Abstract
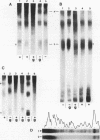
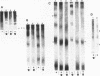
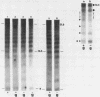
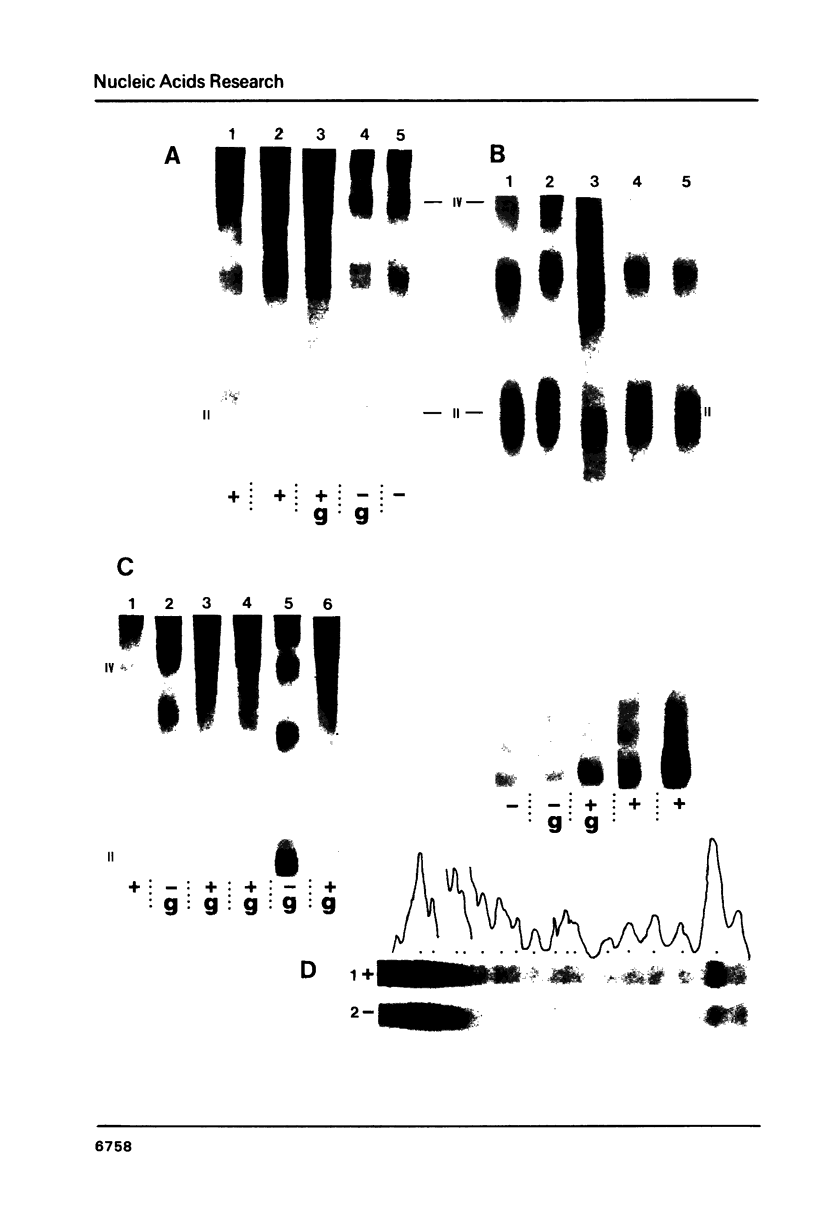
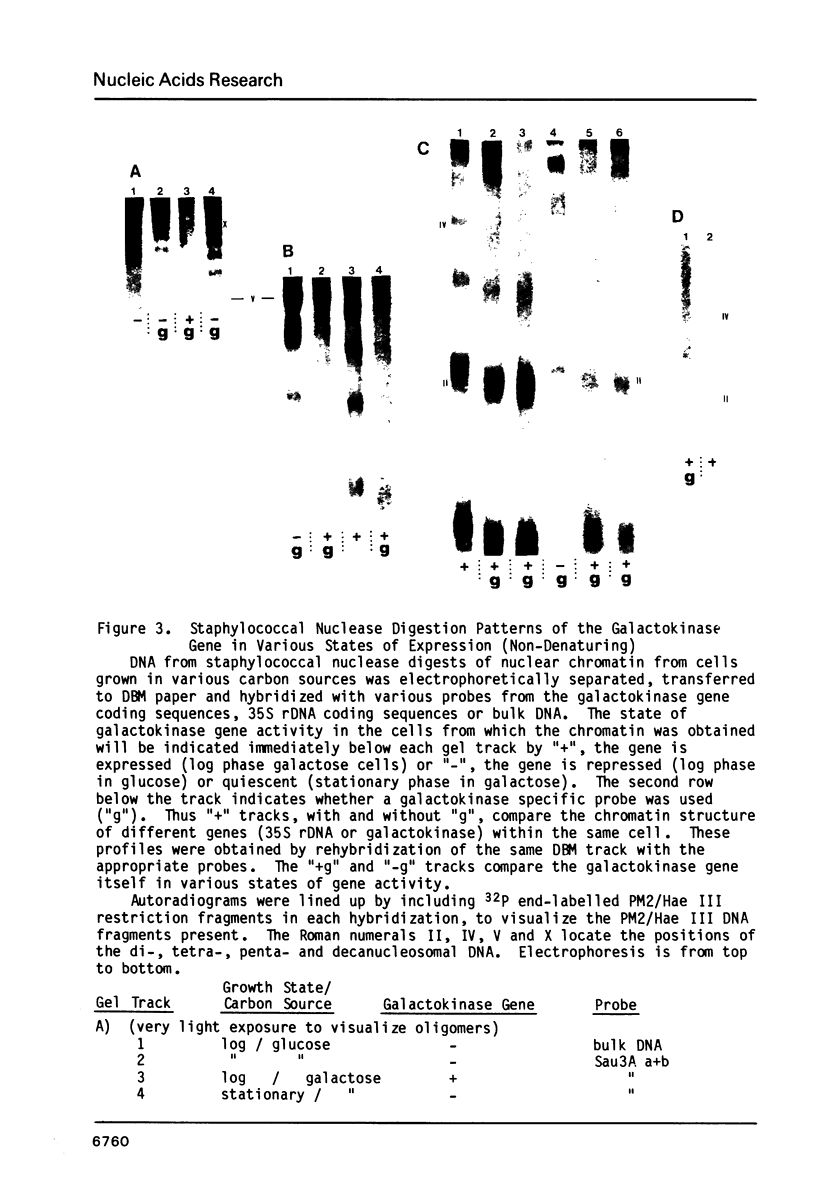
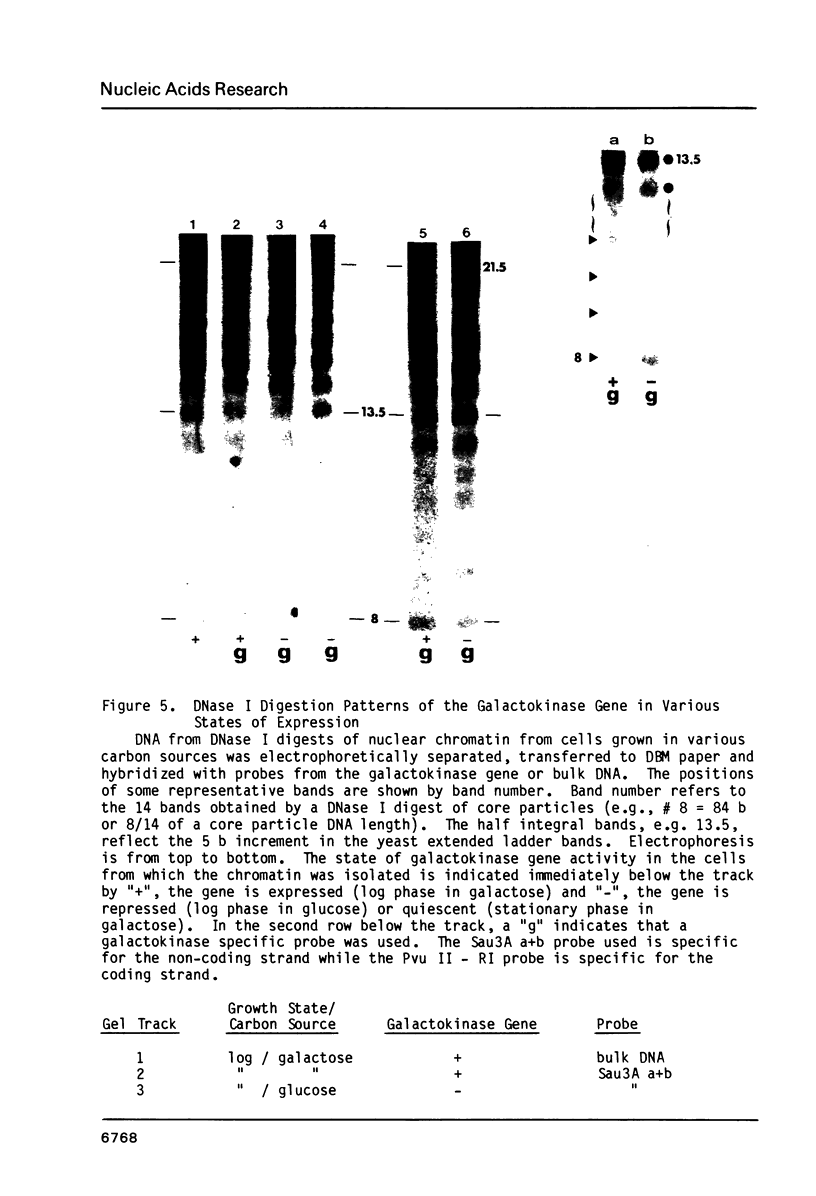
When the yeast galactokinase gene is not active (repressed, not expressed, quiescent), there is an exceptionally regular nucleosome array on coding sequence galactokinase chromatin, as shown by both denaturing and non-denaturing gel analysis of staphylococcal nuclease digests. Expression of the gene results in a limited smearing of the nucleosome repeat peaks and an increase in interpeak DNA, appearing as a regular ladder of DNA bands on denaturing gels. On non-denaturing gels the pattern is more complex and molecular weight dependent. These data suggest an increase in intracore particle DNA accessibility, allowing staphylococcal nuclease to digest throughout the nucleosome in expressed chromatin. Comparison to bulk chromatin and to an operationally inactive gene (35S rDNA) show that the alteration is specific to expressed chromatin. In contrast, DNase I shows no differences in the digestion of the gene specific chromatin in expressed or inactive states.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Bergman L. W., Kramer R. A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Jun 10;258(11):7223–7227. [PubMed] [Google Scholar]
