Abstract
Killer yeasts secrete protein toxins that are lethal to sensitive strains of the same or related yeast species. Among the four types of Saccharomyces killer yeasts already described (K1, K2, K28, and Klus), we found K2 and Klus killer yeasts in spontaneous wine fermentations from southwestern Spain. Both phenotypes were encoded by medium-size double-stranded RNA (dsRNA) viruses, Saccharomyces cerevisiae virus (ScV)-M2 and ScV-Mlus, whose genome sizes ranged from 1.3 to 1.75 kb and from 2.1 to 2.3 kb, respectively. The K2 yeasts were found in all the wine-producing subareas for all the vintages analyzed, while the Klus yeasts were found in the warmer subareas and mostly in the warmer ripening/harvest seasons. The middle-size isotypes of the M2 dsRNA were the most frequent among K2 yeasts, probably because they encoded the most intense K2 killer phenotype. However, the smallest isotype of the Mlus dsRNA was the most frequent for Klus yeasts, although it encoded the least intense Klus killer phenotype. The killer yeasts were present in most (59.5%) spontaneous fermentations. Most were K2, with Klus being the minority. The proportion of killer yeasts increased during fermentation, while the proportion of sensitive yeasts decreased. The fermentation speed, malic acid, and wine organoleptic quality decreased in those fermentations where the killer yeasts replaced at least 15% of a dominant population of sensitive yeasts, while volatile acidity and lactic acid increased, and the amount of bacteria in the tumultuous and the end fermentation stages also increased in an unusual way.
INTRODUCTION
Wild killer Saccharomyces cerevisiae yeasts are widespread in most of the wine regions of the world that have been studied (6, 7, 11, 13, 16, 17, 30, 31, 33, 36, 38, 39, 42, 43, 48). As deduced from the few in-depth studies done to date, the frequency of killer yeasts in a given wine production area or single spontaneous must fermentation seems to be very variable, and the proportion of spontaneous fermentations that contain killer yeasts can be as high as 88%, although this proportion can be much influenced by the fermentation stage, vintage period, or production area (13, 39).
The influence of killer toxins on wine fermentation has been studied for more than 20 years (36, 47), and the relative importance of this influence in commercial winemaking is still a topic of discussion (13). The presence of killer yeasts may become particularly important in wine fermentations conducted by inoculation with selected killer-sensitive strains of Saccharomyces, which may be suppressed by wild killer yeasts during the fermentation (15, 25, 40). Also, in spontaneous must fermentation, replacement of a given dominant population by low-frequency killer strains may result in nutrient limitation, leading to fermentation problems. Any of these chance occurrences may decrease wine quality or even cause stuck or sluggish wine fermentation (22, 25). On the other hand, must inoculation with killer yeast may suppress undesirable wild yeast strains, thus preserving wine quality.
The magnitude of the killer effect in wine fermentation depends on a variety of environmental and genetic factors: the initial ratio of killer to sensitive strains (15, 26), the presence of protein-adsorbing substances (3, 5, 25, 27, 40, 46, 47), the environmental conditions and the growth phase of the sensitive cells (4, 46), the presence of protective neutral yeasts (7), the susceptibility of sensitive strains to the killer toxins of different yeast strains (18), the inoculum size and nitrogen availability (22), and the must treatment and the winemaking procedure (13, 25). This circumstance makes it difficult to draw definitive conclusions about the importance of yeast killer activity in commercial winemaking, leading some authors to suggest that the killer character is of lower technological importance than was previously supposed (13). Some workers have found dominance of the killer strains inoculated at as low a level as 0.01 to 10% of the total S. cerevisiae yeast population (14, 18, 25, 27, 37, 41), whereas others found clear dominance of killer yeast only when inoculated at proportions greater than 50% (26). Considering these contradictory reports and the lack of comprehensive studies on the effect of killer yeast in spontaneous fermentations, further research on the occurrence and effect of killer yeast in the vineyard-winery ecosystem is required for precise quantification and control of the killer activity in winemaking.
S. cerevisiae killer strains secrete protein toxins that are lethal to sensitive strains of the same or related yeast species. They have been grouped into four types, K1, K2, K28, and Klus, based on their killing profiles and lack of cross-immunity. To date, only the K2 and Klus types have been found in winemaking environments (31). Members of each type can kill sensitive yeasts, as well as killer yeasts belonging to the other types. Each killer strain is immune to its own toxin and to toxins produced by strains of the same killer type (31, 34). These killer toxins are genetically encoded by medium-size double-stranded RNA (dsRNA) viruses (M1, M2, M28, and Mlus at 1.8, 1.7, 2.1, and 2.1 to 2.3 kb, respectively). These four toxin-coding M dsRNAs show no sequence homology to each other (31, 35). The M viruses depend on a second, large (4.6-kb) dsRNA helper virus, L-A, which is obviously always present in K1, K2, K28, or Klus yeasts, for maintenance and replication. L-A provides the capsids in which both L-A and M dsRNAs are separately encapsidated (reviewed by Schmitt and Breinig [34]). These viruses, called Saccharomyces cerevisiae viruses (ScVs), belong to the family Totiviridae and are cytoplasmically inherited, spreading horizontally by cell-cell mating or by heterokaryon formation (45).
The aim of the present work was to perform a comprehensive study of the characterization and distribution of killer yeasts in five wine subareas of southwestern Spain, as well as of the population dynamics of killer yeasts in spontaneous must fermentations. The recently discovered Klus-type killer yeast is included in this survey for the first time. The improved precision of the conclusions drawn from the present work concerning the winemaking significance of the killer yeast effect in spontaneous fermentations and on the consequent wine quality may help explain the existence of previous contradictory reports in the literature.
MATERIALS AND METHODS
Yeast strains and culture media.
The yeast strains used in the killer phenotype assays are summarized in Table 1. The representative wine yeast collection contained 1,040 prototrophic and homothallic Saccharomyces sensu stricto (24) yeast clones isolated from 104 spontaneous winery fermentations of grapes collected from vineyards of Extremadura in southwestern Spain. These fermentations were done for six consecutive vintages (2000 to 2005), and the grapes were harvested from five vineyard subareas: Tierra de Barros (TB), Ribera Alta (RA), Ribera Baja (RB), Matanegra (MA), and Northern Cáceres (NC). Three must/wine samples were taken from each fermentation (at the beginning [BF], tumultuous stage [TS], and end of fermentation [EF]) for yeast isolation on yeast extract-peptone-dextrose (YEPD) plates. S. cerevisiae-like colonies were isolated from most samples, except for some at the beginning of fermentation, when the yeast species is usually rare and other microorganisms, such as molds, often overgrow the isolation culture plate. The mitochondrial DNA restriction fragment length polymorphism (mtDNA-RFLP) was analyzed for 20 S. cerevisiae-like isolates from each sample, except for the samples at the beginning of fermentation. A total of 4,160 yeast isolates were analyzed, and 126 different mtDNA-RFLP patterns were found. Ten yeast isolates from each fermentation were selected to make the representative wine yeast collection, which contained 120 yeast clones from MA, 130 from RA, 340 from RB, 350 from TB, and 100 from NC.
