Abstract
Bifidobacterium longum subsp. infantis ATCC 15697 utilizes several small-mass neutral human milk oligosaccharides (HMOs), several of which are fucosylated. Whereas previous studies focused on endpoint consumption, a temporal glycan consumption profile revealed a time-dependent effect. Specifically, among preferred HMOs, tetraose was favored early in fermentation, with other oligosaccharides consumed slightly later. In order to utilize fucosylated oligosaccharides, ATCC 15697 possesses several fucosidases, implicating GH29 and GH95 α-l-fucosidases in a gene cluster dedicated to HMO metabolism. Evaluation of the biochemical kinetics demonstrated that ATCC 15697 expresses three fucosidases with a high turnover rate. Moreover, several ATCC 15697 fucosidases are active on the linkages inherent to the HMO molecule. Finally, the HMO cluster GH29 α-l-fucosidase possesses a crystal structure that is similar to previously characterized fucosidases.
INTRODUCTION
The genus Bifidobacterium is frequently overrepresented in the breast-fed infant colon relative to its appearance in adults, where these organisms are believed to benefit their host through nutrient supplementation, participating in host energy cycling and binding to preferred host receptor molecules otherwise available to pathogens (12). Selective growth of bifidobacteria has been attributed to utilization of oligosaccharides abundant in human milk (10 to 20 g/liter) that present complex structures resistant to infant digestion (17, 35). Approximately 200 species of human milk oligosaccharides (HMOs) have been characterized that are composed of glucose, galactose, N-acetylglucosamine, and often fucose and/or sialic acid residues via several glycosidic linkages (25). The HMO core is typically elongated from a lactosyl reducing end (Galβ1-4Glc) that is linked via β1-3 (or β1-6 in branched molecules) to serial lacto-N-biose I units (Galβ1-3GlcNAc) or lactosamine (Galβ1-4GlcNAc) with a degree of polymerization of ≥4.
As with other fucosylated glycoconjugates, α1-2/3/4 fucosyl moieties often shield HMOs from digestion unless this linkage at the nonreducing terminus is first cleaved. Similarly, acidic HMOs or milk sialyloligosaccharides (MSOs) obstruct enzymatic degradation with sialyl residues via α2-3/6 linkages. Removal of these termini is postulated to initiate bacterial catabolism of HMOs (2, 8, 30). To this end, it has been recently demonstrated that Bifidobacterium longum subsp. infantis ATCC 15697 utilizes milk sialyloligosaccharides via a sialidase encoded within a large gene cluster dedicated to HMO metabolism (30).
Previous research conducted on bifidobacterial metabolism of fucosylated oligosaccharides identified a Bifidobacterium bifidum α1-2-l-fucosidase that exhibited an atypical inverting mechanism (glycoside hydrolase [GH] family 95), termed AfcA (10, 22). Inverting glycoside hydrolases modify anomeric stereochemistry via a single nucleophilic displacement, mechanistically contrasting with retaining enzymes, which maintain the anomeric configuration through catalysis of a second displacement. A second B. bifidum fucosidase, one that hydrolyzes α1-3/4 linkages, was characterized to be active on the purified HMO lacto-N-fucopentaose II {Gal(β1-3)[Fuc(α1-4)]GlcNAc(β1-3)Glc(β1-4)Glc} and lacto-N-fucopentaose III {Gal(β1-4)[Fuc(α1-3)]GlcNAc(β1-3)Glc(β1-4)Glc}. Both fucosidases are secreted and attach to the B. bifidum extracellular surface.
The specific HMOs consumed by B. longum subsp. infantis ATCC 15697 have been previously detailed, and several small-mass, fucosylated oligosaccharides are clearly preferred (16, 17). The physiological basis for fucosylated HMO metabolism is evidenced by several potential α-l-fucosidases, including two residing within the HMO gene cluster; however, the B. longum subsp. infantis ATCC 15697 genome sequence is decidedly more ambiguous with respect to successive steps in fucose metabolism, as genes of the canonical fucIAK pathway have not been detected (29).
We describe here our investigation into fucosylated HMO metabolism, including a temporal glycoprofile to monitor consumption preferences through fermentation. Moreover, we have characterized fucosidases expressed by B. longum subsp. infantis ATCC 15697 representing an essential activity necessarily employed early in the catabolism of these molecules.
MATERIALS AND METHODS
Bacteria and media.
B. longum subsp. infantis ATCC 15697 was routinely grown on modified de Mann, Rogosa, and Sharpe medium supplemented with 2% (wt/vol) lactose and 0.25% (wt/vol) l-cysteine (Sigma-Aldrich, St. Louis, MO). Cells were cultured in a vinyl anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) at 37°C and 5% carbon dioxide, 5% hydrogen, and 90% nitrogen. Chemically competent Escherichia coli BL21 Star and Top10 cells were obtained from Invitrogen (Carlsbad, CA), and recombinant clones were grown in Luria broth supplemented with 50 μg/ml of carbenicillin (Teknova, Hollister, CA) when necessary at 37°C.
RNA extraction.
