Abstract
Transposons are mobile genetic elements bounded by insertion sequences that are recognized by a specific mobilizing transposase enzyme. The transposase may mobilize not only the insertion sequences but also intervening DNA. mariner is a particularly efficient transposon for the random chromosomal integration of genes and insertional mutagenesis. Here, we modify an existing mariner transposon, TnYLB, such that it can easily be genetically manipulated and introduced into Bacillus subtilis. We generate a series of three new mariner derivatives that mobilize spectinomycin, chloramphenicol, and kanamycin antibiotic resistance cassettes. Furthermore, we generate a series of transposons with a strong, outward-oriented, optionally isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter for the random overexpression of neighboring genes and a series of transposons with a promoterless lacZ gene for the random generation of transcriptional reporter fusions. We note that the modification of the base transposon is not restricted to B. subtilis and should be applicable to any mariner-compatible host organism, provided that in vitro mutagenesis or an in vivo species-specific delivery vector is employed.
INTRODUCTION
Transposons are mobile genetic elements made of two repeated insertion sequences (IS) and intervening DNA (10). An enzyme called transposase recognizes, excises, and moves the specific insertion sequences from one piece of DNA to another. Transposons that carry antibiotic resistance cassettes are powerful tools in forward molecular genetics, as they generate insertion mutations with a tightly linked selectable marker (19, 22). To correlate a gene with a particular phenotype, one need only screen a transposon insertion library for a phenotype of interest, backcross the transposon insertion to ensure inseparable linkage, and identify the transposon insertion site.
Bacillus subtilis is a powerful model system for Gram-positive molecular genetics, and historically three different transposon systems have been used to generate random insertional mutations. The first transposon system developed for use in Bacillus subtilis was Tn917, which generated erythromycin-marked insertions but was biased in favor of insertions in the terminus-proximal half of the chromosome (28, 32). The second transposon system was MiniTn10, which generated spectinomycin-marked insertions with greater randomness than Tn917 but tended to insert at so-called hotspots that effectively reduced library insertion diversity (25, 27). The third and most recent transposon was mariner-derived TnYLB, which generated kanamycin-marked insertions at dinucleotide base sequence TA sites in the chromosome (21). As B. subtilis is an organism with low G+C content, TA base sequences are common and TnYLB insertion was efficient, random, and did not seem to be biased toward hotspots.
Whereas TnYLB is an excellent transposon for random insertional mutagenesis, it is not without limitations. First, TnYLB is limited to generating kanamycin resistance-marked insertions. Second, the host must not contain an erythromycin (mls) resistance marker in the chromosome due to a similar marker on the delivery plasmid that could generate spurious integrations by recombination. Third, TnYLB is limited primarily to loss-of-function mutations, as random insertions commonly abolish gene activity. Fourth, the TnYLB transposon lacks convenient restriction sites between the IS elements and therefore is difficult to reengineer for other applications (21).
Here, we take the fundamental principles behind the highly efficient and random insertions of TnYLB and reengineer the system for rapid and convenient genetic modification. A base transposon was designed into which one may clone any genetic constructs of interest. Once built, the modified transposon is moved into a delivery vector for introduction to, and mutagenesis of, any B. subtilis strain of interest. As proof of principle, we generated transposons with kanamycin, spectinomycin, and chloramphenicol resistance cassettes. We further generated a transposon called TnHyJump with an outward-facing, unregulated, high-expression promoter (Physpank) and a transposon called TnLacJump, with a promoterless lacZ gene for generating random β-galactosidase transcriptional reporters (24, 33). The system presented here expands the kind of genetic screens that can be conducted with mariner in terms of antibiotic resistance cassettes, reporters, and many other constructs for use not only in B. subtilis but all mariner-compatible bacteria.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis strains were grown in Luria-Bertani (LB) (10 g tryptone per liter, 5 g yeast extract per liter, 5 g NaCl per liter) broth or on LB plates supplemented with 1.5% Bacto agar at 37°C. In some experiments, cells were grown in 10 ml MSgg biofilm-promoting medium (5 mM potassium phosphate [pH 7], 100 mM morpholinepropanesulfonic acid [MOPS; pH 7], 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg/ml tryptophan, 50 μg/ml phenylalanine, and 50 μg/ml threonine) in 6-well microtiter plates or on MSgg medium solidified with 1.5% Bacto agar and incubated for 3 days at room temperature (4). When appropriate, antibiotics were included at the following concentrations: 100 μg/ml spectinomycin, 5 μg/ml chloramphenicol, 5 μg/ml kanamycin, and 1 μg/ml erythromycin plus 25 μg/ml lincomycin (mls). Isopropyl-β-d-1-thiogalactopyranoside (IPTG; Sigma) was added to the medium at a concentration of 1 mM when appropriate. Bromo-chloro-indolyl-galactopyranoside (X-gal; Sigma) was added to the medium at a concentration of 20 μg/ml when appropriate. Blue/white colony pictures were photographed with a Canon Powershot A620 digital camera.
Strain construction.
All constructs were introduced into the domesticated strain PY79 by natural competence. In some cases the transposon delivery plasmids were transduced to the ancestral 3610 strain background by SPP1-mediated generalized phage transduction (5, 31, 34). All strains used in the present study are listed in Table 1. All plasmids used in the present study are listed in Table 2. All primers used in the present study are listed in Table 3.
Table 1.