Table 1.
S. cerevisiae yeast strains used as references for the killer phenotype assay
| Strain | Genotype/relevant phenotyped | Origin |
|---|---|---|
| EX33 | MATa/α HO/HO [K10 K20 K280 Klus0] | J. A. Regodóna (from wine) |
| EX73 | MATa/α HO/HO L-A M2 [K2+] | J. A. Regodón (from wine) |
| F166 | MATα leu1 kar1 L-A-HNB M1 [K1+] | J. C. Ribasb (from R.B. Wickner) |
| F182 | MATα his2 ade1 leu2-2 ura3-52 ski2-2 L-A M28 [K28+] | J. C. Ribas (from M. Schmitt) |
| EX436 | MATa/α HO/HO L-A Mlus-1 [Klus+] | M. Ramírezc |
| EX122 | MATa/α HO/HO L-A Mlus-2 [Klus+] | M. Ramírez |
| EX198 | MATa/α HO/HO L-A Mlus-3 [Klus+] | M. Ramírez |
| EX229 | MATa/α HO/HO L-A Mlus-4 [Klus+] | M. Ramírez |
| 5x47 | MATa/α his1/+ trp1/+ ura3/+ [K10 K20 K280 Klus0] | J. C. Ribas (from R. B. Wickner) |
J. A. Regodón, Departamento de Química Analítica, Universidad de Extremadura, Badajoz, Spain. Isolated from D. O. Ribera del Guadiana, Spain.
J. C. Ribas, Departamento de Microbiología y Genética, Universidad de Salamanca, Salamanca, Spain.
M. Ramírez, Departamento de Ciencias Biomédicas, Área de Microbiología, Universidad de Extremadura, Badajoz, Spain.
Relevant known phenotypes are in brackets; a superscript 0 indicates the confirmed absence of the given phenotype.
The yeasts isolated from laboratory spontaneous fermentations were also prototrophic and homothallic Saccharomyces sensu stricto yeast clones isolated from 42 spontaneous fermentations made during the 2006 vintage season, with 7 to 9 fermentations from each vineyard subarea. Thirteen fermentations were done at the beginning of the vintage season (25 August to 10 September), 17 in the middle (11 September to 25 September), and 12 at the end (26 September to 5 October). S. cerevisiae-like colonies were isolated from each fermentation: 10 to 15 from the beginning of fermentation (depending on availability), 40 from the tumultuous stage, and 40 from the end of fermentation.
The proportions of non-Saccharomyces, Saccharomyces sensu stricto, and bacteria were determined at the beginning, tumultuous, and end stages for every fermentation. Saccharomyces sensu stricto (24) colonies are easy to distinguish from the other yeast species present in fermenting grape must by their aspect in YEPD agar (white or cream color, buttery, smooth, circular, and prominent) and under the microscope (globose, ellipsoid to elongate in shape, with multilateral budding) and by their ability to sporulate, producing typical asci (tetrads) (20).
All isolated S. cerevisiae-like yeasts were grown in sporulation medium, and the sequence(s) of the 16S ribosome gene(s) of 1 to 10 (depending on the abundance) yeast isolates containing each mtDNA-RFLP pattern was analyzed. All the analyzed yeasts produced more than 50% typical asci (tetrads), and the 18S sequence in each case matched that of one of the four species belonging to the Saccharomyces sensu stricto group (24).
Standard culture media were used for yeast growth (35a) and yeast sporulation (20).
Spontaneous wine fermentations.
For the laboratory vinification trials, the grapes were harvested with sterile material from different vineyards located in the five subareas studied. For white wines, the grapes from different varieties (mostly Cayetana, Pardina, Eva, Cigüente, and Montúa) were crushed, and the fresh white must obtained (19.6 to 22.7 °Brix; pH 3.1 to 4.2) was clarified by settling for 18 h at 12°C. For red wines, the grapes from different varieties (mostly Tempranillo, Cabernet-Sauvignon, Merlot, Garnacha, and Syrah) were destemmed and crushed (21.3 to 26.3 °Brix; pH 3.2 to 4.2). The fresh white must or crushed red grapes were transferred to 5-liter Erlenmeyer flasks for spontaneous fermentation. The vinification process was conducted at 18°C for white wine and at 22°C for red wine. The must density and the °Brix were monitored every day. The flasks were capped hermetically after reducing sugars reached around 1% to avoid oxidation problems. At the end of fermentation, the settled solids were discarded and a centrifuged sample of each wine was taken for the assays. The uncentrifuged wines were stored at 4°C. After 50 days following the end of fermentation, settled solids were again discarded and the wines were returned to storage at 4°C. At 85 days, settled solids were discarded once more and the wines were bottled. At 105 days following the end of fermentation, the sensory characteristics (flavor, color, and odor) of the wines were tested by a panel of 12 expert judges of the official committee of the Ribera del Guadiana wine Protected Designation of Origin. Wines were presented in clear tulip-shaped wine glasses covered with glass petri dishes. A sample of 50 to 70 ml of wine was poured into each glass immediately before evaluation by each judge. The temperatures of the samples were from 10 to 13°C for white wines and from 16 to 18°C for red wines. The judges scored the quality of the wines on a six-point scale (0, very poor; 1, deficient; 2, acceptable; 3, good; 4, very good; and 5, excellent). The preference value for each wine was the mean of the 12 test results. The maximum score possible (60 points) was considered 100% preference.
A similar procedure was used for the winery vinification trials, but using the equipment available in each company. The main modifications were that nonsterile material was used for grape harvest and processing, spontaneous fermentations were done in 1,000- to 5,000-liter stainless steel tanks, and the fermentation temperature control was not as precise as in the laboratory vinification trials (16 to 20°C for white wine and 20 to 25°C for red wine).
Analytical methods for wine fermentations.
Density, °Brix, pH, total acidity, volatile acid, reducing sugars, alcohol, and malic acid were determined according to the European Community (EC) recommended methods (9). Lactic acid was determined using the European Economic Community (EEC) recommended method (10). T15 is the time needed to ferment 15% of the total sugars present in the must, and T100 is the time needed to ferment 100% of the total sugars (28).
Determination of yeast killer activity.
Killer activity was tested on low-pH (pH 4 or 4.7) methylene blue (MB) plates (20) seeded with 100 μl of a 48-h culture of the sensitive strain (29). Depending on the experiments, the strains being tested for killer activity were either loaded as 4-μl aliquots of stationary-phase cultures, patched from solid cultures, or replica plated onto the seeded low-pH MB plates. Then, the plates were incubated for 4 days at 20°C or 28°C.
Total nucleic acid preparation.
