Name | Descriptiona | Source or referenceb |
|---|---|---|
| Strain | ||
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| BL21(DE3) | Protein expression host | Novagen |
| B. subtilis | ||
| 168 | trpC2 | BGSC |
| LD1 | 168 expressing ecSecB | This work |
| LD2 | 168 expressing ecSecB L75Q | This work |
| LD3 | 168 expressing ecSecB E77K | This work |
| LD4 | 168 expressing ecSecB L75Q&E77K | This work |
| Plasmid | ||
| pSJ2, pSJ3 | pET21a derivative, Apr, T7p | 22 |
| pSJ4 | pET30a derivative, Kmr, T7p | 22 |
| pGEX-2T | Apr, tacp | GE Healthcare |
| pAX01 | Integration vector, Apr, Emr, xylAp | 13 |
| pT7Div | Encoding bsSecA | 20 |
| pMA5 | pUB110 derivative, Apr, Kmr, HpaIIp | 8 |
| pOE | pHCMC05 derivative, Apr, Cmr, HpaIIp | This work |
| pSJ3-ecSecA | Encoding ecSecA | This work |
| pSJ3-bsSecA | Encoding bsSecA | This work |
| pSJ3-hiSecA | Encoding hiSecA | This work |
| pSJ3-heSecA | Encoding heSecA comprised of residues 1 to 866 from hiSecA and residues 870 to 901 from ecSecA | This work |
| pSJ3-ehSecA | Encoding ehSecA comprised of residues 1 to 869 from ecSecA and residues 867 to 901 from hiSecA | This work |
| pSJ3-beSecA | Encoding beSecA comprised of residues 1 to 809 from bsSecA and residues 870 to 901 from ecSecA | This work |
| pSJ3-bhSecA | Encoding bhSecA comprised of residues 1 to 809 from bsSecA and residues 867 to 901 from hiSecA | This work |
| pSJ4-hiSecB | Encoding hiSecB | 39 |
| pSJ2-ecSecB | Encoding ecSecB | This work |
| pGEX-2T-ecSecAc | Encoding GST fused to residues 878 to 901 from ecSecA | This work |
| pGEX-2T-hiSecAc | Encoding GST fused to residues 875 to 901 from hiSecA | This work |
| pMA5-ecMalE | Encoding E. coli MalE | This work |
| pMA5-ecMalE11 | Encoding the E. coli MalE11 mutant | This work |
| pMA5-ecPhoA | Encoding E. coli PhoA | This work |
| pAX01-ecSecB | Encoding ecSecB | This work |
| pAX01-ecSecBL75Q | Encoding ecSecB L75Q | This work |
| pAX01-ecSecBE77K | Encoding ecSecB E77K | This work |
| pAX01-ecSecBL75Q&E77K | Encoding ecSecB L75Q&E77K | This work |
| pOE-beSecA | Encoding beSecA comprised of residues 1 to 809 from bsSecA and residues 870 to 901 from ecSecA | This work |
| pOE-bsSecA | Encoding bsSecA | This work |
ecSecB, E. coli SecB; hiSecB, H. influenzae SecB; bsSecA, B. subtilis SecA; ecSecA, E. coli SecA; hiSecA, H. influenzae SecA; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Kmr, kanamycin resistance; T7p, T7 promoter; tacp, tac promoter; xylAp, xylose-inducible promoter; HpaIIp, HpaII promoter; GST, glutathione S-transferase.
BGSC, Bacillus Genetics Stock Center.
Bacterial strain constructions.
B. subtilis LD1 containing the E. coli secB gene under the regulatory control of the xylose-inducible promoter xylAp integrated into the lacA locus of the chromosome was constructed by transforming B. subtilis 168 with integration plasmid pAX01-ecSecB, followed by the selection of erythromycin-resistant colonies. B. subtilis LD2, LD3, and LD4 were constructed by the same procedure using the integration plasmids pAX01-ecSecBL75Q, pAX01-ecSecBE77K, and pAX01-ecSecBL75Q&E77K, respectively.
Plasmid constructions.
The primers used in this study are listed in Table S1 in the supplemental material. For protein purification purposes in E. coli, the various SecA and SecB proteins were expressed in E. coli as fusion proteins containing an N-terminal His8 tag that is cleavable by tobacco etch virus (TEV) protease. To construct pSJ3-ecSecA, the E. coli secA gene was amplified using primers ecSecA-N5/ecSEcA-B3 and E. coli W3110 genomic DNA as the template. The resulting PCR fragment was digested with NdeI/BamHI and ligated into pSJ3 digested with NdeI/BamHI. pSJ3-bsSecA was constructed by amplifying the B. subtilis secA gene using primers bsSecA-B5/bsSecA-X3 and pT7Div (20) as the template. The resulting PCR fragment was digested with BamHI/XhoI and ligated into pSJ3. To construct pSJ3-hiSecA, the H. influenzae secA gene was amplified using primers hiSecA-N5/hiSecA-H3 and H. influenzae genomic DNA as the template. The resulting PCR fragment was digested with NdeI/HindIII and ligated into pSJ3.