B. subtilis strains
| Strain | Genotypea |
|---|---|
| 3610 | Wild type |
| DS7133 | [PY79] pEP4 TnLacJump spec amp mls mariner-Himar1ori(TS)Bs |
| DS7167 | [PY79] srfABΩTnLacJumpspec |
| DS7169 | [PY79] azlBΩTnLacJumpspec |
| DS7487 | [PY79] pEP6 TnLacJump kan amp mls mariner-Himar1ori(TS)Bs |
| DS7996 | [PY79] pEP20TnHyJumpkan amp mls mariner-Himar1ori(TS)BslacA::lacI tet amyE::lacZ cat |
| DS7997 | [PY79] pEP19 TnHyJump spec amp mls mariner-Himar1ori(TS)BslacA::lacItet amyE::lacZ cat |
| DS8042 | [PY79] yesVΩTnHyJumpspec lacA::lacI tet amyE::lacZ cat |
| DS8044 | [PY79] yesQΩTnHyJumpspec lacA::lacI tet amyE::lacZ cat |
| DS8137 | [PY79] pEP26TnHyJumpcat amp mls mariner-Himar1ori(TS)BslacA::lacItet thrC::lacZ spec |
| DS8164 | [PY79] yesWΩTnHyJumpcat lacA::lacI tet thrC::lacZ spec |
| DS8274 | [PY79] pEP25 TnLacJump cat amp mls mariner-Himar1ori(TS)Bs |
| DS8328 | [PY79] yqeYΩTnLacJumpkan |
| DS8329 | [PY79] <yvrMΩTnLacJump kan |
| DS8355 | [PY79] rpsAΩTnLacJumpcat |
| DS8357 | [PY79] ptsGΩTnLacJumpcat |
| DS8359 | [PY79] yeeAΩTnLacJumpcat |
| DS8473 | [3610] pKB178 TnKRM kan mls mariner-Himar1ori(TS)Bs |
| DS8474 | [3610] pEP24 TnKRM cat mls mariner-Himar1ori(TS)Bs |
| DS8546 | [3610] ymdBΩTnKRMkan |
| DS8564 | [3610] galEΩTnKRMkan |
| DS8606 | [PY79] fnrΩTnLacJumpkan |
| DS8677 | [3610] epsKΩTnKRMcat |
| DS8681 | [3610] epsNΩTnKRMcat |
| DS8684 | [3610] slrRΩTnKRMcat |
| DS8725 | [PY79] rrn-23SΩTnLacJump spec |
| DS8742 | [PY79] <yesOΩTnHyJump cat lacA::lacI tet thrC::lacZ spec |
| DS8871 | [PY79] yesXΩTnHyJumpkan lacA::lacI tet amyE::lacZ cat |
| DS8872 | [PY79] yesPΩTnHyJumpkan lacA::lacI tet amyE::lacZ cat |
| DS8879 | [3610] remBΩTnKRMkan |
| DS8994 | [3610] pDP384 TnKRM spec mls mariner-Himar1ori(TS)Bs |
| DS9040 | [3610] epsEΩTnKRMspec |
| DS9045 | [3610] epsMΩTnKRMspec |
| DS9047 | [3610] epsDΩTnKRMspec |
| PY79 | sfpswrA |
parental strain backgrounds are indicated in brackets, transposon insertion site is upstream of the indicated gene.
Table 2.
Plasmids
| Plasmid | Genotype | BGSC no. |
|---|---|---|
| pAC225 | cat amp (18) | |
| pAH54 | spec amp (18) | |
| pDG268 | amyE::lacZamp (2) | |
| pDG1515 | tet amp (13) | |
| pDP383 | TnKRMspec amp | ECE241 |
| pDP384 | TnKRMspec amp mls mariner-Himar1ori(TS)Bs | ECE233 |
| pDR111 | amyE::Physpankspec mls amp (3) | |
| pEP1 | TnLacJumpspec amp | ECE242 |
| pEP3 | TnLacJump kan amp | ECE243 |
| pEP4 | TnLacJump spec amp mls mariner-Himar1ori(TS)Bs | ECE236 |
| pEP6 | TnLacJump kan amp mls mariner-Himar1ori(TS)Bs | ECE235 |
| pEP17 | TnHyJump spec amp | ECE244 |
| pEP18 | TnHyJump kan amp | ECE245 |
| pEP19 | TnHyJump spec amp mls mariner-Himar1ori(TS)Bs | ECE239 |
| pEP20 | TnHyJump kan amp mls mariner-Himar1ori(TS)Bs | ECE238 |
| pEP21 | TnKRMcat amp | ECE246 |
| pEP22 | TnLacJump cat amp | ECE247 |
| pEP23 | TnHyJump cat amp | ECE248 |
| pEP24 | TnKRMcat amp mls mariner-Himar1ori(TS)Bs | ECE234 |
| pEP25 | TnLacJump cat amp mls mariner-Himar1ori(TS)Bs | ECE237 |
| pEP26 | TnHyJump cat amp mls mariner-Himar1ori(TS)Bs | ECE240 |
| pKB157 | IS<kan>IS amp | ECE249 |
| pKB169 | lacA5′tet amp | |
| pKB170 | lacA::tetamp | |
| pKB175 | lacA::lacI tet amp | ECE252 |
| pKB176 | amp mls mariner-Himar1ori(TS)Bs | ECE250 |
| pKB177 | TnKRMkan amp | ECE251 |
| pKB178 | TnKRMkan amp mls mariner-Himar1ori(TS)Bs | ECE232 |
| pMarA | TnYLB-1 A mls mariner-Himar1 ori(TS)Bs (21) | |
| pUC19 | amp |
Table 3.