The procedure for routine dsRNA and mtDNA minipreps was described previously (21). Basically, the cells were suspended in 10 mM Tris-HCI (pH 7.5) buffer containing 0.1 M NaCl, 10 mM EDTA, and 0.2% SDS, and an equal volume of phenol (pH 8.0) was added. The mixture was incubated at room temperature for 30 min with shaking. After centrifugation, the nucleic acids recovered in the aqueous phase were precipitated with isopropanol, washed with 70% ethanol, dried, and dissolved in Tris-EDTA (TE) buffer, pH 8.0.
Nucleic acid analysis for yeast strain typing.
The procedure for mtDNA and virus dsRNA analysis was described previously (21). The samples (4 μl of each) were directly separated in 1× TAE-1% agarose gels (60 to 75 min) for virus dsRNA analysis. Alternatively, 3 μl of each sample, previously digested with RNase A to avoid RNA interference, was digested with RsaI for 2 h at 37°C and separated in 0.5× TBE-0.8% agarose gels (75 to 90 min) for mtDNA-RFLP analysis. Nucleic acids were visualized on a UV transilluminator after ethidium bromide staining of the gels and photographed with a Gel Doc 2000 (Bio-Rad). The data analysis was performed using Diversity Database software (Bio-Rad). The bands were typed by Rf (relative mobility), and band assignment was determined by Rf values plus or minus 2% error.
PCR amplification, sequencing of 18S ribosomal DNA (rDNA) and yeast identification.
PCR was performed directly from the nucleic acid minipreps with the pReTaq Ready-To-Go PCR Beads kit (Amersham Biosciences) and with the 18S rDNA-specific primers EukA (AACCTGGTTGATCCTGCCAGT) and EukB (TGATCCTTCTGCAGGTTCACCTAC) (8, 23). The thermocycler protocol was an initial denaturation step of 95°C for 2 min, followed by 35 cycles of denaturing at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 2 min and a final extension at 72°C for 10 min. The amplification products were purified with the Jetquick PCR purification Spin Kit (Genomed, Löhne, Germany) following the manufacturer's recommendations. The purified rDNA PCR fragment from each isolated microorganism was sent to a sequencing service (Secugen S.L., Madrid, Spain). The 18S rDNA gene sequences were manually edited with the software Chromas v. 1.45 (Technelysium) and were analyzed against those in GenBank using BLAST (1). Sequences with >99% similarity to previously published data available at NCBI (http://ncbi.nlm.nih.gov) were binned into the same species.
Miscellaneous.
DNA manipulations (enzyme digestions, PCR, and electrophoresis) were done following standard methods (32). Most of the enzymes were purchased from Promega or Sigma. Synthetic oligonucleotides were purchased from Biomers.
Data were analyzed for statistical significance by the Kruskal-Wallis and Spearman nonparametric tests. A 5% probability level (P = 0.05) was used to accept or reject the null hypothesis. All the statistical analyses were performed with the software package SPSS version 15.0 for Windows (SPSS, Chicago, IL).
RESULTS
Characterization of the representative wine yeast collection from the southwestern Spain wine production area.
We found 126 different mtDNA-RFLP patterns in our yeast collection and assumed that all the isolated yeasts having the same pattern belonged to the same “presumptive strain” of Saccharomyces sensu stricto. Most presumptive strains were present at low frequency, and a few strains were detected at high frequency. Only 33 out of the 126 strains did not contain any M virus; the M2 virus was found in 88 strains, while the new Mlus virus was found in 27 strains. In addition, for a given yeast strain, we found different situations, as shown in Table 2.
Table 2.
Number of Saccharomyces sensu stricto presumptive strains in the representative yeast collection according to the times they were isolated and the type of M virus they contained
| No. of times isolated | No. of isolates | No. of strainsa | No. of different strains containing the virusb: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| M0 | M2 | Mlus | M0 or M2 | M0 or Mlus | M2 or Mlus | M0, M2, or Mlus | |||
| 1 | 44 | 44 | 20 | 21 | 3 | 0 | 0 | 0 | 0 |
| 2–5 | 135 | 43 | 8 | 15 | 1 | 10 | 3 | 2 | 4 |
| 6–10 | 118 | 15 | 2 | 2 | 0 | 8 | 0 | 2 | 1 |
| 11–50 | 430 | 22 | 3 | 0 | 0 | 10 | 0 | 0 | 9 |
| >50 | 313 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 1,040 | 126 | 33 | 38 | 4 | 28 | 3 | 4 | 16 |
All yeast isolates containing the same mtDNA-RFLP pattern were considered to belong to the same yeast strain.
M0, no M virus was detected.
Of the collection isolates, 59.2% were nonkiller (K−) and 40.8% were killer (K+). Among the nonkiller isolates, 78.8% contained no M virus, while 21.2% contained an M virus. Among the killer isolates, 80.4% were K2 and 19.6% belonged to the recently discovered Klus type (31). Among the collection isolates containing M virus, 23.5% did not show detectable killer activity (20.3% of these isolates had M2 virus, and 3.2% had Mlus virus).
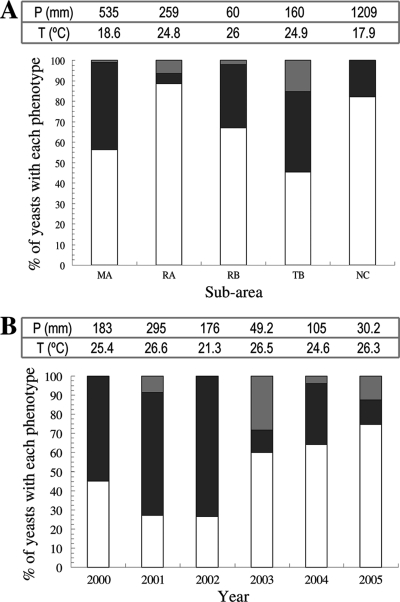
The killer yeasts were found in all the winemaking subareas and in all the vintages analyzed (Fig. 1), although they were more frequent in the TB and MA subareas, which are the greatest wine-producing subareas with the most wineries and vineyards. The Klus yeasts were mostly isolated from the warmer subareas, TB, RA, and RB (mean temperature during the ripening/harvest season, 24.8 to 26°C), but not from the distant, coolest NC subarea (mean temperature, 17.9°C). Usually, an increase in the environmental temperature during the ripening/harvest season means an increase in the must pH, which is indeed what occurred in our case (the Spearman correlation between pH and temperature was 0.624; P = 0.01). The must pH was 3.1 to 3.6 in the cooler subareas (NC and MA) and 3.3 to 4.2 in the warmer subareas (RA, RB, and TB). As an exception, some Klus yeasts were also found in MA (just visible in Fig. 1A), which is not one of the warmer subareas (mean temperature, 18.6°C). In contrast, the K2 strains were found in all five producing subareas, even in NC (Fig. 1A), with higher altitude and lower mean temperatures than the rest of the subareas. The proportion of Klus yeasts was also higher in the warmer ripening/harvest seasons (26.3 to 26.6°C in 2001, 2003, and 2005), while the proportion of K2 killer yeasts was higher in the rainier ripening/harvest seasons (2000, 2001, 2002, and 2004) (Fig. 1B).