- Brady J. N., Lavialle C., Salzman N. P. Efficient transcription of a compact nucleoprotein complex isolated from purified simian virus 40 virions. J Virol. 1980 Aug;35(2):371–381. doi: 10.1128/jvi.35.2.371-381.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Martinson H. G. The roles of H1, the histone core and DNA length in the unfolding of nucleosomes at low ionic strength. Nucleic Acids Res. 1980 Nov 11;8(21):4969–4987. doi: 10.1093/nar/8.21.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman H. M. Repair of overlapping DNA termini. Methods Enzymol. 1980;65(1):63–64. doi: 10.1016/s0076-6879(80)65010-1. [DOI] [PubMed] [Google Scholar]
- Green M. H., Brooks T. L. The sv40 transcription complex. II. Non-dissociation of protein from SV40 chromatin during transcription. Nucleic Acids Res. 1977 Dec;4(12):4279–4289. doi: 10.1093/nar/4.12.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Levitt A., Axel R., Cedar H. Nick translation of active genes in intact nuclei. Dev Biol. 1979 Apr;69(2):496–505. doi: 10.1016/0012-1606(79)90307-5. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Lohr D. Chromatin structure differs between coding and upstream flanking sequences of the yeast 35S ribosomal genes. Biochemistry. 1983 Feb 15;22(4):927–934. doi: 10.1021/bi00273a034. [DOI] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moyne G., Harper F., Saragosti S., Yaniv M. Absence of nucleosomes in a histone-containing nucleoprotein complex obtained by dissociation of purified SV40 virions. Cell. 1982 Aug;30(1):123–130. doi: 10.1016/0092-8674(82)90018-6. [DOI] [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Palter K. B., Foe V. E., Alberts B. M. Evidence for the formation of nucleosome-like histone complexes on single-stranded DNA. Cell. 1979 Oct;18(2):451–467. doi: 10.1016/0092-8674(79)90064-3. [DOI] [PubMed] [Google Scholar]