Primers
| Primer | Sequence |
|---|---|
| 1886 | AGGAGGGATCCGACTCTCTAGCTTGAGGCATCAAAT |
| 1888 | AGGAGGGATCCCCAGCTTGAATTGATACACTAATGCT |
| 1889 | CTCCTCTCGAGTTATTTTTGACACCAGACCAACTGGTAAT |
| 2054 | AATTGTGAGCGGATAACAATTTCACA |
| 2077 | GTCGACCGGCCGGGATCCTACTCGAGCCACATAGATGGCGTCGCTAGTA |
| 2078 | ACTAGTGCTAGCCCCGGGCTCGAGAACAAAGAAAAACACATTTTTTTGTTAAAA |
| 2079 | AGGAGAAGCTTTAACAGGTTGGCTGATAAGTCCCCGGTCTGGTCGACCGGCCGGGATCCTA |
| 2080 | CTCCTGAATTCTTTAGACATCTAAATCTAGGTACTAAAAC |
| 2081 | AGGAGCAATTGATGCTTTAACTACATGCTTTTTAGACA |
| 2082 | CTCCTGGTACCCTGCAGTAACAGGTTGGCTGATAAGTCCCCGGTCTACTAGTGCTAGCCCCGGGCTC |
| 2201 | AGGAGCTCGAGTAGTTCTAGAGCGGCCGCCGCCAC |
| 2202 | CTCCTGCTAGCACGACTCACTATAGGGCGAATTGG |
| 2203 | AGGAGCTCGAGCCCTGGCGAATGGCGATTTTCG |
| 2204 | CTCCTGCTAGCCCCTATGCAAGGGTTTATTGTTTTC |
| 2207 | AGGAGCTCGAGATGGCGTCGCTAGTATTAAATGCAT |
| 2208 | CTCCTGCTAGCTTTAGACATCTAAATCTAGGTACTAAAA |
| 2209 | AGGAGGTCGACGACTCTCTAGCTTGAGGCATCAAA |
| 2224 | AGGAGAGATCTACGCACGTAACAAAAGCAAAATTTATG |
| 2225 | CTCCTCTGCAGTTGCTGTAGTCGAGGCCCTGATA |
| 2226 | AGGAGAAGCTTAGAGGATCCAGGCTCGAGAGCTAGCGATGCTCAGGGGTTTGCGAAGG |
| 2227 | CTCCTGGTACCGTGTGTTTACGACAATTCTCACTTC |
| 2211 | CTCCTGGATCCTAACTCACATTAATTGCGT |
| 2271 | CTCCTGTCGACAATTGTTATCCGCTCACAATTACAC |
| 2312 | CGACGGCCAGTGAATTCGAGCT |
| 2567 | GTACATCCGCAACTGTCCATA |
| 2569 | ATATTCATTCTAATTGGTAATCAGA |
| 2570 | CTAAGTCATAATTTCCGTATATTC |
| 2729 | CCTGCTGTAATAATGGGTAGAA |
| 2757 | TCAGATAGGCCTAATGACTGG |
| 2816 | AGCTGGCACGACAGGTTTC |
| 2817 | AGGCTGCGCAACTGTT |
| 2818 | TCTCCCAATCAGGCTTGAT |
SPP1 phage transduction.
To 0.2 ml of dense culture grown in TY broth (LB broth supplemented after autoclaving with 10 mM MgSO4 and 100 μM MnSO4), serial dilutions of SPP1 phage stock were added and statically incubated for 15 min at 37°C. To each mixture, 3 ml TYSA (molten TY supplemented with 0.5% agar) was added, poured atop fresh TY plates, and incubated at 37°C overnight. Top agar from the plate containing nearly confluent plaques was harvested by scraping into a 50-ml conical tube, vortexed, and centrifuged at 5,000 × g for 10 min. The supernatant was treated with 25 μg/ml DNase before being passed through a 0.45-μm syringe filter and stored at 4°C.
Inverse PCR.
Chromosomal DNA was isolated from the candidate strain, and 1 μg of chromosomal DNA was digested with Sau3AI or TaqI for 1 h. The digestion then was heat inactivated for 20 min, and 0.1 μg of digested DNA was ligated using T4 DNA ligase at room temperature for 1 h. PCR was conducted using each of the ligation reaction mixtures as a template, primers 2567/2818 for transposons containing a kanamycin resistance marker, primers 2569/2570 for transposons containing a spectinomycin resistance marker, primers 2757/2729 for transposons containing a chloramphenicol resistance marker (Table 4), and Phusion DNA polymerase (New England BioLabs). Each PCR was purified (Qiagen PCR purification kit) and sequenced using primer 2567, 2569, or 2757 for kanamycin, spectinomycin, and chloramphenicol resistance cassettes, respectively. The sequence CCAACCTGT marks the end of the mariner transposon insertion sequence. The next two bases will be TA, according to the mechanism of mariner transposition, and mark the beginning of the chromosomal DNA specific to the insertion site. We note that we encountered one strain, DS9040, that contained a transposon insertion for which the first two bases of chromosomal DNA adjacent to the insertion sequence were GA rather than TA. We infer that this particular insertion was a rare event and perhaps represents an infrequent error in Himar1 transposition.
Table 4.
Amplification and sequencing primers for inverse PCR
| Transposon series | Amplification primers | Sequencing primer |
|---|---|---|
| Kanamycin | 2567/2818 | 2567 |
| Spectinomycin | 2569/2570 | 2569 |
| Chloramphenicol | 2757/2729 | 2757 |
Please address all plasmid requests to the Bacillus Genetic Stock Center (www.bgsc.org).
RESULTS AND DISCUSSION
Generating vectors for mariner genetic manipulation and delivery.
mariner-based transposon insertions can be rapidly and efficiently generated in vivo using pMarA, and we used pMarA as the basis for the design of a universal delivery vector (16, 21, 29). The vector pMarA contains transposon TnYLB with a kanamycin resistance cassette internal to the transposon, the gene encoding the hyperactive C9 allele of mariner-Himar1 transposase, an E. coli origin of replication (oriC), a temperature-sensitive B. subtilis origin of replication [ori(Ts)Bs], and an erythromycin (mls) resistance cassette external to the transposon (21). To generate a universal transposon delivery vector, pMarA was digested with PstI to excise TnYLB, and the larger backbone fragment was gel purified and religated to generate pKB176 (Fig. 1A). Once a transposon has been constructed, it can be cloned into the empty pKB176 delivery vector or into pMarA itself by the replacement of TnYLB and introduced to B. subtilis strains.
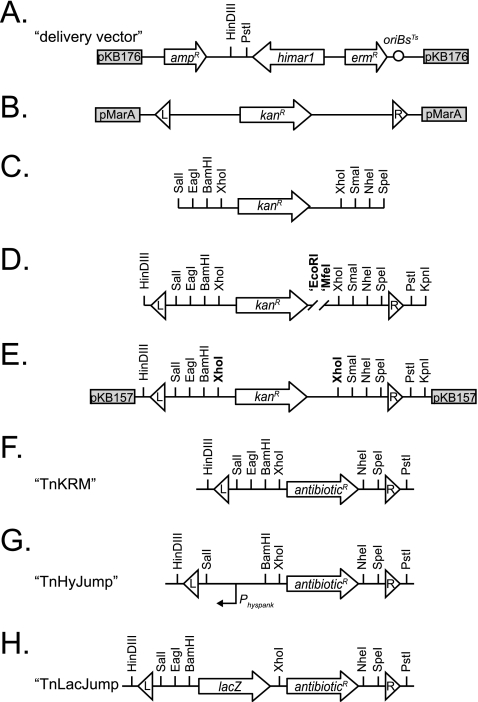
Fig 1.