Fig 1.
Ecological distribution of the Saccharomyces sensu stricto killer yeasts among the five wine-producing subareas (A) and the six vintages (B) analyzed. White, nonkiller yeasts; black, killer K2 yeasts; gray, killer Klus yeasts. T, mean daily temperature during the ripening/harvest season; P, rainfall during the ripening/harvest season.
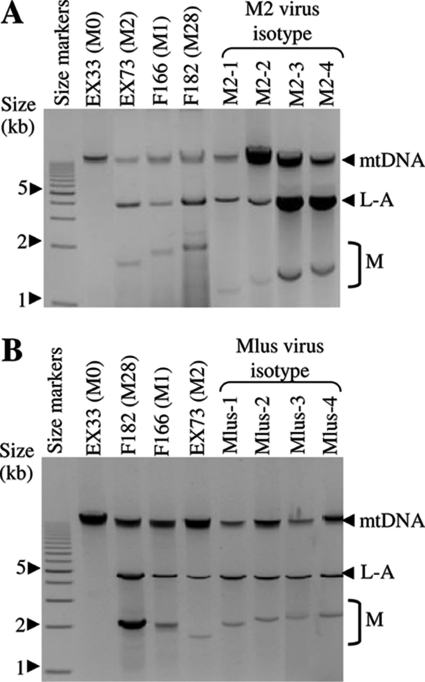
All K2 and Klus yeasts contained L-A (helper) virus plus the corresponding M virus, M2 or Mlus. The L-A dsRNA size was the same in all isolated yeasts, 4.6 kb, while the dsRNA size of M2 or Mlus was variable among the different isolates. However, a given dsRNA size did not vary for a given yeast isolate after 100 cell doublings (29, 31). We identified four isotypes for M2 (M2-1, M2-2, M2-3, and M2-4, at 1.3, 1.5, 1.6, and 1.75 kb, respectively) and another four isotypes for Mlus (Mlus-1, Mlus-2, Mlus-3, and Mlus-4, at 2.1, 2.2, 2.25, and 2.3 kb, respectively) according to the dsRNA size (Fig. 2). No correlation was found between these virus isotypes and the mtDNA-RFLP patterns of the host yeasts.
Fig 2.
Genetic determinants of Klus and K2 toxins. (A and B) Presence of L-A and M dsRNA molecules in K2 (A) and Klus (B) strains. Nucleic acids were obtained from reference killer yeasts K1 (F166), K2 (EX73), K28 (F182), Klus strains (Mlus-1 to Mlus-4), and K2 strains (M2-1 to M2-4) containing different virus isotypes. Samples were separated by agarose gel electrophoresis. The ethidium bromide staining of the gel is shown. The size markers correspond to a 1- to 15-kb molecular ruler (Bio-Rad).
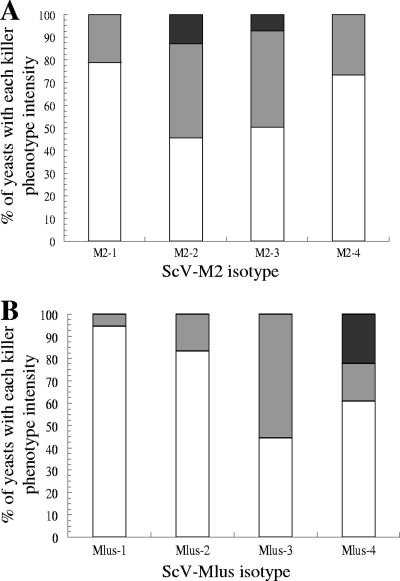
The intensity of the killer phenotype was variable among the different yeast isolates containing the same virus isotype. However, on average, among K2 yeasts, those containing the middle-size dsRNA isotypes (M2-2 at 1.5 kb and M2-3 at 1.6 kb) showed more intense killer activity than the yeasts containing either the smallest (M2-1 at 1.3 kb) or the largest (M2-4 at 1.75 kb) dsRNA isotype. The yeasts containing M2-2 or M2-3 isotypes were also isolated more frequently than those containing M2-1 or M2-4 isotypes (Fig. 3A). Among the Klus yeasts, those containing the largest dsRNA isotype (Mlus-4 at 2.3 kb) showed the most intense killer activity. However, these yeasts were less frequent than the yeasts containing the smallest dsRNA isotype (Mlus-1 at 2.1 kb), which showed the least intense killer activity (Fig. 3B). No correlation was found between the killer activity intensity and the mtDNA-RFLP patterns of the host yeasts.
Fig 3.
Frequencies of yeast isolates containing each virus isotype with each killer phenotype intensity. (A) Yeasts containing M2 virus isotypes. (B) Yeasts containing Mlus virus isotypes. The thicknesses of the growth inhibition halos (killer phenotype intensities) were as follows: white, less than 1 mm; gray, 1 to 2 mm; black, more than 2 mm.
Population dynamics of wild killer yeasts during spontaneous wine fermentation.
We analyzed 42 spontaneous wine fermentations during the 2006 vintage season. No killer yeast was found in 17 of those fermentations (40.5%), although we frequently detected yeasts containing M virus dsRNA without killer activity under our working conditions (data not shown). As many as 25 fermentations (59.5%) contained killer yeast in at least one fermentation stage, in proportions ranging from 2% to 100%. Most killer yeasts were Saccharomyces. Non-Saccharomyces killer (NSK) yeasts were found in two fermentations. They were identified as Candida and Hanseniaspora and coexisted with Saccharomyces killer yeasts. It seemed that the presence of these NSK yeasts increased the fermentation onset time (T15 mean, 7.1 ± 0.4 days) relative to the rest of the fermentations (T15 mean, 3.9 ± 0.3 days).