B. longum subsp. infantis cells were grown on Zhang-Mills-Block 1 (ZMB-1) medium (38), and 2% carbohydrate, such as lactose (Sigma), inulin (raftiline HP; Orafti, Malvern, PA), or purified HMO (gift from J. B. German) was added. Lacto-N-tetraose and lacto-N-neotetraose (V-Labs, Covington, LA) were also added at a final concentration of 0.5%. Growth profiles were monitored by measuring the optical density at 600 nm (OD600) using a PowerWave microplate spectrophotometer (BioTek Instruments, Inc., Winoosky, VT). Each experiment was performed in triplicate. Cells were recovered at exponential phase, pelleted, resuspended in 1 ml of RNAlater (Ambion, Austin, TX), and stored overnight at 4°C and then at −80°C until use. RNA extraction was performed as previously described (7), and RNA was converted to cDNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA).
Quantitative real-time PCR.
Relative quantification of the transcript levels of B. infantis fucosidase genes was determined using quantitative real-time PCR (qRT-PCR). Blon_0393, a gene encoding a cysteinyl-tRNA synthetase, was used as an endogenous control (27). Primers and 5′ nuclease probes were designed and synthesized by TibMolBiol (Adelphia, NJ) (see Table S1 in the supplemental material). qRT-PCR was performed on a 7500 Fast real-time PCR system with TaqMan universal mastermix (Applied Biosystems). Each reaction mixture contained 0.5 μM each primer, with its specific probe at 125 nM, 5 ng of cDNA, and reaction buffer as indicated by the manufacturer. qPCRs were run at 95°C for 10 min, 40 cycles of denaturation at 95°C for 10 s, and annealing and elongation for 1 min at 60°C. Relative transcript levels were normalized to lactose as the basal condition. Threshold cycle data and relative efficiencies were analyzed using the Q-Gene software (www.gene-quantification.de/qgene.zip). Results are expressed as the fold changes in gene expression.
Recombinant cloning and protein expression.
Gene sequences were analyzed using the Integrated Microbial Genomes (IMG) database (19). For each fucosidase gene, primers were designed (see Table S1 in the supplemental material) with modifications necessary for cloning using the pET101 directional TOPO expression kit (Invitrogen). Genomic DNA from B. longum subsp. infantis was obtained as described in reference 7. Fucosidase genes were amplified by PCR using 0.5 μM each forward and reverse primer, 1 ng genomic DNA, 0.2 mM deoxynucleotriphosphate mix (Fermentas, Glen Burnie, MD), and 2 U of Phusion Hot Start high-fidelity DNA polymerase (Finnzymes, Vantaa, Finland) in a 150-μl final volume. PCR was performed using a PTC200 Thermo Cycler (MJ Research, Ramsey, MN), with initial denaturation at 95°C for 2 min and 35 cycles of denaturation at 95°C 1 min, annealing at 58°C for 1 min, extension at 72°C 2 min, and a final extension at 72°C for 7 min. PCR products were gel purified and incorporated into pET101 via the TOPO reaction, and transformation was performed as indicated by the manufacturer. Recombinant BL21 Star clones were confirmed by plasmid sequencing using primers T7prom and T7term. Protein expression was carried out using 100 ml LB broth supplemented with 50 μg/ml carbenicillin, using different optimized growth temperatures and isopropyl-β-d-thiogalactopyranoside (IPTG) concentrations (see Table S1 in the supplemental material). Cells were grown in a shaker at 250 rpm (Innova-4000; New Brunswick Scientific, Edison, NJ) until they reached an OD of 0.6 and were induced with IPTG for 6 h. Cultures were centrifuged at 1,700 × g on an Eppendorf 5804 centrifuge (Hauppauge, NY) for 20 min at 4°C and stored at −80°C. Pellets were reconstituted in Bugbuster protein extraction reagent (EMD Chemicals), using 5 ml of the detergent for 50 ml of culture. Lysozyme (50 μl of a 50-mg/ml stock; Sigma-Aldrich) and DNase I (20 μl of a 10,000-U stock; Roche Applied Sciences) were added, vortexed, and incubated for 20 min at room temperature. The suspension was centrifuged for 20 min at 18,500 × g at 4°C, and the supernatant was applied to 1-ml Bio-Scale Mini Profinity immobilized-metal affinity chromatography cartridges connected to an EP-1 Econo pump (Bio-Rad, Hercules, CA). Protein purification was performed as recommended by the manufacturer. Recombinant proteins were evaluated for molecular mass and purity on 10% SDS-PAGE gels. Finally, imidazole was exchanged for phosphate-buffered saline using Amicon Ultra-15 centrifugal filter units, with a cutoff of 10 kDa (Millipore, Billerica, MA).
Determination of kinetic parameters.
McIlvaine buffer solutions, between pH 4.0 and 8.0, were prepared for optimum pH determinations. Relative activity at different pH values was determined by incubating each fucosidase in a 100-μl reaction volume with 2 mg/ml of 2-chloro-4-nitrophenyl-α-l[scap]-fucopyranoside (CNP-fucose; Carbosynth, Berkshire, United Kingdom) in 96-well microwell plates at 37°C for 10 min. Reactions were stopped by adding an equal volume of 1 M Na2CO3. Absorbance at 405 nm was determined using a Synergy2 microplate reader (Biotek). The optimum temperature for enzymatic activity was determined in McIlvaine buffer at each enzyme's optimum pH, incubating them with CNP-fucose at 4, 20, 30, 37, 45, 55, and 65°C. Relative activity was determined from A405 reads. For determination of kinetic values, substrate concentrations in the range of 0.1 to 4 mM CNP-fucose were coincubated with constant amounts of each enzyme under optimum conditions as determined. The time course for each reaction was determined previously to fit steady-state assumptions, and rate reactions were determined under initial conditions. The amount of CNP generated in each reaction was estimated using a standard curve. Nonlinear regression was used to determine Km and Vmax values, using the Solver program.