The plasmids for the expression of chimeric SecA proteins in E. coli were constructed by megaprimer PCR (31). To construct pSJ3-heSecA, two rounds of PCR were performed. First, a DNA fragment encoding amino acid residues 870 to 901 from the C terminus of E. coli SecA was amplified using primers heSecA-mp5p/ecSecA-B3 and pSJ3-ecSecA as the template. The resulting PCR fragment was used as the megaprimer in the second PCR together with primer hiSecA-N5 and pSJ3-hiSecA as the template. The new PCR fragment was digested with NdeI/BamHI and ligated into pSJ3 digested with the same enzymes to generate pSJ3-heSecA. To construct pSJ3-ehSecA, the megaprimer encoding amino acid residues 867 to 901 from the C terminus of H. influenzae SecA was amplified using primers ehSecA-mp5p/hiSecA-H3 and pSJ3-hiSecA as the template. Next, the megaprimer and ecSecA-N5 were used to amplify the fragment encoding the E. coli-H. influenzae hybrid protein (ehSecA) using pSJ3-ecSecA as the template. The PCR product was restricted with NdeI/HindIII and ligated into NdeI/HindIII-digested pSJ3. To construct pSJ3-beSecA, the megaprimer encoding amino acid residues 870 to 901 from the C terminus of E. coli SecA was amplified using primers beSecA-mp5p/ecSecA-B3 and pSJ3-ecSecA as the template. Next, the megaprimer and bsSecA-B5 were used to amplify the fragment encoding beSecA using pSJ3-bsSecA as the template. The PCR product was restricted with BamHI and ligated into BamHI-digested pSJ3. To construct pSJ3-bhSecA, the megaprimer encoding amino acid residues 867 to 901 from the C terminus of H. influenzae SecA was amplified using primers bhSecA-mp5p/hiSecA-H3 and pSJ3-hiSecA as the template. Next, the megaprimer and bsSecA-B5 were used to amplify the fragment encoding bhSecA using pSJ3-bsSecA as the template. The PCR product was restricted with BamHI/HindIII and ligated into BamHI/HindIII-digested pSJ3.
pSJ4-hiSecB, encoding H. influenzae SecB (hiSecB), was constructed as described previously (39). pJS2-ecSecB was constructed to express SecB in E. coli (ecSecB). First, the E. coli secB gene was amplified by PCR using primers ecSecB-B5/ecSecB-H3 and E. coli W3110 genomic DNA as the template. The resulting PCR fragment was digested with BamHI/HindIII and ligated into pSJ2 digested with the same enzymes.
The construction of pGEX-2T-hiSecAc, which encodes glutathione S-transferase (GST) fused to residues 875 to 901 of hiSecA, has been described previously (43). To construct pGEX-2T-ecSecAc, which encodes GST fused to residues 878 to 901 of E. coli SecA (ecSecA), the DNA fragment encoding residues 878 to 901 of ecSecA was amplified using primers ecSecAc-B5/ecSecAc-E3 and pSJ3-ecSecA as the template. The resulting PCR fragment was digested with BamHI/EcoRI and ligated into pGEX-2T digested with the same enzymes.
To express SecA proteins in B. subtilis, the corresponding secA genes were cloned into pOE, a pHCMC05 derivative (the original spac expression cassette was replaced by the constitutive promoter HpaIIp) that is compatible with pMA5 to coexist in B. subtilis. pOE-beSecA was constructed by amplifying the B. subtilis secA gene using primers bsSecAf/ecSecAr and pSJ3-beSecA as the template. The resulting PCR fragment was digested with KpnI and ligated into KpnI-digested pOE. pOE-bsSecA was constructed identically using primers bsSecAf/bsSecAr and B. subtilis genomic DNA as the template.
To express wild-type and mutant ecSecB proteins in B. subtilis, the corresponding genes were cloned into the integration vector pAX01 (13) under the control of the xylose-inducible xylAp promoter. First, the wild-type secB gene was amplified using primers ecSecBf/ecSecBr and E. coli W3110 genomic DNA as the template. The obtained PCR fragment was cleaved with BamHI and ligated into BamHI-digested pAX01, resulting in pAX01-ecSecB. To introduce the mutation L75Q into ecSecB, overlap extension PCR was used. First, two DNA fragments were amplified using E. coli W3110 genomic DNA as the template and primer pairs ecSecBf/ecSecB and ecSecBL75Qf/ecSecBr, respectively. Subsequently, the full-length gene for ecSecBL75Q was assembled in the second round of PCR using primers ecSecBf/ecSecBr and a mixture of the two PCR fragments described above as the template. The obtained PCR fragment was cleaved with BamHI and ligated into BamHI-digested pAX01, resulting in pAX01-ecSecBL75Q. pAX01-ecSecBE77K and pAX01-ecSecBL75Q&E77K were constructed identically, except that the primer ecScBL75Qf was replaced by ecSecBE77Kf and ecSecBL75Q&E77Kf, respectively.
To express E. coli wild-type MalE in B. subtilis, pMA5-ecMalE was constructed as follows. Primers ecMalEf/ecMalE11mr were used to amplify the malE gene using E. coli genomic DNA as the template. The PCR fragment was digested with NdeI/HindIII and then cloned into pMA5 (8) treated with the same enzymes. To express MalE11 (harboring the mutations K2T and K4T in the signal peptide and E29G in the early mature region) in B. subtilis, pMA5-ecMalE11 was constructed by overlap extension PCR. First, two separate PCRs using E. coli W3110 genomic DNA as the template were performed to amplify the DNA fragment encoding the mutated signal peptide using primers ecMalE11spf/ecMalE11spr and the mutated mature MalE using primers ecMalE11mf/ecMalE11mr, respectively. In the following step, the full-length malE11 gene was assembled using a mixture of the two overlapping PCR fragments described above as the template and primers ecMalE11spf/ecMalE11mr. The resulting PCR fragment was cleaved with NdeI/HindIII and ligated into pMA5 digested with the same enzymes.
To express E. coli alkaline phosphatase (PhoA) in B. subtilis, the phoA gene was amplified using primers ecPhoAf/ecPhoAr and E. coli W3110 genomic DNA as the template. The resulting PCR fragment was cleaved with NdeI/HindIII and ligated into pMA5 digested with the same enzymes, resulting in pMA5-ecPhoA.
Protein expression and purification.
The different His-tagged SecA proteins were expressed in E. coli as described previously (43), with slight modifications. E. coli BL21(DE3) strains transformed with pSJ3-derived plasmids encoding the different SecA proteins were grown at 37°C in LB medium supplemented with 100 mM ZnCl2. Protein production was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C when the cells reached an optical density at 600 nm (OD600) of 0.8. The cells were harvested 20 h after IPTG induction.