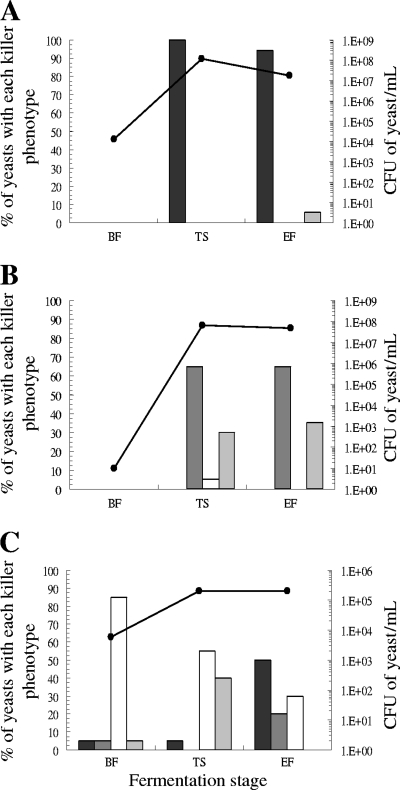
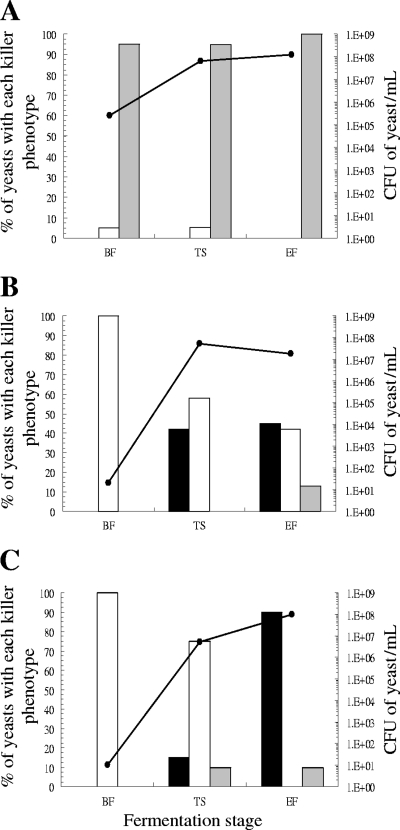
As was the case in the representative wine yeast collection, two types of Saccharomyces killer yeasts were detected: the already known killer K2 and the recently discovered Klus. The K2 yeasts were found in 23 vinifications and the Klus yeasts in 6. We found only K2 killer yeasts in 19 fermentations, only Klus yeasts in two fermentations, and both types of killer yeast in four fermentations. The two types of killer yeast separately dominated seven fermentations with more than 50% of the total yeast population—K2 yeasts in six fermentations and Klus yeasts in one fermentation (Fig. 4A and B, respectively). A fermentation containing both types of killer yeast, K2 and Klus, was unusually slow because the Saccharomyces yeast population was particularly low, around 2 × 105 CFU/ml in tumultuous and end fermentation stages (Fig. 4C).
Fig 4.
Representative examples of the three different killer wild yeast population dynamics found among the spontaneous must fermentations. (A) The dominant yeasts were killer K2. (B) The dominant yeasts were killer Klus. (C) The killer K2 and Klus yeasts codominated at the end of must fermentation. White, nonkiller yeasts sensitive to K2 toxin; light gray, nonkiller yeasts resistant to K2 toxin; dark gray, killer Klus yeasts; and black, killer K2 yeasts. Dots and solid line, CFU of yeast/ml.
The proportion of vinifications containing killer yeasts did not increase over the course of the vintage season. However, the mean proportion of killer yeasts significantly increased in the vinifications made at the end of the vintage season, with a corresponding decrease in the mean proportion of killer-sensitive yeasts (Table 3).
Table 3.
Percentages of killer, sensitive, and resistant Saccharomyces sensu stricto yeasts in the must fermentations made at different periods of the vintage seasonsa
| Yeast phenotype | % at vintage season periodb: |
Pc | ||
|---|---|---|---|---|
| Beginning | Middle | End | ||
| Killer (K2 and Klus) | 12 ± 5.5 | 21 ± 6 | 55 ± 8 | 0.001 |
| Nonkiller resistant to K2 | 39 ± 13 | 60 ± 6.9 | 32 ± 7 | 0.032 |
| Nonkiller sensitive to K2 | 49 ± 11 | 19 ± 5.7 | 13 ± 4.2 | 0.010 |
Kruskal-Wallis nonparametric test to study the effect of the harvest date on the proportions of killer and sensitive yeasts.
The data are the mean values and standard errors of 13, 17, and 12 independent experiments done at the beginning, middle, and end of the vintage season, respectively. The nonkiller yeasts resistant or sensitive to Klus toxin were not considered separately in this analysis because of the difficulty in achieving reliable results due to the weak killer phenotype of Klus yeasts and because of their low frequency in the spontaneous fermentations.
P values obtained by the Kruskal-Wallis nonparametric test to analyze the changes in the proportions of killer yeasts in the must fermentations over the course of the vintage season.
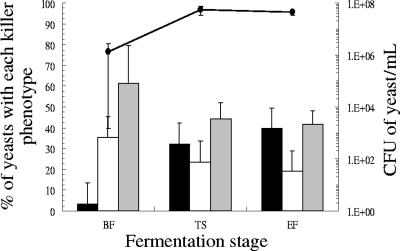
The vinifications were classified into three different types according to the presence/absence of killer yeasts and the effect of the killer phenotype on the killer-sensitive yeast population (Fig. 5): VK-0, those vinifications without killer yeasts (Fig. 5A); VK-L (for low killer effect), vinifications containing killer yeasts whose population did not increase or increased less than 15% during must fermentation (Fig. 5B); and VK-H (for high killer effect), vinifications containing killer yeasts whose population increased more than 15% (usually above 50%) during must fermentation (Fig. 5C). The proportion of each fermentation type was as follows: 40.5% for VK-0, 40.5% for VK-L, and 19% for VK-H. Among the VK-H fermentations, the K2 yeasts were dominant in seven fermentations and the Klus yeasts in one fermentation. Also, in general, the mean proportion of killer yeasts increased in VK-L and VK-H vinifications during must fermentation while the killer-sensitive yeasts decreased (Fig. 6). Nonkiller yeasts containing M virus were found in 11 out of the 17 VK-0 vinifications, although no increase of these yeasts during must fermentation was observed. However, the proportion of nonkiller yeasts containing M virus was greater than the proportion of killer yeasts in 15 out of the 25 VK-L and VK-H vinifications.
Fig 5.
Representative vinification types according to the presence/absence of killer yeasts and the effect of the killer phenotype on the killer-sensitive yeast population. (A) VK-0. (B) VK-L. (C) VK-H. White, nonkiller yeasts sensitive to K2 toxin; gray, nonkiller yeasts resistant to K2 toxin; and black, killer K2 yeasts. Dots and solid lines, CFU of yeast/ml.
Fig 6.
Average changes in the proportion of killer yeasts during the VK-L and VK-H fermentations. White, nonkiller yeasts sensitive to K2 toxin; grey, nonkiller yeasts resistant to K2 toxin; and black, killer K2 and Klus yeasts. Dots and solid lines, CFU of yeast/ml. The data are the mean values of 17 VK-L and 8 VK-H fermentations. The error bars indicate standard errors of the mean.
The microbial populations were more complex in the VK-H vinifications than in the VK-0 or VK-L ones, containing an unusually large amount of bacteria in the tumultuous and end fermentation stages. As a result, the VK-H fermentation kinetics were significantly slower (high T15 and T100) and the wines had less malic acid, more volatile acidity, more lactic acid, and poorer organoleptic quality (marginally significant) than those of VK-0 or VK-L vinifications (Table 4).
Table 4.