Genetic maps of transposon construction and final architecture. (A) Map of the delivery vector pKB176, into which completed transposons are cloned and introduce to B. subtilis. (B to E) Maps of the intermediates in transposon assembly. All subsequent constructs were cloned into pKB157 as shown in panel E. (F to H) Generalized maps of the completed transposons for which the actual antibiotic resistance maker has been replaced by the term antibioticR. Arrows indicate genes. Boxes indicate the rest of the plasmid backbone and plasmid name. Circle indicates the temperature-sensitive origin of replication in B. subtilis [ori(TS)Bs]. Triangles indicate mariner insertion sequences (IS elements) labeled L (left) and R (right) to provide an arbitrary orientation. Vertical lines indicate restriction endonuclease sites. The bent arrow represents a promoter.
Our next goal was to design a transposon consisting of two flanking mariner IS elements and an intervening polylinker for convenient genetic modification. The transposon was built in a series of four steps using a combination of PCR and molecular cloning (Fig. 1B to E). First, the kanamycin resistance (Kanr) cassette from TnYLB was PCR amplified with primers 2077/2078 using pMarA as a template (Fig. 1B). The resulting PCR product incorporated three unique restriction sites on each end of the kanamycin resistance cassette flanked by two innermost XhoI restriction sites (Fig. 1C). The kanamycin resistance cassette shown in Fig. 1C then was used as a template for a second round of separate PCRs using primer pairs 2079/2080 and 2081/2082 to add the left and right mariner IS elements, respectively (Fig. 1D). The transposon was assembled by digesting the left arm with HindIII and EcoRI, digesting the right arm with MfeI and KpnI, and simultaneously ligating both arms into the HindIII/KpnI sites of plasmid pUC19 to generate plasmid pKB157 (Fig. 1E). Note that the EcoRI and MfeI sites have compatible overhangs which, when ligated, destroy each site in the final product (Fig. 1D and E). All subsequent genetic manipulations of the mariner transposon were generated in pKB157 and then cloned into the delivery vector pKB176 for introduction to B. subtilis.
mariner transposons with modified antibiotic cassettes.
We constructed three transposons, TnKRM spec, TnKRM cat, and TnKRM kan, containing antibiotic resistance cassettes for spectinomycin, chloramphenicol, and kanamycin, respectively (Fig. 1F). To build the transposons, the genes encoding spectinomycin, chloramphenicol, and kanamycin resistance were PCR amplified from templates pAH54 (using primers 2203/2204), pAC225 (using primers 2201/2202), and pKB157 (using primers 2207/2208), respectively (18). Each of the PCR products was digested with XhoI and NheI and separately ligated into the XhoI and NheI sites of pKB157 to generate plasmids pDP383, pEP21, and pKB177. Note that pKB157 and pKB177 both contain the kanamycin resistance cassette between the mariner insertion elements, but pKB177 has compatible restriction sites that enable subsequent constructs to be built in parallel with its sibling plasmids pDP383 (carrying a spectinomycin resistance cassette) and pEP21 (carrying a chloramphenicol resistance cassette). To generate transposons for use in B. subtilis, each of the transposon-containing plasmids was used as the DNA template for PCR amplification using primers 2054/2312. The resulting PCR products were digested with HindIII and PstI and ligated into the HindIII and PstI sites of pKB176 to generate the transposon delivery vectors pDP384, pEP24, and pKB178, respectively.
To introduce the transposon delivery vectors to B. subtilis, each vector was concatemerized by propagation in recA+ E. coli TG1 and transformed into the naturally competent B. subtilis laboratory strain PY79 by mls selection at the permissive temperature of 30°C for the extrachromosomal maintenance of the replicon. Phage lysates then were made of strains harboring the desired transposon delivery vector with the generalized transducing phage SPP1, such that the vectors could be transduced to any B. subtilis strain of interest (5). For mutagenesis, cells were grown for 10 h at 25°C, serially diluted, plated onto plain LB and selective LB agar plates, and incubated overnight at 42°C. Growth at 42°C is nonpermissive for the maintenance of the delivery vector, and thus antibiotic resistance primarily arises from the chromosomal insertion of the transposon. Transposition frequencies were measured for each of the modified TnKRM series of transposons by dividing the number of CFU grown on antibiotic by the number of CFU grown in the absence of antibiotic (Table 5). The spurious integration of the transposon and delivery vehicle into the chromosome occurred at 10- to 100-fold lower frequency than transposition (Table 5).
Table 5.
Transposition frequency
| Delivery plasmid | Transposon | Transposition frequencya | Delivery plasmid integration frequencyb |
|---|---|---|---|
| pMarA | TnYLB kan | 2.1 × 10−2 | 4.2 × 10−4 |
| pKB178 | TnKRM kan | 2.1 × 10−2 | 1.5 × 10−4 |
| pDP384 | TnKRM spec | 1.1 × 10−1 | 7.0 × 10−4 |
| pEP24 | TnKRM cat | 6.8 × 10−2 | 1.8 × 10−4 |
| pEP6 | TnLacJump kan | 1.2 × 10−2 | 4.7 × 10−4 |
| pEP4 | TnLacJump spec | 1.5 × 10−2 | 1.8 × 10−4 |
| pEP25 | TnLacJump cat | 2.6 × 10−2 | 3.9 × 10−4 |
| pEP20 | TnHyJump kan | 4.3 × 10−2 | 5.6 × 10−4 |
| pEP19 | TnHyJump spec | 8.2 × 10−2 | 9.0 × 10−4 |
| pEP26 | TnHyJump cat | 3.4 × 10−1 | 5.2 × 10−4 |
Transposition frequency calculated as the number of colonies resistant to the antibiotic within the transposon at 42°C divided by the total number of colonies plated in the absence of antibiotic.