Thin-layer chromatography.
The activities of α-fucosidases against fucosylated substrates were studied using the optimum conditions determined as described above, in 10-μl reaction mixtures under optimum conditions for different times. 2′-Fucosyl lactose (2′FL), 3′-fucosyl lactose (3′FL), and H-disaccharide (Fucα1-2Gal) were purchased from V-labs. Aliquots of the reaction mixtures were inactivated at 95°C for 5 min and spotted in thin-layer chromatograph (TLC) glass-silica gel plates (Sigma). A mixture of ethyl acetate, acetic acid, and water in a 2:2:1 ratio was used as solvent. After drying, plates were sprayed with 0.5% α-naphthol and 5% H2SO4 in ethanol. Plates were dried and revealed at 150°C for 10 min.
Relative substrate preferences for HMO cluster fucosidases.
Equimolar concentrations of 2′-fucosyl lactose, 3′-fucosyl lactose, H-disaccharide, and Lewis a and Lewis x (V-labs) were coincubated with 5 μg of either Blon_2335 or Blon_2336 for 10 min at 37°C. Reactions were inactivated by incubation at 95°C for 5 min. The galactose assay kit (Biovision, Mountain View, CA) was used to quantify galactose concentrations present in each sample (in an equimolar ratio to released fucose), after overnight incubation with lactase (2′FL and 3′FL) and a mix of 2 μg of B. longum subsp. infantis enzymes Blon_2016 and Blon_2334 (known to cleave Galβ1-3GlcNAc and Galβ1-4GlcNAc [data not shown]) for Lewis a and Lewis x reactions. Fluorescence was quantified using a standard curve in a Synergy 2 microplate reader.
Glycoprofiling.
Aliquots of supernatant were spiked with an internal standard consisting of deuterated HMOs as described previously (35, 36). Each sample was desalted by solid-phase extraction employing graphitic carbon Top Tip cartridges (Glygen, Columbia, MD) and was evaporated to dryness under vacuum. The dried samples were prepared for liquid chromatography-mass spectrometry (LC-MS) analysis by dissolving the sample into 18 MΩ water to a final concentration of 20 ppm based on the amount of the internal standard added to each sample. Analytical triplicates were prepared from each sample of supernatant, and each triplicate was analyzed three times by LC-MS analysis.
LC-MS analysis was performed with an Agilent HPLC-Chip/TOF accurate mass time-of-flight mass spectrometer equipped with both an Agilent 1200 series capillary flow high-performance LC (HPLC) pump for sample loading and an Agilent 1200 series nanoflow HPLC pump for chromatographic separation. All chromatography was performed on an Agilent glycan chip, a microfluidic platform incorporating an enrichment/concentration column, nano-LC column, and nanospray emitter. Both the enrichment and nano-LC column were packed with porous graphitized carbon. A binary solvent system was employed consisting of 3% aqueous acetonitrile plus 0.1% formic acid (solvent A) and 90% aqueous acetonitrile plus 0.1% formic acid (solvent B). Chromatographic separation of the HMOs was achieved using a previously optimized gradient consisting of the following solvent changes: 0 to 2.5 min 0% B, 2.5 to 20 min 16% B, 20 to 30 min 44% B, 30 to 35 min 100% B, 35 to 45 min 100% B, 45.01 to 65 min 0% B (24). The flow rate was held constant at 300 nL/min during the entirety of the gradient. All analyses were performed in the positive ion mode with the source conditions being adjusted to provide for maximum signal with minimal source fragmentation. Each analysis was processed postacquisition by summing the spectra across the chromatographic range. The m/z values and intensities in these summed spectra were then exported as text files and used to calculate the percent consumption of the fucosylated HMOs.
The percent consumption levels of the fucosylated HMOs were calculated based on a previously described method (26, 36). Briefly, the percent consumption of a fucosylated HMO was calculated using the ratio of the intensities of the HMO in the supernatant and the corresponding HMO from a control sample (D/H ratio). The D/H ratio for each fucosylated HMO of interest in both the supernatant and control samples had to be adjusted for overlap of the isotopic envelope of the nondeuterated HMO. This was accomplished by using the following equation, D/H = [(ID/IH) − IH(k)/IH], where ID and IH are the intensities of the monoisotopic peak of deuterated HMO standard and HMO, respectively. The weighting factor k is used to remove the contribution of the HMO isotopic envelope from that of the deuterated standard. All weighting factors were calculated using the IonSpec Exact Mass Calculator software (version 9.0.15). Once the D/H ratio for each supernatant and control sample was determined, the percent consumption was then calculated using the following equation: 1 − {[(D/H)sample]/[(D/H)control]} × 100.
Fucosidase digestion of purified HMO.