Must fermentation and wine parameters of the three types of spontaneous laboratory vinificationsa
| Parameter | Value for vinification typeb: |
Pc | ||
|---|---|---|---|---|
| VK-0 | VK-L | VK-H | ||
| T15 (days) | 3.5 ± 0.6 | 3.7 ± 0.6 | 5.5 ± 0.5 | 0.032 |
| T100 (days) | 11 ± 1.5 | 11 ± 1.9 | 19 ± 3.0 | 0.026 |
| Preference (%) | 53 ± 5.2 | 58 ± 3.1 | 44 ± 3.4 | 0.056 |
| Total acidity (g/liter) | 6.8 ± 1.3 | 5.4 ± 0.2 | 8.1 ± 1.5 | 0.064 |
| Volatile acidity (g/liter) | 0.6 ± 0.2 | 0.6 ± 0.1 | 1.9 ± 0.7 | 0.006 |
| Malic acid (g/liter) | 1.1 ± 0.2 | 1.3 ± 0.2 | 0.5 ± 0.1 | 0.032 |
| Lactic acid (g/liter) | 0.8 ± 0.2 | 0.8 ± 0.2 | 2.4 ± 1.1 | 0.039 |
| No. of bacteria-BF (105 CFU/ml) | 0.5 ± 0.4 | 0.4 ± 0.2 | 0.2 ± 0.1 | 0.916 |
| No. of bacteria-TS (106 CFU/ml) | 0.9 ± 0.6 | 0.3 ± 0.1 | 4.4 ± 1.7 | 0.000 |
| No. of bacteria-EF (106 CFU/ml) | 0.4 ± 0.4 | 0.2 ± 0.1 | 2.9 ± 0.9 | 0.000 |
Kruskal-Wallis nonparametric test to study the effect of the changes in the killer/sensitive yeast proportion on winemaking.
The data are the mean values and standard errors of 17 VK-0, 17 VK-L, and 8 VK-H independent experiments.
P values obtained by the Kruskal-Wallis nonparametric test to analyze the differences in the fermentation and wine parameters between the three vinification types.
DISCUSSION
Characterization of the representative wine yeast collection.
Our results indicate the existence of great biodiversity among the S. cerevisiae strains in the spontaneous wine fermentations, because most presumptive strains appeared at low frequency in the yeast representative collection.
The M viruses were widespread among the Saccharomyces wine yeasts. Most presumptive strains contained these viruses, and most strains free of M virus were isolated just once (Table 2). We would possibly have been able to find M virus in all yeast strains if they had been isolated more frequently, as was the case for all strains isolated more than 50 times and almost all strains isolated more than 10 times. Also, it should be considered that M viruses are more difficult to detect than L-A virus, and they could actually be present in some of the strains that we observed as free of M virus. Therefore, one may suspect that all wine yeast strains may potentially become infected by any M virus (M2 or Mlus). Moreover, different isolates of the same yeast strain can contain different M viruses and show different killer phenotypes (nonkiller, killer K2, or Klus) (Table 2), as has previously been reported (13).
Most collection isolates were nonkiller, although between a fifth and a quarter contained an M virus. For some reason, some yeast isolates containing M virus either are unable to produce enough active killer toxin or their killer activity was not detectable under our laboratory working conditions. If the latter possibility is assumed, the proportion of killer yeast isolates may be greater than we estimated. Among the killer isolates, most were K2, with the minority being Klus, similar to the case among the 126 yeast strains. This lower frequency of Klus yeasts than K2 killer yeasts seemed surprising in principle, because K2 killer yeasts are clearly sensitive to Klus yeasts, while Klus yeasts are mostly resistant to K2 killer yeasts under laboratory test conditions. A partial explanation of these findings may be that the intensity of the Klus phenotype is fairly low compared to the K2 phenotype (31).
The killer yeasts were also widespread in all the winemaking subareas and vintages analyzed (Fig. 1). The only exception was the absence of Klus yeasts in the coolest subarea (NC). They appeared especially in the warmer subareas (TB, RA, and RB) in the warmest vintages, which corresponded to the highest must pHs. This could be because under these conditions, Klus strains may have some ecological advantage, given that they show the most intense phenotype at a higher pH (4.7) and temperature (28 to 30°C) than the K2 strains (pH 4; 20°C) (31). The exception where some Klus yeasts are also found in MA, which is not one of the warmer subareas, may be because this subarea is located very close to the warmer ones (TB, RA, and RB) and it is known that there is a frequent commercial transfer of grapes from the vineyards of any these subareas to the wineries of the others, which would facilitate the spread of any given killer yeast strain among the four subareas.
While all L-A dsRNAs had the same size, four different dsRNA sizes (virus isotypes) were found for each M virus, M2 or Mlus (Fig. 2). The middle-size isotypes of M2 dsRNA (M2-2 and M2-3) were the most frequent among K2 yeasts (Fig. 3A), probably because they encoded the most intense K2 killer phenotype. However, the smallest dsRNA isotype of the Mlus virus (Mlus-1) was the most frequent among Klus yeasts, although it encoded the less intense Klus killer phenotype. On the other hand, the largest dsRNA isotype of Mlus (Mlus-4; 2.3 kb), which showed the most intense Klus killer activity, was not the most frequent among Klus yeasts (Fig. 3B). This could be due to a decrease in the encapsidation or replication efficiency of this long Mlus dsRNA isotype (44), which indeed is the longest of all the Saccharomyces M viruses. The Mlus-1 virus may have a less pronounced decrease in its replication or encapsidation efficiency because its dsRNA is smaller than that of the other Mlus isotypes and closer to the dsRNA size of the M2 virus isotypes. Alternatively, given the interference effects on killer toxin activity reported to exist in grape must, it is possible that killer yeasts containing Mlus-1 were more killer than those containing Mlus-4 virus under winemaking conditions, the opposite of what we observed in the laboratory killer phenotype test. However, we have no evidence for this situation, essentially because its analysis would be very difficult to perform considering all the possible environmental conditions at the winery; there are many variable factors in must fermentation that may affect in different ways each killer toxin produced by each yeast strain and coded by each virus isotype.
The absence of correlation between these virus isotypes or the killer activity intensity and the host yeast strains (mtDNA-RFLP patterns) indicates that a given virus is not in any way exclusive to a given yeast strain. As yeast mating between haploid spore clones from tetrads of different yeast strains is rare under winemaking conditions (2), this result raises the possibility that these viruses, contrary to previous suggestions (45), may follow an external, natural, but as yet undiscovered route of infection to pass from a given yeast strain to a different one without the need for yeast mating.
Population dynamics of wild killer yeasts during spontaneous wine fermentation.