Delivery plasmid integration frequency calculated as the number of colonies resistant to mls resistance antibiotic carried on the delivery plasmid outside the transposon at 42°C divided by the total number of colonies plated in the absence of antibiotic.
Mutagenizing a population of B. subtilis cells with transposons is a simple and efficient method for conducting forward genetic screens. To determine the usefulness of our newly designed transposons, a forward genetic screen was designed to generate mutants defective in biofilm formation in the undomesticated B. subtilis strain 3610. Biofilms manifest as robust floating pellicles in liquid that are correlated with rough colonies of complex architecture on agar surfaces (4). To screen for biofilm-defective mutants, three strain derivatives of 3610 containing either TnKRM spec, TnKRM cat, or TnKRM kan (DS8473, DS8474, and DS8994) were mutagenized and plated on LB agar containing the corresponding antibiotic. Mutations that conferred a smooth-colony phenotype, which is correlated with biofilm defects, were isolated. SPP1 phage lysates were generated on candidate colonies, and the transposon insertions were backcrossed into 3610 to ensure inseparable linkage to the smooth-colony phenotype and the transposon insertion.
Wild-type B. subtilis 3610 forms rough colonies on complex LB medium, but biofilm formation is most robust on the defined medium MSgg (4). Each candidate mutant was toothpick inoculated on MSgg solid medium to assay colonies of complex architecture, and 10 μl of an overnight LB broth culture was inoculated into the wells of 6-well microtiter plates containing 10 ml MSgg broth to assay pellicle formation. Both plates and broth were incubated at room temperature for 3 days. Wild-type cells formed colonies of complex architecture on MSgg solid agar, but each of the candidate transposon mutants exhibited smooth-colony morphology (Fig. 2). Similarly, the wild type formed a robust floating pellicle in liquid MSgg media, but many of the smooth mutants exhibited either incomplete or shattered and sunken pellicles, which is indicative of a biofilm defect (Fig. 2).
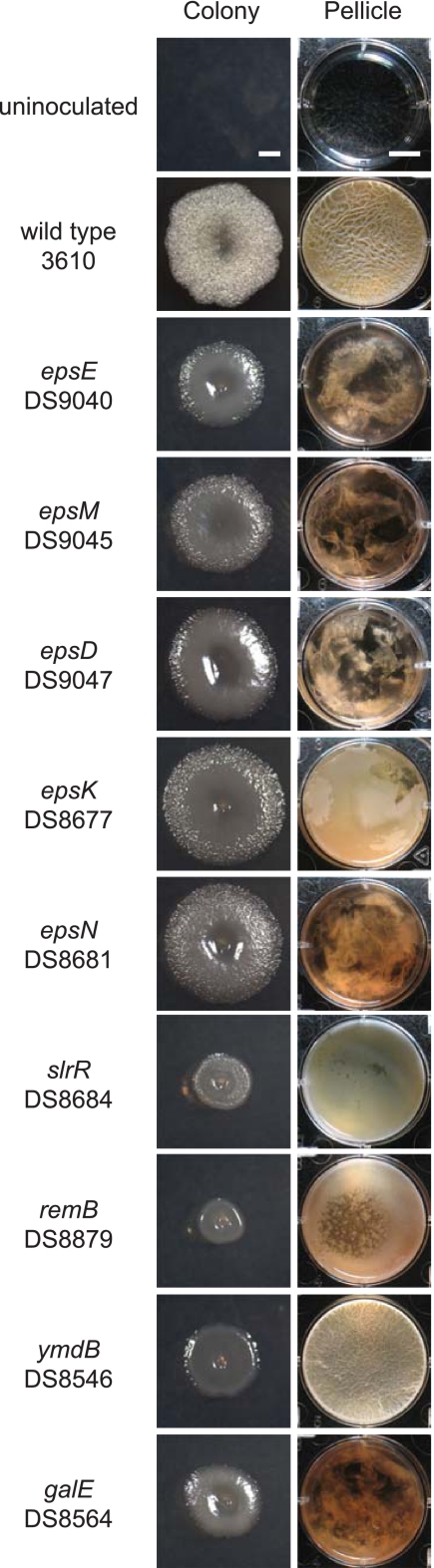
Fig 2.
Mutants defective for biofilm formation. The pellicle column depicts microtiter wells (6-well plate) in which cells have been grown in MSgg medium for 3 days at 25°C. Scale bar is 1 cm. The colony column depicts 10× images of individual colonies grown on MSgg medium for 3 days at 25°C. Scale bar is 1 mm.
Transposon insertion sites were identified by inverse PCR, and each transposon that conferred a biofilm defect disrupted a gene that had been previously reported to be required for biofilm formation (Table 6). Mutations in the eps operon, including the epsD, epsE, epsK, epsM, and epsN genes, impaired biofilm formation, likely due to the disruption of the assembly of the extracellular polysaccharide (EPS) component of the biofilm matrix (4, 14). The mutation of galE, which encodes GalE (the UDP-glucose-4-epimerase), likely abolished the synthesis of a sugar precursor necessary for EPS synthesis (23). The mutation of remB disrupted RemB, a protein that activates extracellular matrix production (30). The mutation of slrR disrupted the transcription factor SlrR, which activates biofilm gene expression (8, 9, 20). Finally, the mutation of ymdB disrupted YmdB, a recently identified phosphoesterase shown to be required for complex colony architecture formation (12). Interestingly, although the mutation of ymdB resulted in a smooth-colony phenotype, it did not abolish or alter pellicle formation. We infer that either a secondary suppressor mutation quickly arises to restore pellicle formation to the ymdB mutant or the ymdB mutation is the first mutation identified that abolishes complex colony architecture without disrupting pellicle formation, suggesting that the two phenotypes in some cases are genetically separable. We conclude that the mutagenesis screens using the TnKRM series of transposons were successful in isolating biofilm-defective mutants.
Table 6.