The recombinant SecA proteins for isothermal titration calorimetry (ITC) were purified by two rounds of nickel affinity chromatography followed by gel filtration chromatography. In brief, the His-tagged SecA proteins purified from the first nickel affinity chromatography were digested with TEV protease overnight at 4°C to remove the His tags. The cleaved His tags, further protein contaminants, and the His-tagged TEV protease were removed by the second nickel affinity chromatography. Proteins from the flowthrough were pooled, concentrated, and run over a Superdex 200 16/60 column (GE Healthcare) with 25 mM Tris-HCl, pH 7.6, 50 mM KCl, 10 mM MgCl2, 10 μM ZnCl2, and 1 mM dithiothreitol (DTT). Protein concentrations were determined by measuring the OD280. SecB proteins and GST fused to the C-terminal 22 amino acids of SecA proteins (SecAc) were expressed and purified as described previously (39, 43).
In vitro SecB-SecA binding assay.
The interaction between His-tagged SecA and SecB proteins was analyzed by a pulldown assay as described previously (43). To analyze the interaction between GST-SecAc and SecB proteins, purified GST-hiSecAc (2.5 mg ml−1) or GST-ecSecAc (2.5 mg ml−1) and hiSecB (14 mg ml−1) or ecSecB (12.5 mg ml−1) were used at the indicated concentrations. Thirty μl GST-hiSecAc or GST-ecSecAc, 9 μl hiSecB or ecSecB, and 61 μl phosphate-buffered saline (PBS) buffer (150 mM NaCl, 16 mM Na2HPO4, pH 7.3, 1 mM DTT) were mixed and incubated at room temperature for 10 min. Thirty μl of glutathione agarose beads (equilibrated with PBS already) was added, and the mixture was left on ice for another 10 min. The beads were sedimented by centrifugation for 5 min at 5,000 × g at 4°C, followed by three washing steps with 200 μl PBS buffer. The elution of the bound proteins from the beads was done by resuspending the beads in 50 μl 50 mM Tris-HCl, pH 8.0, containing 10 mM reduced glutathione. The beads were centrifuged down, and the resulting supernatant was analyzed subsequently by 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
ITC.
ITC was performed at 20°C with a Microcal (Piscataway, NJ) ITC200 calorimeter calibrated according to the manufacturer's instructions. All of the protein samples were dialyzed against the ITC buffer (25 mM Tris-HCl, pH 7.4, 50 mM NaCl) before use. The sample cell (200 μl) was loaded with B. subtilis SecA (bsSecA; 32 μM), ecSecA (13.3 μM), or beSecA (35 μM). ecSecB (376 μM for bsSecA, 251 μM for ecSecA, and beSecA) in the syringe was added in a sequence of 20 to 30 injections of 1.2 (for bsSecA), 2 (for ecSecA), or 1.33 μl (for beSecA) at 3-min intervals. The data were analyzed with the Origin 7.0 software supplied with the instrument.
Protein localization and Western blotting.
B. subtilis whole-cell extracts and supernatant fractions were prepared as described previously (21), with minor modifications. From 1 ml culture, cells and supernatant were separated by centrifugation. The cell pellet was washed once with lysis buffer (10 mM Tris-HCl, pH 8.0, 25 mM MgCl2, 200 mM NaCl) and resuspended in 100 μl lysis buffer. For the disruption of the cells, lysozyme was added to a final concentration of 5 mg ml−1, and the mixture was incubated for 30 min at 37°C. The proteins in the supernatant were precipitated by adding 10% trichloroacetic acid for 12 h at 4°C. The distribution of MalE- or PhoA-derived polypeptides in the corresponding fractions was analyzed by SDS-PAGE and Western blotting using MalE- or PhoA-specific antibody and the Western blotting detection kit (Amersham Biosciences) as described previously (2). The chemiluminescent protein bands were recorded using a charge-coupled device (CCD) camera and the image analyzing system Fujifilm LAS-1000 (Fuji Photo Film).
Enzyme assay.
PhoA activity was determined as described previously (7).
RESULTS
The extreme carboxyl terminus (C tail) of SecA is an important SecB binding specificity determinant.