Recombinant enzymes were used without further purification. HMO standards—LNFP I, LNFP III, and LNDFH I—were obtained from Dextra Laboratory (Earley Gate, United Kingdom). In a 0.2-ml PCR tube, 3 μl nanopure water was added, followed by 1 μl HMO standard sample and 1 μl enzyme solution (the mole ratio of protein to OS was about 1:100 to 1:200). The reaction mixture was incubated at 37° C in a water bath. The reaction was monitored by matrix-assisted laser desorption ionization–Fourier transform infrared spectroscopy-ion cyclotron resonance MS (MALDI–FT-ICR MS) at 1 h and 3 h. The HiRes MALDI–FT-ICR apparatus (IonSpec, Irvine, CA) has an external MALDI source with a pulsed 355-nm Nd:YAG laser, a hexapole ion guide, an ultrahigh vacuum system maintained by two turbo pumps, one cryopump, and a 7.0-T shielded superconducting magnet. DHB (2,5-dihydroxybenzoic acid) was used as the matrix (8 mg/160 μl in 50% acetonitrile-water [vol/vol]) in the positive ion mode. The HMO solution (0.5 μl) was spotted on a 100-sample stainless steel probe followed by adding 0.25 μl, 0.01 M NaCl solution as a cation dopant, and 0.5 μl matrix solution. The sample was dried in the vacuum chamber before placing it into the ion source.
Protein crystallization and X-ray structure determination.
For structural characterization of Bifidobacterium longum subsp. infantis ATCC 15697 fucosidases, in particular Blon_2336, SeMet-labeled Blon_2336 was screened for crystallization conditions using the hanging drop vapor diffusion technique. Diffraction-quality crystals appeared under the condition of 0.8 M succinic acid, pH 7.0, at 16°C. Prior to X-ray data collection, crystals were treated in paratone N oil and were flash-frozen directly in liquid nitrogen.
A set of single-wavelength anomalous diffraction (SAD) data was collected near the selenium absorption peak at a temperature of 100 K from a single selenomethionine (SeMet)-labeled crystal. The data were obtained at the 19ID beamline of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory, using the program SBCcollect. The intensities were integrated and scaled with the HKL3000 suite (20) (see Table S2 in the supplemental material). Two Blon_2336 molecules (monomers A and B) with a total 20 methionine residues were expected in one asymmetric unit. Twenty heavy atom sites (potentially Se) were located using the program SHELXD (28), and 19 selenium sites were used for phasing with the program MLPHARE (4). After map averaging and density modification (DM) (4), a partial model of 800 residues (84% of two Blon_2336 molecules) with 506 side chains was built via 3 cycles of Arp/warp model building (3). All of the above programs are integrated within the program suite HKL3000 (20). Further model-building efforts to complete the structure were performed manually using the program COOT (5). In and outside the active site of the catalytic domain, there were extra electron densities that appeared similar to a short peptide with a tyrosine residue pointing into the ligand-binding pocket. Only a tyrosine residue was built into the densities for structural refinement purposes. The nature of the short peptide-like ligand remains unknown. The final model was refined using the program Phenix.refine (21) (see Table S2). In the final model, 17 residues between A236 and V255 in monomer A and 11 residues between E241 and T254 in monomer B are missing due to the lack of electron densities.
Protein structure accession number.
The crystal structure was deposited in the Protein Data Bank (PDB:3MO4).
RESULTS
The B. longum subsp. infantis ATCC 15697 temporal glycoprofile of preferred neutral HMOs.
We have previously demonstrated that B. longum subsp. infantis ATCC 15697 prefers small-mass oligosaccharides secreted early in lactation (17). Several of these HMOs are fucosylated and represent the most abundant species within the aggregate pool. Recently, we monitored oligosaccharide consumption during a fermentation to resolve substrate preferences while controlling for spurious degradation, which revealed that bacteria are capable of utilizing sialylated milk oligosaccharides (30). Using a similar approach, FT-ICR MS was employed to ascertain at what point during fermentation B. longum subsp. infantis ATCC 15697 utilizes neutral compositions from a mixed population of purified HMOs (i.e., <200 molecular species).
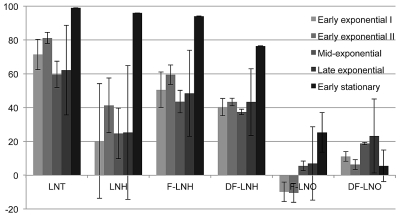
Six low-molecular-mass neutral HMOs were examined, with fermentation culture supernatants sampled at various cell physiological stages corresponding to increasing optical densities (OD600) of approximately 0.2, 0.3, 0.6, 0.75, and 1.0 (Fig. 1). The six HMO species corresponded to those that are consumed by B. longum subsp. infantis efficiently and have been termed preferred (i.e., lacto-N-tetraose [LNT], lacto-N-hexaose [LNH], F-LNH, and DF-LNH), with two additional compositions that are not utilized well and are referred to as nonpreferred (i.e., fucosylated lacto-N-octaose [F-LNO] and DF-LNO). While fucosylated LNT (F-LNT) possesses a comparable mass and is readily consumed, it was excluded due to an extremely low abundance within our HMO pool (<0.7%) (17). We defined consumption conservatively as ≥35% elimination from the culture supernatant.
Fig 1.
Temporal glycoprofile of abundant neutral HMO consumption by B. longum subsp. infantis ATCC 15697. HMO composition results are shown for a representative isomer signifying a characteristic oligosaccharide composition. All samples were analyzed at least in duplicate. LNT, lacto-N-tetraose-like composition; LNH, lacto-N-hexaose-like composition; F-LNH, fucosylated lacto-N-hexaose-like composition; DF-LNH, difucosylated lacto-N-hexaose-like composition; F-LNO, fucosylated lacto-N-octaose-like composition; DF-LNO, difucosylated lacto-N-hexaose-like composition.