Transposon insertions
| Strain | Transposon | Insertion tag | Phenotype | Genea | Function |
|---|---|---|---|---|---|
| DS9040 | TnKRM spec | GACAAGAAT | Smooth | epsE | EPS synthesis, flagellar clutch protein |
| DS9045 | TnKRM spec | TATTGATGA | Smooth | epsM | EPS synthesis putative acetyltransferase |
| DS9047 | TnKRM spec | TACATCCGC | Smooth | epsD | EPS synthesis putative glycosyltransferase |
| DS8677 | TnKRM cat | TACGCATTT | Smooth | epsK | EPS synthesis putative flippase |
| DS8681 | TnKRM cat | TAGATTGAC | Smooth | epsN | EPS synthesis putative aminotransferase |
| DS8684 | TnKRM cat | TATATTGGA | Smooth | slrR | Regulator of EPS synthesis |
| DS8879 | TnKRM kan | TATATTCTG | Smooth | remB | Regulator of EPS synthesis |
| DS8546 | TnKRM kan | TATGTGAAA | Smooth | ymdB | Putative metallophosphatase |
| DS8564 | TnKRM kan | TAGCAGTCG | Smooth | galE | UDP-glucose-4-epimerase |
| DS8042 | TnHyJump spec | TATACAGCA | Blue | yesV | Unknown |
| DS8044 | TnHyJump spec | TATACTGCG | Blue | yesQ | Putative rhamnogalacturonan permease |
| DS8164 | TnHyJump cat | TAGTTCGTT | Blue | yesW | Putative rhamnogalacturonan lyase |
| DS8742 | TnHyJump cat | TACATTTTA | Blue | <yesO | intergenic |
| DS8871 | TnHyJump kan | TATAGATAC | Blue | yesX | Putative rhamnogalacturonan lyase |
| DS8872 | TnHyJump kan | TACACCTGC | Blue | yesP | Putative rhamnogalacturonan permease |
| DS7167 | TnLacJump spec | TACATTGCA | Blue | srfAB | Surfactin synthetase |
| DS8725 | TnLacJump spec | TATAACGGT | Blue | rrn-23S | 23S rRNA geneb |
| DS7169 | TnLacJump spec | TATAGTCAT | White | azlB | Transcriptional regulator |
| DS8357 | TnLacJump cat | TACCTTGGC | Blue | ptsG | Glucose phosphotransferase system enzyme II |
| DS8355 | TnLacJump cat | TATCTAAAC | Blue | rpsA | 30S ribosomal protein S1 |
| DS8359 | TnLacJump cat | TAATGAAGG | White | yeeA | Putative type II restriction methylase |
| DS8329 | TnLacJump kan | TACAAGATT | Blue | <yvrM | Intergenic/unannotated open reading frame |
| DS8328 | TnLacJump kan | TAAAGTTCA | Blue | yqeY | Unknown |
| DS8606 | TnLacJump kan | TACCTTGGT | White | fnr | Transcriptional regulator |
< indicates “upstream of.”
Sequence conservation in 23S rRNA makes it impossible to distinguish which of the 10 23S rRNA genes contains the insertion.
Construction of TnHyJump, a transposon carrying an outward-oriented inducible promoter.
Transposon insertions most commonly result in null loss-of-function mutations in the gene into which they have inserted. We set out to build a transposon which, when integrated, would cause the high-level expression of the adjacent gene(s) for use in artificial-expression “gene-trap” screens (33). To build transposons with an outward-oriented, strong, constitutive promoter, the Physpank promoter was PCR amplified using primers 1886/2271 and pDR111 as the DNA template (3). The PCR product then was digested with BamHI and SalI and ligated into the BamHI and SalI sites of pDP383, pEP21, and pKB177, resulting in pEP17, pEP23, and pEP18, respectively. To clone the TnHyJump transposons into the delivery vector, pEP17, pEP23, and pEP18 were PCR amplified using primers 2054/2312, digested with HindIII and PstI, and ligated into the HindIII and PstI sites of pKB176, generating pEP19, pEP26, and pEP20 (Fig. 1F).
The Physpank promoter may be controlled by the addition of IPTG to the culture medium when the lacI gene encoding the LacI repressor protein is present in the genome. To control the expression of the Physpank promoter on the TnHyJump transposons, a vector was constructed in which the lacI gene of E. coli and a tetracycline resistance cassette was integrated between the arms of the B. subtilis lacA gene. The insertion of lacI at lacA serves a dual purpose: first, it disrupts the lacA gene that encodes an endogenous LacZ homolog in B. subtilis which, when misregulated, can confound blue/white β-galactosidase screens (11), and second, the construct expresses the LacI repressor protein to regulate the IPTG-inducible expression of the Physpank promoter in trans. To build a lacA::tet integration vector, the region containing the 5′ end of the lacA gene and adjacent upstream DNA was PCR amplified with primers 2224/2225 using 3610 chromosomal DNA as a template, digested with BglII and PstI, and cloned into the BglII/PstI sites of pDG1515 (13) carrying the tetracycline resistance gene to generate pKB169. The region containing the 3′ end of the lacA gene and adjacent downstream DNA was PCR amplified with primers 2226/2227 using 3610 chromosomal DNA as a template, digested with HindIII and KpnI, and cloned into the HindIII/KpnI sites of pKB169 to generate pKB170. Finally, the lacI gene was PCR amplified using primer pair 2209/2211 with plasmid pDR111 as a template, digested with SalI and BamHI, and cloned into the BamHI/SalI sites of pKB170 to generate pKB175. Thus, when pKB175 is integrated into the chromosome, lacA will be disrupted by the insertion of the lacI gene and a tetracycline resistance marker.
To demonstrate proof of principle with the TnHyJump transposons, B. subtilis strains DS7996, DS7997, and DS8137 were generated containing separate TnHyJump transposon delivery vectors, a promoterless lacZ gene encoding β-galactosidase integrated at the amyE locus, and the lacI gene integrated into the lacA locus. The promoterless lacZ in the chromosome was included in an attempt to isolate and identify TnHyJump insertions that inserted upstream of, and in the same orientation as, lacZ and thereby direct the transcription of the lacZ gene to turn colonies a blue color on media containing X-gal and the appropriate antibiotic selection. The inserted lacI gene enabled the control of the Physpank promoter such that blue colony color, if any, should be dependent on the presence of IPTG.