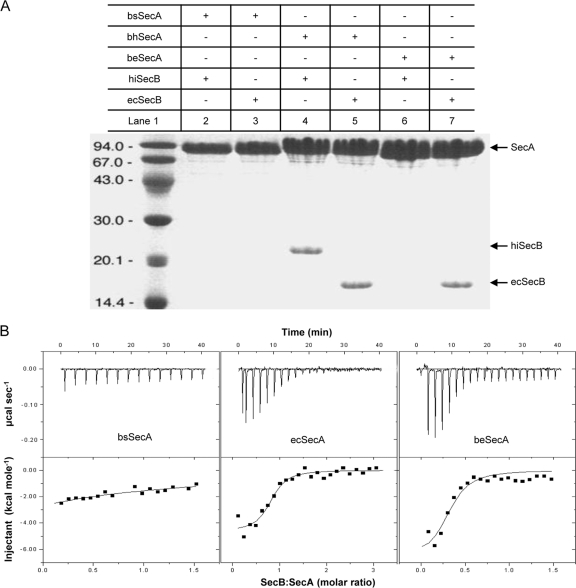
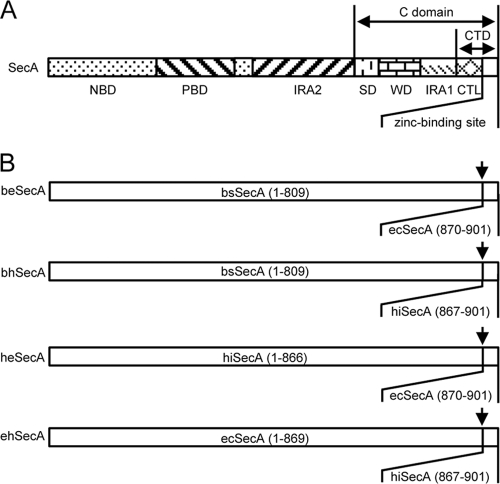
As the first step for the establishment of a functional SecB-SecA protein targeting pathway in B. subtilis, we analyzed whether the full-length B. subtilis SecA protein (bsSecA) is able to interact with SecB from E. coli (ecSecB) or from H. influenzae (hiSecB) in an in vitro binding assay. In contrast to the C-terminal 22 amino acids of bsSecA (bsSecAc) that could bind ecSecB in the context of a GST-bsSecAc fusion (37), no such interaction between a His-tagged version of full-length bsSecA and ecSecB or hiSecB could be detected (Fig. 1A, lanes 2 and 3). Next, two hybrid SecA proteins were constructed in which the C-terminal 32 amino acids of bsSecA were replaced by the corresponding residues of SecA from E. coli (resulting in beSecA) or H. influenzae (resulting in bhSecA), respectively (Fig. 2). Strikingly, the beSecA and the bhSecA hybrid proteins gained the ability to interact with ecSecB (Fig. 1A, lanes 5 and 7). Furthermore, bhSecA (but not beSecA) also could bind hiSecB (Fig. 1A, lanes 4 and 6). A similar scenario was observed when the C tail was swapped between the SecA proteins of E. coli (ecSecA) and H. influenzae (hiSecA). Also in this case, the C tail derived from ecSecA conferred the binding of H. influenzae-E. coli SecA (heSecA) (Fig. 2) to ecSecB but not hiSecB, whereas the C tail from hiSecA promoted the binding of ehSecA (Fig. 2) to both ecSecB and hiSecB (see Fig. S1 in the supplemental material). Interestingly, the C tails alone (SecAc) did not show any SecB preference, since both ecSecAc and hiSecAc efficiently interacted with ecSecB and hiSecB (see Fig. S2 in the supplemental material). These results suggested that the structural details of the conserved zinc-containing C tail strongly influence the specificity as well as the binding affinity between the respective full-length SecA proteins and SecB proteins from different species.
Fig 1.
beSecA hybrid protein efficiently binds ecSecB. (A) Analysis of SecA-SecB interactions by an in vitro pulldown assay. The SecA and SecB proteins analyzed in each experiment are indicated by a plus sign above the respective lanes. SecA, position of the corresponding SecA proteins; hiSecB, SecB from H. influenzae; ecSecB, SecB from E. coli; bsSecA, SecA from B. subtilis; beSecA, B. subtilis-E. coli SecA hybrid protein; bhSecA, B. subtilis-H. influenzae SecA hybrid protein. Lane 1, molecular size markers. (B) Determination of SecA-SecB dissociation constants by isothermal titration calorimetry (ITC). ITC assays of ecSecB interacting with bsSecA, ecSecA, and beSecA were performed at 20°C in triplicate for each interaction. One of the respective experiments is shown for each interaction. Integration of the raw data (top of panel B) yielded the heat in kcal/mol of SecB injected versus the SecB-to-SecA molar ratio (bottom of panel B). The dissociation constant we determined for the complex of ecSecA and ecSecB (Kd = 0.8 ± 0.3 μM) is slightly different from the value (Kd = 1.7 ± 0.2 μM) described previously (26), which most likely is due to differences in the temperature and the buffers used for ITC between the two experiments and the scatter in our data from the ITC200 titration.
Fig 2.
SecA hybrid proteins. (A) Overall domain structure of SecA. The functional SecA domains and subdomains are indicated as defined previously (15, 25). NBD, nucleotide binding domain; PBD, preprotein binding domain; IRA2, intramolecular regulator of ATPase 2; SD, scaffold domain, WD, wing domain; IRA1, intramolecular regulator of ATPase 1; CTL, C-terminal linker; CTD, C-terminal domain. (B) Schematic representation of SecA hybrid proteins used in this study. The numbers in parentheses correspond to the amino acids that are derived from the indicated parental SecA proteins. The respective fusion sites are indicated by an arrow.
To further confirm our hypothesis, the binding affinities of ecSecB to three different SecA proteins were determined by isothermal titration calorimetry (ITC). The dissociation constant for the binding of ecSecB to bsSecA was in the submillimolar range (Kd = 0.17 ± 0.03 mM) (Fig. 1B, left), which is 200-fold higher than the dissociation constant observed for the binding between ecSecB and ecSecA (Kd = 0.8 ± 0.3 μM) (Fig. 1B, middle). The binding affinity between ecSecB and beSecA (Kd = 2.2 ± 0.4 μM) (Fig. 1B, right) was comparable to that between ecSecB and ecSecA and significantly higher than the affinity between ecSecB and bsSecA. The determined affinity constants are consistent with the results obtained in the in vitro binding assay described above.
Coexpression of ecSecB and beSecA in B. subtilis improves secretion of an inefficiently exported heterologous model protein.