No killer yeasts were found in as many as 40.5% of the fermentations analyzed, although again, nonkiller yeasts containing M virus dsRNA were frequently detected in those fermentations. This again indicates that the proportion of must fermentations containing killer yeasts may be greater than we have estimated if the killer activity for some killer yeasts is not detectable under our laboratory killer test conditions. Saccharomyces killer yeasts were found in at least one fermentation stage in most wine fermentations (59.5%), although this frequency was lower than the previously reported 88% (39). Non-Saccharomyces killer yeasts (Candida and Hanseniaspora) were found in only two fermentations. Although they did not dominate the fermentation and coexisted with Saccharomyces killer yeasts, their presence seemed to increase the fermentation onset time, as has previously been reported (19).
As in the representative collection of yeasts, the killer K2 yeasts were more frequent than the Klus yeasts. In the fermentations where they coexisted, the Klus yeasts did not displace the K2 yeasts to dominate the fermentation (Fig. 4C), probably because, as mentioned above, the Klus killer activity is fairly low (31) and the toxin is partially inactivated by binding to the particles suspended in the must (25). This may also be the reason why the killer yeasts did not dominate all the wine fermentations in which they appeared. However, both types of killer yeasts separately dominated various fermentations. This was expected, because both killer toxins should be active under the present wine fermentation conditions (pH 3.1 to 4.2 and 18 to 22°C). Indeed, the antagonism between the two types of Saccharomyces killer yeasts plus the effects of both on the sensitive yeasts may keep yeast populations unusually low (roughly 2 × 105 CFU/ml in the tumultuous and end fermentation stages) and slow down the fermentation kinetics (Fig. 4C).
The mean proportion of killer yeasts increased in the vinifications at the end of the harvest season, and the mean proportion of killer-sensitive yeasts decreased (Table 3). However, no increase in the proportion of vinifications containing killer yeasts was found over the course of the vintage season. It seems that the ecological advantage of killer yeasts is efficient enough to increase their frequency during the spontaneous fermentations, but not in the surrounding vineyard-winery environment containing the yeasts that inoculate the spontaneous wine fermentations.
The fact that nonkiller yeasts containing M virus were found in many vinifications indicates the presence of mutated viruses unable to produce sufficiently active killer toxins under must fermentation conditions. Indeed, nonkiller yeasts containing M virus were present in most VK-0 vinifications and did not increase during must fermentation. However, when killer yeast activity was actually detected, as in the VK-L and VK-H vinifications, the mean proportion of killer yeasts increased during must fermentation while that of the sensitive yeasts decreased (Fig. 6), as has previously been reported (16). These results indicate that there is good agreement between the ability to detect the killer phenotype under our laboratory working conditions and the killer efficiency of the killer toxins during must fermentation.
The presence of killer yeasts during wine fermentation did not in itself detract from the fermentation kinetics parameters or the wine's quality. In fact, although the fermentation kinetics was slightly better in the VK-0 than in the VK-L vinifications, the quality of the VK-L wines was slightly better than that of the VK-0 wines (Table 4). These results may indicate that the presence of killer yeasts is of low technological importance in winemaking, as has previously been suggested (13). However, the results are quite different if the effect of the killer phenotype on the sensitive yeast population is sufficiently large, as was the case with the VK-H vinifications (Fig. 5C). The VK-H vinifications, which represented 19% of the cases, contained an unusually large amount of bacteria in tumultuous and end fermentation stages, their fermentation kinetics was significantly slower (greater values of T15 and T100), and the wines had less malic acid, more volatile acidity, more lactic acid, and poorer organoleptic quality (preference) than the VK-0 and VK-L wines (Table 4). The main cause of this situation is major interference between the yeast populations (killer versus sensitive strains), which detracts from the total yeast population growth and favors bacterial growth, as is frequent in sluggish and stuck winery fermentations in warm regions, such as Extremadura. As a practical conclusion, therefore, special care should be taken in commercial winemaking to avoid any major effect of the killer phenotype. The winemaker needs to avoid any major killing effect of both the inoculated killer yeasts on putative wild, dominant, sensitive yeasts and putative wild killer yeasts on the inoculated, dominant, sensitive yeasts.
ACKNOWLEDGMENTS
This work was funded by grants 2PR04B003 and GR10088-AGA005 from the Extremadura Regional Government and in part by grant AGL2011-25711 from the Spanish Ministry of Education and Science. M.M. was the recipient of a studentship from the Extremadura Regional Government.
Footnotes
Published ahead of print 18 November 2011
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Ambrona J, Ramírez M. 2007. Analysis of homothallic Saccharomyces cerevisiae strain mating during must fermentation. Appl. Environ. Microbiol. 73:2486–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barre P. 1980. Role du facteur killer dans la concurrence entre souches de levures. Bull. O. I. V. 593–594:560–567 [Google Scholar]
- 4. Bussey H. 1972. Effects of yeast killer factors on sensitive cells. Nat. New Biol. 235:73–75 [DOI] [PubMed] [Google Scholar]
- 5. Carrau FM, Neirotti EN, Giogia O. 1993. Stuck wine fermentation: effect of killer/sensitive yeast interactions. J. Ferment. Bioeng. 76:67–69 [Google Scholar]
- 6. Coratza G, Musmanno RA, Cresti S, Vagnoli P, Di Maggio T. 1992. L'evoluzione della popolazione di lieviti durante la fermentazione. Vignevini 9:24–27 [Google Scholar]
- 7. da Silva GA. 1996. The occurrence of killer, sensitive, and neutral yeasts in Brazilian Riesling Italico grape must and the effect of neutral strains on killing behaviour. Appl. Microbiol. Biotechnol. 46:112–121 [DOI] [PubMed] [Google Scholar]
- 8. Díez B, Pedrós-Alió C, Marsh TL, Massana R. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. European Community 1999. Commission regulation (EC) no. 761/99 of 12 April 1999 amending regulation (EEC) no. 2676/90 determining community methods for the analysis of wines. Off. J. European Communities 99:4 [Google Scholar]
- 10. European Community 1990. Métodos de análisis comunitarios aplicables en el sector del vino. Official report of the European Community L, no. 2676. 272:191 [Google Scholar]
- 11. Frezier V, Dubourdieu D. 1992. Ecology of yeast strain Saccharomyces cerevisiae during spontaneous fermentation in Bordeaux winery. Am. J. Enol. Vitic. 43:375–380 [Google Scholar]
- 12.Reference deleted.