Interestingly, the LNT-like composition (Galβ1-3/4GlcNAcβ1-3Galβ1-4Glc) was consumed early in fermentation and to a greater degree than other neutral oligosaccharides. LNT fermentation terminated at the commencement of stationary phase, with near-complete elimination from the culture supernatant (98.97 ± 0.13% [mean ± standard error of the mean]).
With respect to oligosaccharides of increasing degrees of polymerization (DPs), the next two preferred neutral oligosaccharides, LNH (3 Gal:2 GlcNAc:1 Glc) and F-LNH (3 Gal:2 GlcNAc:1 Glc:1 Fuc), were consumed efficiently and approached extinction at 95.98 ± 0.10% and 94.13 ± 0.09% elimination, respectively. LNH and F-LNH utilization levels, however, appeared to occur at a later stage than LNT, achieving >60% elimination only at stationary phase. Of interest, LNH was not preferred over F-LNH, despite similar defucosylation prior to metabolism. This suggests a differential transport efficiency between these two oligosaccharides, which differ by one fucosyl residue.
DF-LNH differs from F-LNH by an additional fucosyl moiety, and with the initiation of stationary phase is consumed to a lesser extent than the other three preferred oligosaccharides (∼76%). Structural constraints presented by the difucosylated molecule likely contribute to this potentially deleterious effect on transport efficiency.
The clearest differences in utilization were observed, as expected, in the “nonpreferred” lacto-N-octaose series (F-LNO, 4 Gal:3 GlcNAc:1 Glc:1 Fuc) and (DF-LNO, 4 Gal; 3 GlcNAc:2 Fuc:1 Glc), which may represent the DP limit to which B. longum subsp. infantis ATCC 15697 efficiently transports HMOs. Translocation of intact molecules is considered critical for B. longum subsp. infantis milk oligosaccharide metabolism, as their HMO-active glycoside hydrolases are localized intracellularly (1, 2, 29, 30).
Of the preferred oligosaccharides, LNT is favored, as it disappeared earlier from the recovered fermentate than LNH, F-LNH, and DF-LNH, although the large intersample variation in early fermentation (e.g., LNH utilization) precludes a definitive conclusion. This error may be the result of the difficulty in obtaining biological replicates identical in the amount of arbitrarily defined phases. Similar variability has been observed in previous glycoprofiling studies of bifidobacterial consumption of HMOs (16, 18).
Identification of α-fucosidases in the ATCC 15697 genome.
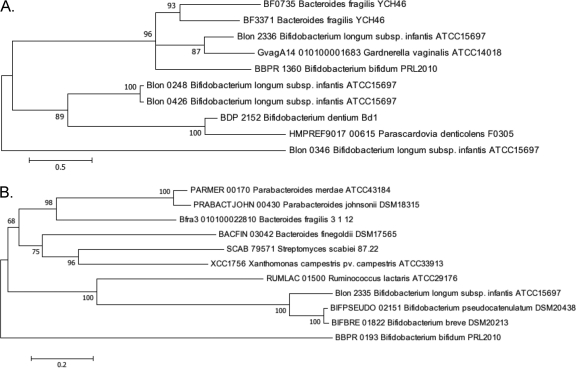
The ATCC 15697 predicted proteome was scanned for evidence of fucosidase sequence signatures. An examination of GH29 (pfam01120) α-fucosidases revealed the presence of a GH29 α-fucosidase (Blon_2336) that localized to a cluster of genes linked with HMO utilization. This α1-3/4 fucosidase exhibited 33% amino acid identity to the fucosidase domain of the B. bifidum NCIMB 41171 AfcB fucosidase (BbifN4_010100008741). Interestingly, the Blon_2336 sequence diverged less from the B. bifidum AfcB than from other ATCC 15697 fucosidases (Fig. 2). Clearly, this does not match the organismal phylogeny and suggests different functions for these fucosidases and/or lateral gene transfer. Moreover, Blon_2336 was predicted to be expressed intracellularly, as it does not possess the N-terminal signal sequence, C-terminal transmembrane region, or the LPXTG motif displayed by the larger, membrane-anchored B. bifidum AfcB (478 versus 1,468 amino acids [aa]). Both AfcB and Blon_2336 include a carbohydrate-binding module 32 (CBM32) domain (PF00754), which has been shown to ligate lactosyl/galactosyl residues for the Micromonospora viridifaciens sialidase (23), lactosamine (Galβ1-3/4GlcNAc) in association with a Clostridium perfingens β-hexosaminidase (6) and found in the LNT-cleaving lacto-N-biosidase (EC 3.2.1.140) characterized for B. bifidum (34). This strongly suggests that the CBM32 domain binds HMO epitopes and may be common to HMO-active glycoside hydrolases, consistent with Blon_2336 integration within the HMO cluster.
Fig 2.
Phylogenetic relationships of fucosidases encoded by select bacteria. Branch lengths are in the same units (number of amino acid substitutions per site) as those of the evolutionary distances used to construct the tree. The evolutionary history was inferred by the maximum likelihood method, followed by 100 bootstrapped replicates. The organism and loci are listed for those fucosidases found in glycoside hydrolase family 29 (A) and glycoside hydrolase family 95 (B).