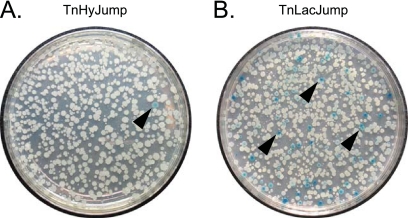
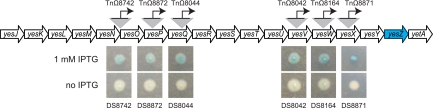
Cells mutagenized with the TnHyJump transposon were plated on LB agar plates containing the antibiotic appropriate for the TnHyJump used, the chromogenic β-galactosidase substrate X-gal, and 1 mM IPTG, and they were grown overnight at 42°C, the nonpermissive temperature for the transposon delivery vector. Blue colonies were found at a low frequency (Fig. 3A). The rare blue colonies were isolated and restreaked onto selective media containing X-gal with and without the inducer IPTG. In all cases, colonies had a more intense blue color in the presence of IPTG, and colonies had a less intense blue color in the absence of IPTG (Fig. 4). None of the IPTG-dependent blue colonies contained transposon insertions upstream of the promoterless lacZ gene that was integrated at the amyE locus. Rather, each transposon was inserted within the large 25-kb yes putative operon, such that the Physpank promoter was oriented to express the downstream yes genes. The most distal insertion that resulted in blue colony color in the presence of X-gal was located within yesX, suggesting that β-galactosidase activity originated from one of the three colinear genes expressed downstream, yesY, yesZ, and yetA. Of these three genes, the most likely candidate to encode β-galactosidase activity is yesZ, encoding YesZ, a putative β-galactosidase (26). We infer that yesZ encodes a cryptic β-galactosidase, the native regulation and expression of which is unknown. We speculate that insertions were favored in front of yesZ rather than in front of the artificially integrated lacZ gene, because yesZ appears to be preceded by 25 kb of genes with which it may be cotranscribed and is thus a much larger target. We conclude that the TnHyJump transposons insert efficiently and express downstream genes when the Physpank promoter is activated.
Fig 3.
Primary plates for blue/white β-galactosidase screens. (A) A petri plate with colonies of cells mutagenized with TnHyJump. (B) A petri plate with colonies of cells mutagenized with TnLacJump. Both plates contain antibiotic and the chromogenic LacZ substrate X-gal. Black carets point to representative blue colonies.
Fig 4.
TnHyJump transposons inserted in the yes operon that induce blue colony color. Genetic map of the yes operon. Arrows indicate genes. Gray triangles indicate transposon insertions. Bent arrows indicate promoters and the direction of transcription. The yesZ gene is indicated in blue, as it is predicted to encode YesZ, a putative β-galactosidase. Pictures are images of colonies grown on LB containing X-gal and either the presence or absence of IPTG.
Construction of TnLacJump, a transposon carrying a promoterless lacZ gene.
Transposons are useful for inserting other genes into the genome besides antibiotic markers. For instance, a promoterless lacZ gene may be integrated within a transposon to generate randomly inserted transcriptional reporter fusions (6, 7, 17, 24). To begin building transposons with a promoterless lacZ gene, the lacZ gene was PCR amplified using primers 1888/1889 and the lacZ plasmid pDG268 as the DNA template (2). The PCR product then was digested with BamHI and XhoI and ligated into the BamHI and XhoI sites of pDP383, pEP21, and pKB177, resulting in pEP1, pEP22, and pEP3, respectively. To insert the TnLacJump derivatives into the delivery vector, pEP1, pEP22, and pEP3 were used as templates for PCR using primers 2054/2312 to generate a product containing the IS elements, the lacZ gene, and the antibiotic resistance cassette. The PCR product was digested with HinDIII and PstI and ligated into the HinDIII and PstI sites of the delivery vector pKB176, generating pEP4, pEP25, and pEP6 (Fig. 1H).
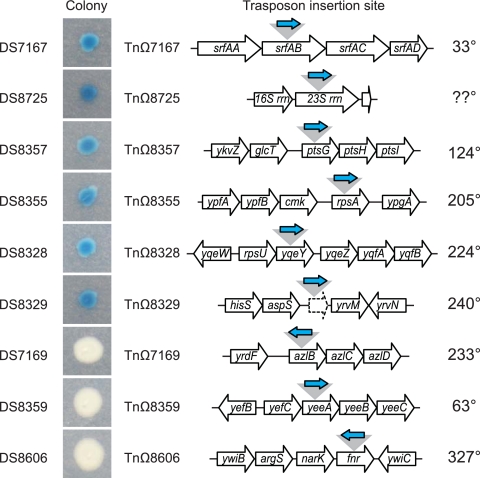
To demonstrate proof of principle with the TnLacJump transposons, strains containing a TnLacJump delivery vector (DS7487, DS7133, and DS8274) were mutagenized by plating on LB agar plates containing X-gal and the appropriate antibiotic at the nonpermissive temperature of 42°C. The rationale of the screen was that the promoterless lacZ in the transposon should generate blue colonies on X-gal only if the TnLacJump transposon had inserted itself within a transcriptionally active region and in the same orientation as an upstream endogenous promoter. Blue colonies of various color intensity were common (Fig. 3B). The identification of the transposon insertion site revealed that all transposons conferring a blue colony color were colinear within the gene in which they were inserted (Fig. 5 and Table 6). Two insertions require additional comment. One transposon, TnΩ8725, was inserted within 1 of the 10 23S rRNA genes in the B. subtilis genome, and due to the high level of sequence identity between the homologous genes, we were unable to identify which specific 23S rRNA gene was disrupted. Nonetheless, the 23S rRNA insertion exhibited the most intense blue colony color of all of the insertions, which is consistent with integration within one of the highly expressed rRNA operons. Another transposon, TnΩ8329, was integrated in an intergenic region upstream of, and colinear with, the gene yvrM. We infer that the insertion lies downstream of an active promoter, and we note that the lacZ insertion appears to disrupt a small, previously unannotated open reading frame colinear with yvrM. We conclude that the TnLacJump transposons are an efficient method for generating insertional lacZ reporters in B. subtilis.