- 13. Gutiérrez AR, Epifanio S, Garijo P, López R, Santamaría P. 2001. Killer yeasts: incidence in the ecology of spontaneous fermentation. Am. J. Enol. Vitic. 52:352–356 [Google Scholar]
- 14. Hara S, Limura I, Otsuka K. 1980. Breeding of useful killer wine yeasts. Am. J. Enol. Vitic. 31:28–33 [Google Scholar]
- 15. Heard GM, Fleet GH. 1987. Occurrence and growth of killer yeasts during wine fermentation. Appl. Environ. Microbiol. 53:2171–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hidalgo P, Flores M. 1994. Occurrence of the killer character in yeasts associated with Spanish wine production. Food Microbiol. 11:161–167 [Google Scholar]
- 17. Izgü F, Altinbay D, Yücelis A. 1997. Identification and killer activity of a yeast contaminating starter cultures of Saccharomyces cerevisiae strains used in the Turkish baking industry. Food Microbiol. 14:125–131 [Google Scholar]
- 18. Jacobs CJ, Van Vuuren HJJ. 1991. Effects of different killer yeasts on wine fermentations. Am. J. Enol. Vitic. 42:295–300 [Google Scholar]
- 19. Jolly NP, Augustyn OPH, Pretorius IS. 2003. The effect of non-Saccharomyces yeasts on fermentation and wine quality. S. Afr. J. Enol. Vitic. 24:55–62 [Google Scholar]
- 20. Kaiser C, Michaelis S, Mitchell A. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Maqueda M, Zamora E, Rodríguez-Cousiño N, Ramírez M. 2010. Wine yeast molecular typing using a simplified method for simultaneously extracting mtDNA, nuclear DNA and virus dsRNA. Food Microbiol. 27:205–209 [DOI] [PubMed] [Google Scholar]
- 22. Medina K, Carrau FM, Giogia O, Bracesco N. 1997. Nitrogen availability of grape juice limits killer yeast growth and fermentation activity during mixed-culture fermentation with sensitive commercial yeast strains. Appl. Environ. Microbiol. 63:2821–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499 [DOI] [PubMed] [Google Scholar]
- 24. Naumova ES, Bulat SA, Mironenko NV, Naumov GI. 2003. Differentiation of six sibling species in the Saccharomyces sensu stricto complex by multilocus enzyme electrophoresis and UP-PCR analysis. Antonie Van Leeuwenhoek J. Microbiol. 83:155–166 [DOI] [PubMed] [Google Scholar]
- 25. Pérez F, Ramírez M, Regodón JA. 2001. Influence of killer strains of Saccharomyces cerevisiae on wine fermentation. Antonie Van Leeuwenhoek J. Microbiol. 79:393–399 [DOI] [PubMed] [Google Scholar]
- 26. Petering JE, Symons MR, Landgridge P, Henschke PA. 1991. Determination of killer yeast activity in fermenting grape juice by using a marked Saccharomyces wine yeast strain. Appl. Environ. Microbiol. 57:3232–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Radler F, Schmitt M. 1987. Killer toxins of yeasts: inhibitors of fermentation and their adsorption. J. Food Prot. 50:234–238 [DOI] [PubMed] [Google Scholar]
- 28. Ramírez M, Regodon JA, Pérez F, Rebollo JE. 1999. Wine yeast fermentation vigor may be improved by elimination of recessive growth-retarding alleles. Biotechnol. Bioeng. 65:212–218 [DOI] [PubMed] [Google Scholar]
- 29. Ramírez M, et al. 2004. Genetic instability of heterozygous hybrid populations of natural wine yeasts. Appl. Environ. Microbiol. 70:4686–4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Regodón JA, Pérez F, Vald ME, De Miguel C, Ramírez M. 1997. A simple and effective procedure for selection of wine yeast strains. Food Microbiol. 14:247–254 [Google Scholar]
- 31. Rodríguez-Cousiño N, et al. 2011. A new wine Saccharomyces cerevisiae double-stranded RNA virus encoded killer toxin (Klus) with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 77:1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Sangorrín MP, Lopes CA, Giraudo MR, Caballero AC. 2007. Diversity and killer behaviour of indigenous yeasts isolated from the fermentation vat surfaces in four Patagonian wineries. Int. J. Food Microbiol. 119:351–357 [DOI] [PubMed] [Google Scholar]
- 34. Schmitt MJ, Breinig F. 2006. Yeast viral killer toxins: lethality and self-protection. Nat. Rev. Microbiol. 4:212–221 [DOI] [PubMed] [Google Scholar]
- 35. Schmitt MJ, Tipper DJ. 1995. Sequence of the M28 dsRNA: preprotoxin is processed to an α/β heterodimeric protein. Virology 213:341–351 [DOI] [PubMed] [Google Scholar]
- 35a. Sherman F. 1991. Getting started with yeast, p 3–21 In Guthrie C, Fink GR. (ed), Guide to yeast genetics and molecular biology. Methods in enzymology, vol. 194 Academic Press; New York, NY: [DOI] [PubMed] [Google Scholar]
- 36. Shimizu K. 1993. Killer yeasts, p 243–263 In Fleet G. H. (ed), Wine microbiology and biotechnology. Harwood Academic Publishers, Newark, NJ [Google Scholar]
- 37. Tredoux HG, Tracey RP, Tromp A. 1986. Killer factor in wine yeasts and its effect on fermentation. S. Afr. J. Enol. Vitic. 7:105–112 [Google Scholar]
- 38. Ubeda-Iranzo JF, Briones-Perez AI, Izquierdo-Canas PM. 1998. Study of the oenological characteristics and enzymatic activities of wine yeasts. Food Microbiol. 15:399–406 [Google Scholar]
- 39. Vagnoli P, Musmanno RA, Cresti S, Di Maggio T, Coratza G. 1993. Occurrence of killer yeasts in spontaneous wine fermentations from the Tuscany region of Italy. Appl. Environ. Microbiol. 59:4037–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Vuuren HJJ, Jacobs CJ. 1992. Killer yeasts in the wine industry: a review. Am. J. Enol. Vitic. 43:119–128 [Google Scholar]
- 41. Van Vuuren HJJ, Wingfield BD. 1986. Killer yeasts. Cause of stuck fermentations in a wine cellar. S. Afr. J. Enol. Vitic. 7:3521–3529 [Google Scholar]
- 42. Vazquez F, Toro ME. 1994. Occurrence of killer yeasts in Argentine wineries. World J. Microbiol. Biotechnol. 10:358–359 [DOI] [PubMed] [Google Scholar]
- 43. Versavaud A, Courcoux P, Roulland C, Dulau L, Hallet JN. 1995. Genetic diversity and geographical distribution of wild Saccharomyces cerevisiae strains from the wine-producing area of Charentes, France. Appl. Environ. Microbiol. 61:3521–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wickner RB. 1993. Double-stranded RNA virus. J. Biol. Chem. 268:3797–3800 [PubMed] [Google Scholar]
- 45. Wickner RB. 1991. Yeast RNA virology: the killer systems, p 263–296 The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Habor Laboratory Press, Cold Spring Habor, NY [Google Scholar]
- 46. Woods RD, Bevan EA. 1968. Studies on the nature of killer factors produced by S. cerevisiae. J. Gen. Microbiol. 51:115–126 [DOI] [PubMed] [Google Scholar]
- 47. Young TW. 1987. Killer yeasts, p 131–164 In Rose A. H., Harrison J. S. (ed), The yeasts. Academic Press, London, United Kingdom [Google Scholar]
- 48. Zagorc T, et al. 2001. Indigenous wine killer yeasts and their application as a starter culture in wine fermentation. Food Microbiol. 18:441–451 [Google Scholar]