Directly adjacent to Blon_2336 is an encoded GH95 α-fucosidase (Blon_2335) predicted to cleave α1-2 linkages by an atypical inverting mechanism. This gene has a much shorter sequence than the homologous B. bifidum afcA (BbifN4_010100000509), with the two enzymes predicted to be comprised of 782 and 1,959 amino acid residues, respectively. These two enzymes exhibited 26% identity along their alignable region, which corresponded solely to the B. bifidum AfcA GH95 catalytic domain. In contrast, an N-terminal signal peptide, C-terminal anchor, or an immunoglobulin-like domain was not identified in the ATCC 15697 AfcA. These three elements, in concert, allocate the B. bifidum AfcA to its extracellular surface.
Two additional GH29 α-l-fucosidase-encoding open reading frames, Blon_0248 and Blon_0426, appear elsewhere on the ATCC 15697 chromosome and are highly homologous at the nucleotide level, sharing 98% identity along their length (1,350 bp), with divergence occurring at the 3′ end. These paralogs are 95% identical along the length of their deduced 449-amino-acid sequence, moderately diverging at the C terminus external to their GH29 α-l-fucosidase domain (amino acids 4 to 360) and hinting at similar catalytic activities. As previously reported (29), Blon_0426 and an associated permease appear to have arisen from a recent duplication event that may have displaced elements of arabinose metabolism in ATCC 15697. This is one of several features by which B. longum subsp. infantis metabolism was redirected from plant oligosaccharides toward mammalian sugars (31). Both Blon_0248 and Blon_0426 are predicted to be intracellular, as is the Bifidobacterium dentium ATCC 27679 homolog (BIFDEN_00927), although its encoded protein only shares 28% identity with the ATCC 15697 Blon_0248 protein (33). The phylogenetic relationship between currently sequenced bifidobacteria GH29 α-l-fucosidases is presented in Fig. 2.
Finally, there is an additional gene that has been annotated as a β-galactosidase (COG1874) that also possesses a pfam01120 α-fucosidase domain at the 5′ terminus (Blon_0346). This gene has a β-galactosidase trimerization domain (pfam08532), which suggests this enzyme may be active as a trimer polypeptide. As indicated in Fig. 2, this enzyme is quite divergent from other α-fucosidases, including those found in ATCC 15697.
α-Fucosidase expression profiles.
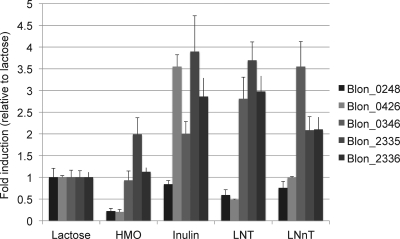
In order to link genetic structure with an encoded enzyme's substrate, and to determine the extent to which a given substrate induces expression, the expression profiles of all five ATCC 15697 α-l-fucosidases were examined (Fig. 3). With the exception of Blon_0248, all fucosidases were induced ≥2-fold, relative to lactose, when ATCC 15697 was grown on the polyfructan inulin, which is devoid of fucosyl residues. A similar pattern has been observed in HMO-interacting solute-binding proteins induced in bifidobacteria subsisting on inulin (7). Interestingly, our complex mix of HMOs only moderately induced (∼2-fold) Blon_2335, while it repressed Blon_0248 and Blon_0426. This appears to indicate a carbon catabolite repression by one or more of the molecules present in the HMO mix. This repression may be due to the abundance of lacto-N-tetraose, because when cells were grown on this substrate, Blon_0248 and Blon_0426 were similarly repressed; however, the other three fucosidases were induced, while ATCC 15697 consumed this single-isomer HMO. LNT's structural isomer, lacto-N-neotetraose (LNnT), which differs only by a terminal β1-4 galactosyl linkage, displayed a different expression pattern. Accordingly, LNnT induces the same three enzymes, with Blon_2335 and Blon_2336 induced to a lesser extent. However, repression of Blon_0248 and Blon_0426 is not evident, as they appeared to be transcribed to a similar degree as with cells grown on lactose.
Fig 3.
ATCC 15697 fucosidase gene expression during carbohydrate fermentation. Gene expression was calculated relative to levels when grown on lactose as the sole carbon source. Averages from three independent experiments are shown, and bars represent standard errors of the means.
Biochemical characterization of ATCC 15697 α-fucosidases.
In order to evaluate the activity of B. longum subsp. infantis ATCC 15697 α-fucosidases, these enzymes were cloned and expressed in a heterologous host. Table S3 in the supplemental maerial indicates optimal expression conditions in addition to pH and temperature optima. All five fucosidases were most active at 37°C and possessed pH optima between 6 and 7.5 (Table S3). The Km values of the fucosidases were <1.0, with the exception of Blon_2335, which had a much lower affinity for CNP-fucose (Km = 33.03 ± 2.807 mM) than the other enzymes (Table 1). Blon_0346, Blon_0426, and Blon_2335 have relatively higher turnover rates (>3.5 s−1) than the other two enzymes, which in turn lead to high efficiency ratios (Kcat/Km) for Blon_0346 and Blon_0426.
Table 1.
Biochemical attributes of ATCC 15697 α-fucosidasesa
| Fucosidase | Vmax (nmol/s) | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Blon_0248 | 0.0017 ± 0.0010 | 0.131 ± 0.010 | 0.110 ± 0.026 | 833.31 ± 134.64 |
| Blon_0346 | 0.0603 ± 0.0101 | 0.115 ± 0.005 | 9.452 ± 1.582 | 81,977.97 ± 10,060.60 |
| Blon_0426 | 0.0089 ± 0.0007 | 0.180 ± 0.030 | 4.481 ± 0.329 | 24,951.71 ± 1,686.06 |
| Blon_2335 | 0.1130 ± 0.0325 | 33.03 ± 2.807 | 3.898 ± 1.122 | 119.87 ± 44.15 |
| Blon_2336 | 0.0092 ± 0.0008 | 0.709 ± 0.149 | 0.285 ± 0.024 | 407.73 ± 51.34 |
Values are means ± standard errors of the means.