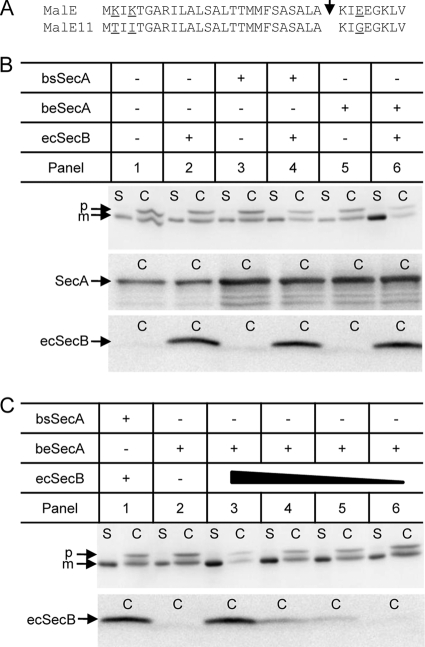
Since the results described above have shown that the beSecA hybrid protein can bind to ecSecB in vitro, we investigated whether the coexpression of these two proteins in B. subtilis results in a functional SecB-SecA protein-targeting pathway in vivo. For the expression of ecSecB in B. subtilis, its gene was cloned under the control of a xylose-inducible promoter and subsequently integrated into the lacA locus in the chromosome. In the corresponding xylose-induced cells, ecSecB can be detected in the cell (C) fraction (Fig. 3B). For the expression of bsSecA or beSecA in B. subtilis, a plasmid-based expression system containing the respective secA genes under the control of a constitutive promoter (HpaIIp) was used. The expression of plasmid-encoded SecA proteins in addition to the chromosomally encoded bsSecA resulted in increased amounts of total SecA protein in the C fraction (Fig. 3B). As a model Sec protein, we chose a variant of the SecB-dependent E. coli maltose-binding protein (MalE11) that possesses three amino acid alterations to convert the protein into a less efficient Sec substrate (Fig. 3A). When MalE11 was expressed in B. subtilis, small amounts of secreted mature MalE protein could be detected in the supernatant (S) fraction (Fig. 3B, panel 1). In the C fraction of the same strain, a mature-sized form of MalE was present which might correspond to the translocated and processed MalE that has not been released from the cell wall or, alternatively, to a nonexported degradation product of the MalE11 precursor in which the signal peptide has been clipped off by cytosolic proteases. Importantly, however, a significant amount of full-length MalE11 precursor was found to accumulate in the C fraction, suggesting that MalE11 export in B. subtilis occurred by an inefficient and most likely posttranslational mode of membrane translocation.
Fig 3.
Improvement of MalE11 secretion in B. subtilis by coexpression of ecSecB and beSecA. (A) Amino acid alterations present in the inefficiently secreted MalE11 protein. MalE, primary sequence of wild-type MalE corresponding to the signal peptide and the early mature region. MalE11, MalE mutant protein possessing mutations K2T and K4T in the signal peptide and mutation E29G in the early mature protein. The corresponding mutations result in a reduction of the positive net charge of the n-region of the signal peptide and in a more positively charged early mature protein domain, both of which are known to slow down the kinetics and overall efficiency of MalE membrane translocation in E. coli (28). The respective amino acid positions are underlined. The arrowhead indicates the signal peptidase cleavage site. (B) MalE11 secretion is enhanced in the presence of ecSecB and beSecA. Cellular (C) and supernatant (S) fractions of B. subtilis expressing the SecA and SecB proteins as indicated above the respective panels were subjected to SDS-PAGE and Western blotting using MalE antibodies (upper). In addition, the cellular fractions were immunoblotted using B. subtilis SecA antibodies (middle) or E. coli SecB antibodies (lower). p, MalE11 precursor protein; m, mature MalE11 protein. (C) Secretion stimulation of MalE11 in the presence of beSecA correlates to ecSecB availability. The amount of ecSecB, as symbolized by the solid triangle, in the cell was modulated by varying the concentration of the inducer xylose in the growth medium. The xylose concentrations used were 0.5% (panel 3), 0.1% (panel 4), 0.05% (panel 5), and 0% (panel 6), respectively. MalE11-derived protein products in the C and S fractions, as well as the amount of ecSecB in the C fractions of the respective cells, were analyzed by immunoblotting as described for panel B.
Expression of ecSecB, bsSecA, or beSecA alone had no effect on the secretion of MalE11 in B. subtilis (Fig. 3B, compare panel 1 to panels 2, 3, and 5). In contrast, the coexpression of ecSecB with bsSecA resulted in a slight increase in MalE11 secretion (Fig. 3B, panel 4), indicating that the weak interaction between bsSecA and ecSecB allowed a slightly improved targeting of the MalE11 precursor to the translocase. Strikingly, however, the coexpression of ecSecB and beSecA with MalE11 resulted in a clear improvement of MalE11 secretion, since significantly more mature MalE protein was present in the S fraction and the amount of accumulated precursor in the C fraction was decreased (Fig. 3B, panel 6). Furthermore, the modulation of the amount of ecSecB in the cell by varying the concentration of the inducer xylose clearly showed that the beSecA-dependent stimulation of MalE11 secretion correlated with ecSecB availability (Fig. 3C, panels 4 to 6). Taken together, these findings suggest that a functional SecB-mediated protein-targeting pathway is operational in B. subtilis cells coexpressing ecSecB and the SecB-binding-proficient beSecA hybrid protein.
Improved secretion of MalE11 in B. subtilis is dependent on the SecB-targeting function.
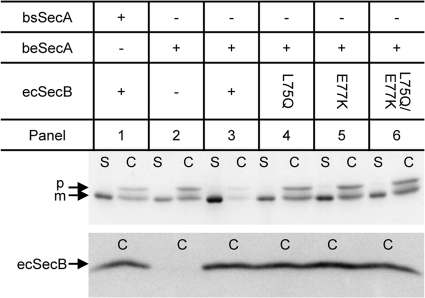
To further support our view that the stimulation of MalE11 secretion by the coexpression of ecSecB and beSecA was due to a functional SecB-mediated protein-targeting pathway, we analyzed the effect of SecB mutations (L75Q and E77K) that are known to cause a marked reduction in the affinity of SecB for SecA but do not affect its substrate binding ability (10, 23, 38). As shown in the lower part of Fig. 4, the SecB mutant proteins ecSecBL75Q, ecSecBE77K, and ecSecBL75Q&E77K were expressed at a level comparable to that of the wild-type ecSecB protein (compare panel 3 to panels 4 to 6). In contrast to the wild-type ecSecB, these mutants no longer promoted an improved secretion of MalE11 when coexpressed with beSecA (Fig. 4, panels 3 to 6), as indicated by a reduction of the amount of mature MalE in the S fraction and a concomitant increase in the amount of accumulated MalE11 precursor in the cytosol. From these results, we conclude that the ecSecB-beSecA interaction-dependent protein-targeting pathway delivers ecSecB-bound MalE11 precursor to beSecA protein and thereby improves MalE11 secretion in B. subtilis.
