Abstract
Bcep22-like phages are a recently described group of podoviruses that infect strains of Burkholderia cenocepacia. We have isolated and characterized a novel member of this group named DC1. This podovirus shows many genomic similarities to BcepIL02 and Bcep22, but it infects strains belonging to multiple Burkholderia cepacia complex (BCC) species.
TEXT
Isolation and characterization of bacteriophages that infect members of the Burkholderia cepacia complex (BCC)—a group of at least 17 species of multidrug-resistant opportunistic pathogens (reviewed in references 18 and 24)—are critical to the development of a phage therapy protocol for these organisms. Five different BCC-specific phages have been shown to be active against Burkholderia cenocepacia in invertebrate or mammalian infection models (3, 17, 22). Three myoviruses (KS4-M, KS12, and KS14) and one siphovirus (KS9) were found to be effective in Galleria mellonella, increasing larva survival at various multiplicities of infection following administration of a lethal dose of B. cenocepacia (22, 17). Similarly, in C57BL/6 mice infected with B. cenocepacia AU0728, the podovirus BcepIL02 was shown to decrease both bacterial density and inflammatory cytokine release (3). BcepIL02 was recently identified as a member of a new phage type, the Bcep22-like phages. To date, this phage type contains only two viruses: the 62,714-bp BcepIL02 (FJ937737) and the 63,882-bp Bcep22 (AY349011) (5). However, we have recently characterized a third member of this podovirus group, a broad-host-range Bcep22-like phage named DC1 (vB_BceP_DC1) (10).
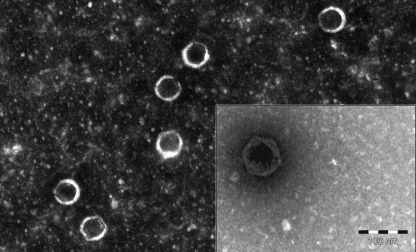
DC1 was isolated from an extract of soil used to cultivate a Dracaena sp. in Edmonton, Canada using Burkholderia cepacia LMG 18821 as a host (22). When plated with LMG 18821 in soft agar overlays on half-strength Luria-Bertani (½ LB) solid medium, DC1 forms mainly clear plaques with a diameter of 1 to 2 mm. Transmission electron microscopy of DC1 virions (performed as described previously [16]) indicates that it is a member of the Podoviridae family (Fig. 1). While the originally described Bcep22-like phages were reported to specifically infect B. cenocepacia (3, 5), the relatively broader host range of DC1 is a significant advantage with respect to clinical use. In contrast to Bcep22 and BcepIL02, which infect B. cenocepacia PC184 (BcepIL02), AU0728 (BcepIL02), and AU1054 (both phages) (3, 5), the DC1 host range includes B. cepacia LMG 18821, B. cenocepacia C6433, PC184, and CEP511, and Burkholderia stabilis LMG 18870 (22). The B. cepacia and B. stabilis strains are CF isolates, while the B. cenocepacia strains are CF epidemic isolates (19). The efficiency of plating for DC1 on each of these strains is similar (within one order of magnitude compared to LMG 18821).
Fig 1.
Transmission electron micrographs of phage DC1 stained with 2% phosphotungstic acid.
DC1 DNA was isolated using the GENECLEAN Turbo Kit (Qbiogene, Irvine, CA) following guanidine thiocyanate lysis of polyethylene glycol (PEG)-precipitated high-titer phage lysates. The complete genome sequence was determined using pyrosequencing (454 Life Sciences, Branford, CT) with PCR cloning (CloneJET PCR cloning kit; Fermentas, Burlington, ON) to fill the contig gaps. Annotation and sequence analysis were performed using GeneMark.hmm-P (15), BLAST (1), EMBOSS matcher (20), TMHMM (9), LipoP (8), tRNAscan-SE (21), HHpred (23), and CoreGenes (25, 12, 13). Comparison plots were prepared using PROmer and Circos (4, 11).