α-Fucosidase activity on purified HMOs and linkage specificities.
A mass spectrometry-based approach was employed to determine the extent to which ATCC 15697 fucosidases are active on HMO linkages (see Fig. S1 in the supplemental material). Purified lacto-N-fucopentaose I (LNFP I) and LNFP III were incubated with the fucosidases to assay α1-3 and α1-2 linkages, respectively. Blon_0346 did not exhibit any activity on purified HMOs. Blon_0248 displayed a defucosylation of the α1-3 LNFP III after 1 h. Its paralog, Blon_0426, incompletely defucosylated LNFP III during the course of a 3-hour incubation with the enzyme. Blon_2335, a GH95 enzyme predicted to cleave α1-2 linkages, does indeed hydrolyze this bond in LNFP I after 1 h. It also partially cleaves α1-3 linkages. Finally, the other HMO cluster Blon_2336 cleaves α1-3 linkages inherent to LNFP III and partially degrades α1-2 linkages.
Thin-layer chromatography was employed to confirm the MS data on small-chain oligosaccharides (DP, ≤3) (Table 2). Blon_2335 had activity on all three substrates tested, including 2-fucosyllactose, 3-fucosyllactose, and H-2 disaccharide (Fucα1-2Gal). Consistent with the prediction that Blon_2335 is an α1-2 fucosidase, it exhibited stronger cleavage of the two α1-2 substrates. The other HMO cluster enzyme, Blon_2336, cleaved 3-fucosyllactose, consistent with its role as an α1-3/4 fucosidase. None of the other three enzymes displayed activities on these substrates. The one exception was Blon_0346, which had minor activity toward the H-2 disaccharide. The relative activities on these substrates (see Table S4 in the supplemental material) indicate that Blon_2335 has a preference for α1-2 linkages and Blon_2336 is more active on α1-3/4 linkages.
Table 2.
TLC assay results for α-fucosidase linkage specificitya
| α-Fucosidase | Linkage specificity to: |
||
|---|---|---|---|
| 2′FL | 3′FL | Fuca1-2Gal | |
| Blon_0248 | − | − | − |
| Blon_0426 | − | − | − |
| Blon_0346 | − | − | + |
| Blon_2335 | +++ | ++ | +++ |
| Blon_2336 | − | +++ | − |
TLC signals were scored qualitatively: −, no signal; +, weak; ++, moderate; +++, strong.
Structural characterization of Blon_2336.
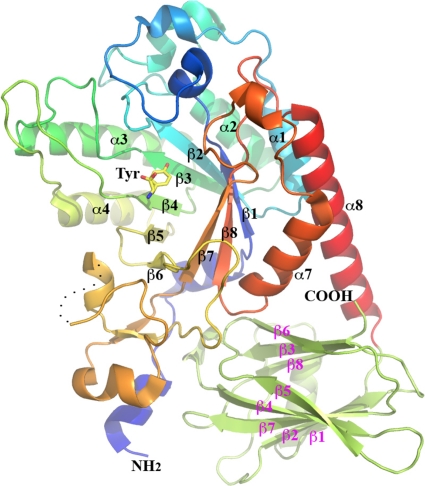
To gain insight into the three-dimensional architecture of the HMO cluster GH29 fucosidase, Blon_2336 was crystallized and its structure was solved. Blon_2336 is monomeric, and each molecule consists of a catalytic domain of a (β/α)8-barrel fold and a β-sandwich domain, which was earlier predicted to be a CBM32 domain (Fig. 4) (13).
Fig 4.
Ribbon drawing of the overall structure of Blon_2336. The N-terminal catalytic domain is shown by the various colors, from the blue at the N terminus to red at the C terminus. The β-strands and α-helices that form the (β/α)8 barrel are labeled in black. The C-terminal carbohydrate-binding domain is shown in lime green, with 9 β-strands labeled in magenta. A tyrosine from a possible short peptide at the active site of the catalytic domain (see description in the text) is drawn in stick format. A dashed curved line indicates a missing part in the final structural model (see the text). The N and C termini of Blon_2336 are also labeled.
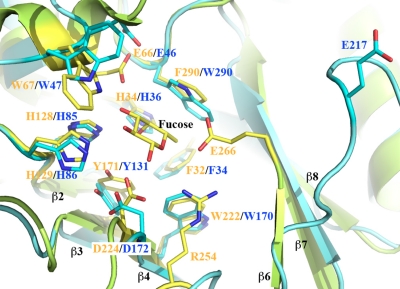
The catalytic domain of Blon_2336 can be structurally aligned well with the other four α-l-fucosidases, including one from Thermotoga maritime (TM0306) (32) and three from Bacteroides thetaiotaomicron (BT2192 [PDB:3EYP], BT2970 [PDB:3GZA], and BT3798 [14]). The root mean square deviation (RMSD) values of the pairwise structural alignments (secondary structural matching [SSM] fitting, Cα only) between Blon_2336 and other four were 1.46 to 1.75 Å, with 244 to 293 aa of 333 aa (Blon_2336) aligned. This indicates that the overall structures of these α-l-fucosidases are quite conserved, although the primary sequence identities between Blon_2336 and others are 23 to 41%. The structural alignments of Blon_2336 with either TM0306 or BT3798 provide valuable information on the active site and potential catalytic residues of Blon_2336, for which extensive studies of their complexes with ligands and inhibitors have been reported (14, 32).