Fig 4.
Stimulation of MalE11 secretion requires the ecSecB targeting function. Cellular (C) and supernatant (S) fractions of B. subtilis coexpressing beSecA together with wild-type ecSecB and targeting-function-deficient mutant proteins (ecSecBL75Q, ecSecBE77K, or ecSecBL75Q&E77K) as indicated above the respective panels were subjected to SDS-PAGE and Western blotting using MalE antibodies (upper part). In addition, the cellular fractions were immunoblotted using E. coli SecB antibodies (lower part). p, MalE11 precursor; m, mature MalE11 protein.
The B. subtilis ecSecB-beSecA targeting pathway improves the secretion of a SecB-independent heterologous model protein.
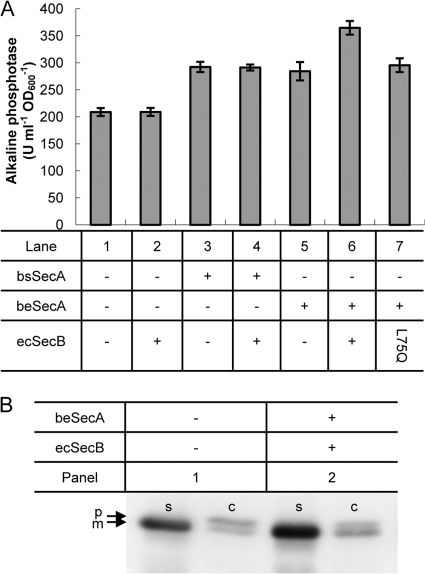
Unlike MalE, the E. coli alkaline phosphatase PhoA is considered to be exported in a SecB-independent manner, at least under normal growth conditions (16, 18). To analyze whether the artificially introduced SecB-mediated targeting pathway can be beneficial for the secretion of normally SecB-independent proteins, the PhoA precursor was coexpressed with different combinations of ecSecB, bsSecA, and beSecA in B. subtilis, and the amount of secreted PhoA in the culture supernatant was determined by measuring alkaline phosphatase activity and by Western blot analysis. As shown in Fig. 5A, without the coexpression of Sec components, the basal level of PhoA activity of 220 U ml−1 OD600−1 was found in the culture supernatant of the respective cells (lane 1). Although the expression of ecSecB alone did not result in an improvement in PhoA secretion (lane 2), the overexpression of either bsSecA (lane 3) or beSecA (lane 5) alone increased the secretion of PhoA by approximately 30%. When ecSecB and bsSecA were coexpressed, no further improvement was observed (lane 4). In contrast, the presence of ecSecB together with beSecA increased the PhoA secretion amount up to 60%, indicating that the targeting of ecSecB-bound PhoA precursor to beSecA also contributes significantly to the total increase in PhoA activity. This notion was further supported by the finding that the coexpression of beSecA with the ecSecB(L75Q) mutant protein did not confer an additional increase of PhoA activity above the 30% observed when only beSecA was expressed (lane 7).
Fig 5.
Improved secretion of PhoA by B. subtilis in the presence of the ecSecB-beSecA targeting pathway. (A) PhoA activity in the supernatants of B. subtilis cells expressing different combinations of SecA and SecB proteins as indicated below the respective lanes. The averages of two replicates for each strain and standard deviations are presented. (B) PhoA-derived polypeptides in the cellular (C) and supernatant (S) fractions of B. subtilis cells lacking (panel 1) or possessing (panel 2) the ecSecB-beSecA protein targeting pathway. p, PhoA precursor; m, mature PhoA.
Western blot analysis of the S and C fractions of B. subtilis cells that expressed PhoA either with (Fig. 5B, panel 2) or without (Fig. 5B, panel 1) the coexpression of the ecSecB-beSecA targeting pathway clearly support the beneficial effect of the artificial targeting pathway on PhoA secretion. An increased amount of mature PhoA was detected in the S fraction of cells that coexpressed ecSecB and beSecA compared to the control cells. Interestingly, the amounts of accumulated pre-PhoA precursor in the C fractions were almost identical in both strains, suggesting that the additional amount of secreted PhoA that is observed in the presence of ecSecB-beSecA reflects a fraction of the PhoA precursor that has been rescued from proteolytic degradation by interaction with the targeting pathway.
DISCUSSION
In the present study, we have introduced a functional SecB-dependent protein-targeting pathway in the Gram-positive model bacterium B. subtilis. The replacement of the C-terminal 32 amino acids of bsSecA by the corresponding part of ecSecA resulted in a beSecA hybrid protein that, in contrast to the unaltered bsSecA, possessed a high binding affinity for ecSecB. The coexpression of ecSecB together with the beSecA hybrid in B. subtilis improved the secretion of two heterologous model proteins (MalE11 and PhoA) into the culture supernatant in an ecSecB-beSecA interaction-dependent manner. From a biotechnological view, the construction of an artificial protein-targeting pathway in a Gram-positive bacterium can be considered a novel strategy for the improvement of this industrially important class of microorganisms with respect to their use as host systems for the secretory production of heterologous proteins.
Multiple interactions involved in SecB-SecA complex formation have been extensively studied in the E. coli system. One of the contact areas identified is between the negatively charged flat ß-sheet of the SecB tetramer and the zinc-containing C-terminal 22 residues of SecA (9–11, 17, 43). Another existing contact area is between the C-terminal 13 residues of SecB and the N-terminal 11 residues of SecA (30). Interestingly, the latter seems to negatively contribute to the formation of the SecB-SecA complex, which is thought to play a crucial role in transferring SecB-bound precursors via SecA to the SecYEG translocon (30).