Fig 5.
TnLacJump transposons insertion sites and orientation. Colony indicates a colony of the indicated strain grown on LB agar containing the chromogenic LacZ substrate X-gal. Blue color indicates that β-galactosidase is being expressed, and therefore lacZ in the transposon is being transcribed. Transposon insertion site indicates the location of the transposon insertion and the surrounding genetic neighborhood. Open arrows indicate genes. The gray triangle indicates the location of TnLacJump insertion. The blue arrow indicates the orientation of the lacZ gene. The position of the insertion in the chromosome, given in degrees is indicated at the right.
White colonies also were obtained from the TnLacJump transposon mutagenesis (Fig. 3B). Two of the white colonies contained transposon insertions TnΩ7169 and TnΩ8606 that were oriented in the opposite direction from the disrupted gene and thus were not transcribed by an upstream promoter. One of the white colonies contained the transposon insertion TnΩ8359, in which the lacZ was inserted within, and colinear with, the gene (yeeA) that was disrupted. We infer that there may be a promoter upstream of yeeA that directs yeeA transcription but that the putative yeeA promoter was not expressed under the conditions of the screen. We conclude that TnLacJump insertions only generate functional reporters when inserted colinearly with an upstream promoter and only under conditions in which the upstream promoter is active.
In sum, we have designed a series of three transposons that include not only different antibiotic markers but also either a strong outward-oriented promoter for overexpression screens or a promoterless lacZ gene for the random generation of transcriptional reporter fusions. Furthermore, we demonstrate that all nine of the transposons mobilize at high frequency, and we demonstrate proof of principle for insertional mutagenesis, the inducible expression of a cryptic β-galactosidase YesZ, and the successful generation of random functional transcriptional reporters. Finally, we note that the base transposon in pKB157 can be modified to suit the needs of bacteria besides B. subtilis, which, when delivered by in vitro mutagenesis (1) or by a species-specific in vivo strategy, should be useful for generating any transposon of interest in any model organism (15).
ACKNOWLEDGMENTS
We are grateful to Anna Bree and Eric Vanderpool for technical and intellectual contributions to the screens. We are grateful to Malcolm Winkler for suggesting the strategy of redesigning mariner for conducting “gene-trap” and “promoter-trap” screens. We are grateful to Yoann Le Breton and Bill Haldenwang for sharing the pMarA TnYLB mariner transposon system from which all constructs presented herein were derived.
This work was funded by NIH grant GM093030 to D.B.K.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Akerley BJ, et al. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 95:8927–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antoniewski C, Savelli B, Stragier P. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 4. Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Canosi U, Lüder G, Trautner TA. 1982. SPP1-mediated plasmid transduction. J. Virol. 44:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casadaban MJ, Chou J. 1984. In vivo formation of gene fusions encoding hybrid betagalactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc. Natl. Acad. Sci. U. S. A. 81:535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casadaban MJ, Cohen SN. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chai Y, Kolter R, Losick R. 2009. Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol. Microbiol. 74:876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu F, et al. 2008. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol. Microbiol. 68:1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig NL, Craigie R, Gellert M, Lambowitz AM. (ed.) 2002. Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- 11. Daniel RA, Haiech J, Denizot F, Errington J. 1997. Isolation and characterization of the lacA gene encoding β-galactosidase in Bacillus subtilis and a regulator gene, lacR. J. Bacteriol. 179:5636–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deithmaier C, et al. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J. Bacteriol. 193:5997–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guérout-Fléury A, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 14. Guttenplan SB, Blair KB, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6:e1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hayes F. 2003. Transposon-based strategies for microbial functional genomics and proteomics. Annu. Rev. Genet. 37:3–29 [DOI] [PubMed] [Google Scholar]
- 16. Horinouchi S, Weisblum B. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hughes KT, Maloy SR. 2007. Use of operon and gene fusions to study gene regulation in Salmonella. Methods Enzymol. 421:140–158 [DOI] [PubMed] [Google Scholar]
- 18. Kain J, He GG, Losick R. 2008. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J. Bacteriol. 190:6749–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kleckner N, Bender J, Gottesman S. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139–180 [DOI] [PubMed] [Google Scholar]
- 20. Kobayashi K. 2008. SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis. Mol. Microbiol. 69:1399–1410 [DOI] [PubMed] [Google Scholar]
- 21. Le Breton Y, Mohapatra NP, Haldenwang WG. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maloy SR. 2007. Use of antibiotic-resistant transposons for mutagenesis. Methods Enzymol. 421:11–17 [DOI] [PubMed] [Google Scholar]
- 23. Nesper J, et al. 2001. Characterization of Vibrio cholerae 01 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perkins JB, Youngman PJ. 1986. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 83:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petit M, Bruand C, Jannière L, Ehrlich D. 1990. Tn10 derived transposons active in Bacillus subtilis. J. Bacteriol. 172:6736–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shipkowski S, Brenchley JE. 2006. Bioinformatic, genetic, and biochemical evidence that some glycoside hydrolase family 42 β-galactosidases are arabinogalactan type I oligomer hydrolases. Appl. Env. Microbiol. 72:7730–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steinmetz M, Richter R. 1994. Easy cloning of Mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tomich PK, An FY, Clewell DB. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Villafane R, Bechhofer DH, Narayanan CS, Dubnau D. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winkelman JT, Blair KM, Kearns DB. 2009. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J. Bacteriol. 191:3981–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 14:1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. U. S. A. 80:2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zagorec M, Steinmetz M. 1991. Construction of a derivative of Tn917 containing an outward-directed promoter and its use in Bacillus subtilis. J. Gen. Microbiol. 137:107–112 [DOI] [PubMed] [Google Scholar]
- 34. Zeigler DR, et al. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]