The DC1 genome is 61,847 bp in length, has a 66.2% GC content, and is predicted to encode 73 proteins and one tRNA (see Table S1 in the supplemental material). BLASTN and EMBOSS matcher analysis of the complete genome sequence indicates that it is most closely related to BcepIL02 (79.5% identity) and Bcep22 (73.1% identity). Using CoreGenes analysis to assess phage protein relatedness (12, 13), 52 matches were found between the proteins of DC1 (n = 73) and Bcep22 (n = 81) (see Table S2 in the supplemental material). Although both DC1 gp56 and gp59 (tail fiber proteins) are closely related to Bcep22gp65, the program tallies only gp56 as a match, so the true total is 53 (Table S2), resulting in a 65.43% similarity value between these two phages. Based on the recommended CoreGenes genus-level threshold of 40% (12, 13), it is evident that Bcep22-like phages (including DC1) not only comprise a new phage type as previously suggested (5) but in fact constitute an entirely novel and distinct podovirus genus.
Predicted DC1 genes show similarity to the majority of both BcepIL02 and Bcep22 genes (see Table S2 in the supplemental material), including those encoding the tyrosine integrase (BcepIL02), RecT/nuclease pair, transcriptional repressor, serine tRNA, replication proteins, PagP (BcepIL02), methyltransferase/endonuclease pair (BcepIL02), capsid morphogenesis and DNA packaging proteins, CsrA, multiple tail fiber proteins, acyltransferase, PAPS reductase, large multidomain protein, and lysis proteins (although, based on TMHMM analysis, we predict that gp68 is the putative antiholin and that the putative holin gp70 contains only one transmembrane domain) (5). Two of these proteins are predicted to be involved in lysogeny: the integrase gp4 and the repressor gp8. Interestingly, with regard to phenotypic similarities between all three phages, we have also observed evidence of unstable lysogeny in DC1 hosts (5) (although the nature of this phenomenon requires further investigation). Three proteins similar to BcepIL02 and Bcep22 conserved proteins have been assigned putative functions based on HHpred analysis (with a 95% probability value cutoff): transcriptional regulators gp9 and gp18 and recombination protein gp16 (see Table S1 in the supplemental material).
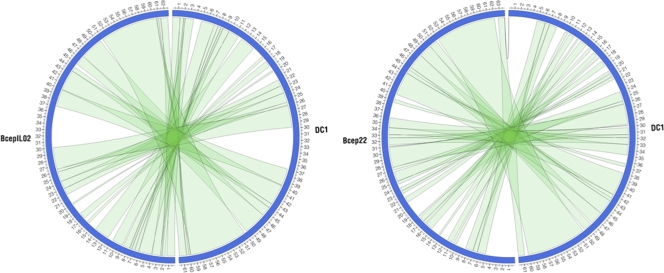
Like many phages, DC1 has a mosaic structure in which regions of strong similarity to BcepIL02 and Bcep22 are interspersed with regions of minimal to no similarity throughout its genome. This mosaicism is evident in the PROmer/Circos plots comparing these three phages (Fig. 2). Based on BLASTP analysis, DC1 lacks homologs of 15 BcepIL02 proteins and 26 Bcep22 proteins (see Table S2 in the supplemental material). The majority of these proteins have no assigned functions. However, DC1 lacks a homolog of a putative transcriptional regulator, DNA ligase, and Rz1-like lysis protein of BcepIL02 and a putative serine recombinase, HNH endonuclease (two proteins), methyltransferase, transposase, transcriptional regulator, and pectin lyase-like protein of Bcep22 (5). The only DC1 proteins with low E-value BLASTP matches to proteins not found in either BcepIL02 or Bcep22 are gp5, gp17, and gp30. Of these, only gp17 has been assigned a putative function (see Table S1 in the supplemental material).
Fig 2.
PROmer/Circos comparisons of DC1 and BcepIL02 (left) or Bcep22 (right). The scale (in kbp) is shown on the periphery. Green ribbons connect regions of protein-level similarity involving the same strand on both genomes. No matches involving opposite strands were detected. PROmer parameters (default): breaklen = 60, maxgap = 30, mincluster = 20, minmatch = 6.