Since SecB is absent from B. subtilis and other Gram-positive bacteria, the targeting of Sec substrates to the translocase in these organisms is thought to be mediated mainly, if not exclusively, by the SRP pathway (14, 34, 41, 42). Nevertheless, the region encompassing the C-terminal 22 residues of the bsSecA is highly homologous to the counterpart of ecSecA that has a high affinity to ecSecB (see Fig. S3 in the supplemental material). Although the isolated bsSecA C-terminal tail can bind ecSecB in the context of a GST fusion protein (37), the full-length bsSecA protein did not form a stable complex with ecSecB in our in vitro binding assay. This suggests that the C tail is not freely accessible for SecB binding or that other unfavorable contacts in bsSecA, such as the extreme N-terminal domain (30), prevent the formation of a stable ecSecB-bsSecA complex. However, the coexpression of ecSecB together with bsSecA in B. subtilis resulted in a slight improvement of MalE11 secretion, indicating that a weak interaction between ecSecB and bsSecA can occur in vivo, thereby allowing an inefficient SecB-dependent targeting of MalE11 precursors to bsSecA.
In contrast, the beSecA hybrid protein can form a stable complex with ecSecB in vitro, and the observed binding affinity closely resembles that between ecSecB and its cognate interacting partner ecSecA. Thus, the few amino acid differences that exist between the C tails of ecSecA and bsSecA (see Fig. S3 in the supplemental material) preclude an efficient ecSecB-dependent targeting to bsSecA, indicating that the structural details of the extreme C tail of SecA are of crucial importance for the overall SecB-SecA binding affinity. Moreover, the coexpression of ecSecB and beSecA strongly stimulates MalE11 secretion in vivo due to an efficient delivery of the ecSecB-bound substrate to beSecA, a notion that was confirmed by the observation that the increased secretion stimulation disappeared when the wild-type ecSecB was replaced by substrate-binding-proficient but SecA-binding-deficient ecSecB mutant proteins.
Compared to that of MalE11, the secretion of wild-type MalE in B. subtilis was more efficient. In the cellular fraction of the corresponding strain, only small amounts of MalE precursor accumulated, indicating that MalE was effectively targeted to the translocase by the B. subtilis in-house targeting systems. Therefore, MalE secretion only marginally benefited from the presence of the implemented SecB-dependent targeting pathway in addition to the slight positive effect by coexpressing ecSecB alone (see Fig. S4 in the supplemental material, compare panels 2 and 6) (5). Interestingly, the yield of secreted PhoA, which in its native host E. coli does not rely on the SecB-dependent targeting pathway under normal conditions (16, 18), was significantly increased in B. subtilis containing the artificial SecB-dependent targeting pathway. Since the increase of secreted PhoA was not paralleled by a complementary decrease of accumulated pre-PhoA precursor, the most reasonable explanation is that the artificially introduced targeting pathway rescued a fraction of PhoA from proteolysis by cytoplasmic proteases.
In general, when the secretion of a heterologous protein by B. subtilis is attempted, the nascent precursor protein faces different potential fates in the foreign environment, such as aggregation, degradation, folding into an export-incompetent state, or successful targeting to the Sec translocase. Although the kinetic partitioning between all those possibilities and, as a consequence, a putative beneficial effect of the SecB-dependent targeting pathway is not predictable in advance for any desired target protein, our results nevertheless show that the implementation of a SecB-dependent targeting pathway adds a promising new tool to the repertoire of strategies aimed at the improvement of the secretory production of heterologous proteins by B. subtilis and, most likely, other SecB null bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (30570028, to S.Y.), Shanghai Municipal Natural Science Foundation (05ZR14137, to S.Y.), Dupont Young Professor Award 2006 (to S.Y.), National Program on Key Basic Research of China (2009CB918600, to J.Z.), Key Program of Shanghai Municipal Natural Science Foundation (08JC1412700, to J.Z.), Hundred Talents Program (to J.Z.), and ExpressO (w0805wb003c, to R.F.). L.D. is a recipient of a DAAD-CAS joint fellowship.
S.Y. first conceived of the possibility of establishing the SecB-mediated targeting pathway in B. subtilis by exploiting hybrid SecA proteins.
We are very grateful to Juan Wang for purifying all SecA and SecB proteins for in vitro assay and to Yongsuo Chen at GE Healthcare for assistance in the ITC experiments. We thank Michael Bott for his support during the 2-year stay of L.D. in the laboratory of R.F. We thank Lefu Lan for critically reading the manuscript.