- Piñon R., Salts Y. Isolation of folded chromosomes from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2850–2854. doi: 10.1073/pnas.74.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Bavykin S. G., Mirzabekov A. D. Primary organization of the nucleosome core particles. Sequential arrangement of histones along DNA. J Mol Biol. 1980 May 25;139(3):491–517. doi: 10.1016/0022-2836(80)90143-6. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Melchior W., Jr, Felsenfeld G. DNAase I, DNAase II and staphylococcal nuclease cut at different, yet symmetrically located, sites in the nucleosome core. Cell. 1978 Jul;14(3):611–627. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. Transcription by eukaryotic RNA polymerases A and B of chromatin assembled in vitro. Eur J Biochem. 1979 Aug 1;98(2):317–327. doi: 10.1111/j.1432-1033.1979.tb13191.x. [DOI] [PubMed] [Google Scholar]
- Williamson P., Felsenfeld G. Transcription of histone-covered T7 DNA by Escherichia coli RNA polymerase. Biochemistry. 1978 Dec 26;17(26):5695–5705. doi: 10.1021/bi00619a015. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Dattagupta N., Hogan M., Crothers D. M. Structural changes of nucleosomes in low-salt concentrations. Biochemistry. 1979 Sep 4;18(18):3960–3965. doi: 10.1021/bi00585a018. [DOI] [PubMed] [Google Scholar]
- Zayetz V. W., Bavykin S. G., Karpov V. L., Mirzabekov A. D. Stability of the primary organization of nucleosome core particles upon some conformational transitions. Nucleic Acids Res. 1981 Mar 11;9(5):1053–1068. doi: 10.1093/nar/9.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]