Although DC1 has certain drawbacks with respect to its potential for use in phage therapy (i.e., genes for lysogeny and a putative lipid A palmitoyltransferase [5]), it also has three key advantages. First, the DC1 host range is relatively broad, infecting clinical strains of multiple BCC species. It remains to be determined if amino acid differences between the tail fiber proteins of DC1 and those of BcepIL02 and Bcep22 are responsible for the expanded host range of DC1 (as DC1 gp56, gp57, gp59, and gp60 exhibit 46 to 96% identity with the tail fiber proteins of BcepIL02 and Bcep22 [see Table S2 in the supplemental material]). Second, DC1 is closely related to the only phage shown to date to be active against the BCC in a mammalian infection model, BcepIL02 (3). Finally, all three Bcep22-like phages encode putative CsrA-like proteins. F116, a podovirus active against Pseudomonas aeruginosa biofilms, encodes a similar regulator (2, 6). In Escherichia coli, CsrA expression is inhibitory to biofilm development by means of both decreased formation and increased dispersion (7). When Lu and Collins (14) engineered an M13 phage derivative to express CsrA in E. coli, host susceptibility to ofloxacin increased. If the action of CsrA in Burkholderia is analogous to that in E. coli, Bcep22-like phages in vivo could potentially induce not only direct killing but also reduced biofilm development and increased antibiotic susceptibility.
Together with the findings of Gill et al. (5), we can conclude that Bcep22-like phages have a wide geographic distribution and a potentially broad range of hosts within the BCC. Since a member of this group has already been shown to be active against the BCC in vivo (3), isolation of related phages with expanded host ranges is important for BCC phage therapy development.
Nucleotide sequence accession number.
The DC1 sequence has been deposited in GenBank under accession number JN662425.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Danielle Carpentier for isolating DC1, Kimberley Seed and Amanda Goudie for preliminary DC1 characterization, Sarah Routier for electron microscopy and unstable lysogen isolation, and Ashraf Abdu for unstable lysogen isolation.
J.J.D. gratefully acknowledges funding from Cystic Fibrosis Canada and the Canadian Institutes of Health Research to the CIHR Team on Aerosol Phage Therapy. K.H.L. thanks Cystic Fibrosis Canada, Alberta Innovates—Health Solutions, the Killam Trusts, and the Natural Sciences and Engineering Research Council of Canada for studentship funding.
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrne M, Kropinski AM. 2005. The genome of the Pseudomonas aeruginosa generalized transducing bacteriophage F116. Gene 346:187–194 [DOI] [PubMed] [Google Scholar]
- 3. Carmody LA, et al. 2010. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J. Infect. Dis. 201:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gill JJ, et al. 2011. Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J. Bacteriol. 193:5300–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanlon GW, Denyer SP, Olliff CJ, Ibrahim LJ. 2001. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 67:2746–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson DW, et al. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Juncker AS, et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 10. Kropinski AM, Prangishvili D, Lavigne R. 2009. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 11:2775–2777 [DOI] [PubMed] [Google Scholar]
- 11. Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lavigne R, Seto D, Mahadevan P, Ackermann H-W, Kropinski AM. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406–414 [DOI] [PubMed] [Google Scholar]
- 13. Lavigne R, et al. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu TK, Collins JJ. 2009. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U. S. A. 106:4629–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lynch KH, Stothard P, Dennis JJ. 2010. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genomics 11:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lynch KH, Seed KD, Stothard P, Dennis JJ. 2010. Inactivation of Burkholderia cepacia complex phage KS9 gp41 identifies the phage repressor and generates lytic virions. J. Virol. 84:1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156 [DOI] [PubMed] [Google Scholar]
- 19. Mahenthiralingam E, et al. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 21. Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seed KD, Dennis JJ. 2009. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 53:2205–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vandamme P, Dawyndt P. 2011. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 34:87–95 [DOI] [PubMed] [Google Scholar]
- 25. Zafar N, Mazumder R, Seto D. 2002. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.