Footnotes
Published ahead of print 23 November 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Bechtluft P, Nouwen N, Tans SJ, Driessen AJ. 2010. SecB–a chaperone dedicated to protein translocation. Mol. Biosyst. 6:620–627 [DOI] [PubMed] [Google Scholar]
- 2. Blaudeck N, Kreutzenbeck P, Müller M, Sprenger GA, Freudl R. 2005. Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient Tat-dependent protein translocation in the absence of TatB. J. Biol. Chem. 280:3426–3432 [DOI] [PubMed] [Google Scholar]
- 3. Breukink E, et al. 1995. The C terminus of SecA is involved in both lipid binding and SecB binding. J. Biol. Chem. 270:7902–7907 [DOI] [PubMed] [Google Scholar]
- 4. Collier DN. 1994. Escherichia coli signal peptides direct inefficient secretion of an outer membrane protein (OmpA) and periplasmic proteins (maltose-binding protein, ribose-binding protein, and alkaline phosphatase) in Bacillus subtilis. J. Bacteriol. 176:3013–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collier DN. 1994. Expression of Escherichia coli SecB in Bacillus subtilis facilitates secretion of the SecB-dependent maltose-binding protein of E. coli. J. Bacteriol. 176:4937–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cross BC, Sinning I, Luirink J, High S. 2009. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 10:255–264 [DOI] [PubMed] [Google Scholar]
- 7. Darmon E, et al. 2006. A disulfide bond-containing alkaline phosphatase triggers a BdbC-dependent secretion stress response in Bacillus subtilis. Appl. Environ. Microbiol. 72:6876–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dartois V, Coppee JY, Colson C, Baulard A. 1994. Genetic analysis and overexpression of lipolytic activity in Bacillus subtilis. Appl. Environ. Microbiol. 60:1670–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fekkes P, de Wit JG, Boorsma A, Friesen RH, Driessen AJ. 1999. Zinc stabilizes the SecB binding site of SecA. Biochemistry 38:5111–5116 [DOI] [PubMed] [Google Scholar]
- 10. Fekkes P, et al. 1998. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol. Microbiol. 29:1179–1190 [DOI] [PubMed] [Google Scholar]
- 11. Fekkes P, van der Does C, Driessen AJ. 1997. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 16:6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grudnik P, Bange G, Sinning I. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782 [DOI] [PubMed] [Google Scholar]
- 13. Härtl B, Wehrl W, Wiegert T, Homuth G, Schumann W. 2001. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 183:2696–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirose I, et al. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65–75 [DOI] [PubMed] [Google Scholar]
- 15. Hunt JF, et al. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297:2018–2026 [DOI] [PubMed] [Google Scholar]
- 16. Kim J, Kendall DA. 1998. Identification of a sequence motif that confers SecB dependence on a SecB-independent secretory protein in vivo. J. Bacteriol. 180:1396–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimsey HH, Dagarag MD, Kumamoto CA. 1995. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J. Biol. Chem. 270:22831–22835 [DOI] [PubMed] [Google Scholar]
- 18. Kononova SV, Khokhlova OV, Zolov SN, Nesmeyanova MA. 2001. Effect of export-specific cytoplasmic chaperone, protein SecB, on secretion of Escherichia coli alkaline phosphatase. Biochemistry (Moscow) 66:803–807 [DOI] [PubMed] [Google Scholar]
- 19. Mandon EC, Trueman SF, Gilmore R. 2009. Translocation of proteins through the Sec61 and SecYEG channels. Curr. Opin. Cell Biol. 21:501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McNicholas P, Rajapandi T, Oliver D. 1995. SecA proteins of Bacillus subtilis and Escherichia coli possess homologous amino-terminal ATP-binding domains regulating integration into the plasma membrane. J. Bacteriol. 177:7231–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meissner D, Vollstedt A, van Dijl JM, Freudl R. 2007. Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria. Appl. Microbiol. Biotechnol. 76:633–642 [DOI] [PubMed] [Google Scholar]
- 22. Moore BA. 2007. Ph.D. thesis University of Michigan, Ann Arbor, MI [Google Scholar]
- 23. Murén EM, Suciu D, Topping TB, Kumamoto CA, Randall LL. 1999. Mutational alterations in the homotetrameric chaperone SecB that implicate the structure as dimer of dimers. J. Biol. Chem. 274:19397–19402 [DOI] [PubMed] [Google Scholar]
- 24. Natale P, Brüser T, Driessen AJ. 2008. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim. Biophys. Acta 1778:1735–1756 [DOI] [PubMed] [Google Scholar]
- 25. Papanikolau Y, et al. 2007. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J. Mol. Biol. 366:1545–1557 [DOI] [PubMed] [Google Scholar]
- 26. Patel CN, Smith VF, Randall LL. 2006. Characterization of three areas of interactions stabilizing complexes between SecA and SecB, two proteins involved in protein export. Protein Sci. 15:1379–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pohl S, Harwood CR. 2010. Heterologous protein secretion by Bacillus species from the cradle to the grave. Adv. Appl. Microbiol. 73:1–25 [DOI] [PubMed] [Google Scholar]
- 28. Puziss JW, Strobel SM, Bassford PJ., Jr 1992. Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations. J. Bacteriol. 174:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Randall LL, Hardy SJ. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59:1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Randall LL, Henzl MT. 2010. Direct identification of the site of binding on the chaperone SecB for the amino terminus of the translocon motor SecA. Protein Sci. 19:1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Sardis MF, Economou A. 2010. SecA: a tale of two protomers. Mol. Microbiol. 76:1070–1081 [DOI] [PubMed] [Google Scholar]
- 33. Schallmey M, Singh A, Ward OP. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1–17 [DOI] [PubMed] [Google Scholar]
- 34. Tjalsma H, et al. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Sluis EO, Driessen AJ. 2006. Stepwise evolution of the Sec machinery in Proteobacteria. Trends Microbiol. 14:105–108 [DOI] [PubMed] [Google Scholar]
- 37. van Wely KH, Swaving J, Klein M, Freudl R, Driessen AJ. 2000. The carboxyl terminus of the Bacillus subtilis SecA is dispensable for protein secretion and viability. Microbiology 146:2573–2581 [DOI] [PubMed] [Google Scholar]
- 38. Woodbury RL, et al. 2000. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions. J. Biol. Chem. 275:24191–24198 [DOI] [PubMed] [Google Scholar]
- 39. Xu Z, Knafels JD, Yoshino K. 2000. Crystal structure of the bacterial protein export chaperone SecB. Nat. Struct. Biol. 7:1172–1177 [DOI] [PubMed] [Google Scholar]
- 40. Yuan J, Zweers JC, van Dijl JM, Dalbey RE. 2010. Protein transport across and into cell membranes in bacteria and archaea. Cell. Mol. Life Sci. 67:179–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zanen G, et al. 2006. Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics 6:3636–3648 [DOI] [PubMed] [Google Scholar]
- 42. Zanen G, et al. 2005. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J. 272:4617–4630 [DOI] [PubMed] [Google Scholar]
- 43. Zhou J, Xu Z. 2003. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nat. Struct. Biol. 10:942–947 [DOI] [PubMed] [Google Scholar]
- 44. Zhou J, Xu Z. 2005. The structural view of bacterial translocation-specific chaperone SecB: implications for function. Mol. Microbiol. 58:349–357 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.