Figure 5 shows a structural alignment of Blon_2336 with TM0306 in complex with fucose (32). Although the sequence identity of the catalytic domains of Blon_2336 and TM0306 is only about 26%, the residues that form their active sites are highly conserved. The residue D224 in TM0306 is the catalytic nucleophile. Its corresponding residue in Blon_2336 is D172. The residue E266 in TM0306 is the catalytic acid/base, which is located after the β6-strand. The region between the β6 and β7 strands is one of the most diversified regions in the α-l-fucosidases in terms of length and structural conformation. However, residue E217 of Blon_2336, shortly after the end of the β6 strand, is the most likely catalytic acid/base of Blon_2336 (Fig. 5). The equivalent amino acids of E217 in BT2192 and BT2970 are E243 and E240, respectively. Even in their apo-forms, these two residues in the BT2192 and BT2970 structures point to their active sites. Upon ligand binding, the E217 of Blon_2336 can flip around to point its side chain toward the active site, as does the catalytic acid/base found in TM0306 and BT3798 complex structures. The conformation of E217 of Blon_2336 observed in this structure may represent one of the possible conformations in its apo-forms. The structural flexibility of part of a catalytic site has been widely observed and assists in the recognition and recruitment of substrate. The function of residue R254 of TM0306, or of its equivalent, R262 of BT3798, is unknown. In the active site, this is the only residue that appears to have no counterpart in Blon_2336 or in BT2192 or BT2970.
Fig 5.
Structural alignment of the active sites of Blon_2336 and TM0306 in complex with fucose. Blon_2336 is shown in cyan, and TM0306 is shown in green-yellow. All active site residues and the fucose from the TM0306/fucose complex are drawn in stick format. The residues from Blon_2336 and TM0306 are labeled in blue and orange, respectively. Equivalent residues from two molecules are paired together. Some visible strands from the (β/α)8 barrel are labeled in black. The tyrosine shown in Fig. 4 is approximately at the position of the fucose in this figure, and it is not shown for the sake of clarity.
The C-terminal domain, as predicted, is a β-sandwich domain. A Dali structural homology search (http://www2.ebi.ac.uk/dali/) using the C-terminal domain resulted in more than 700 hits, including different chains of the same molecules. All of the top 45 hits except 2 were CBM32 domains, with similar scores (Z, 14.4 to 16.7; RMSD, 1.5 to 2.4 Å; sequence identity, 12 to 25%). They included BT2192 and BT2970, which in fact were not outstanding in the top hits list. Interestingly, the C-terminal domain of Blon_2336 has no significant structural homology with that of BT3798 and TM0306.
DISCUSSION
That LNT is consumed early, and to a greater degree, is significant, as LNT is invariably the most abundant oligosaccharide in breast milk and is consumed by a diversity of bifidobacteria, whereas larger HMOs are efficiently utilized by B. longum subsp. infantis and select infant-associated phylotypes (16, 17). LNT also represents the smallest HMO molecule, with a DP of 4, and as such it is a core structure incorporated within larger HMO species. It is possible that LNT is favored over higher-DP oligosaccharides earlier in fermentation, as it is linear and does not present a second β-linkage (1-6) to its terminal lactosyl, as do HMOs with DPs of ≥6, such as LNH and F-LNH. It is tempting to speculate that this β1-6 N-acetylglucosaminyl linkage inherent to branched HMOs may hinder translocation, albeit with minimal consequence to final consumption.
B. longum subsp. infantis VIII-240 was previously characterized (15) as having a relatively strong α1-2 fucolytic activity, a trait now known to be encoded by Blon_2335. This gene was first identified as afcA in B. bifidum (2, 11, 22). In addition to B. bifidum and B. longum subsp. infantis, the Bifidobacterium breve DSM20213 and Bifidobacterium pseudocatenulatum DSM20438 genomes include afcA homologs, BIFBRE_01822 and BIFPSEUDO_02151, respectively, that are highly identical (both 77%) to the ATCC 15697 afcA. It is remarkable that these four infant-type bifidobacteria possess afcA homologs and that other bifidobacteria that usually colonize adults have not been found to possess one. Further investigation is needed to determine if afcA is a key gene linked to infant gut colonization.
Of the four afcA+ bifidobacteria, only B. bifidum secretes its AfcA, advancing a competitive strategy predicated on the import of extracellular hydrolysis products of HMOs (2, 34). In contrast, all five ATCC 15697 fucosidases are likely found in the cytosol, further distinguishing B. longum subsp. infantis from B. bifidum by the need to efficiently transport oligosaccharides (29–31). Accordingly, several ABC transporters appear in the ATCC 15697 HMO cluster with solute-binding proteins known to target milk oligosaccharide substrates (7, 29). While the biology underlying specific fucosylated oligosaccharide transport has yet to be elucidated, the ATCC 15697 genome includes several transporters that may facilitate environmental scavenging when soluble fucose is encountered. Likely candidates include two deduced proteins assigned to the fucose permease COG0738 (Blon_2307 and Blon_0962) and two that are genetically linked to fucosidases (Blon_0247 and Blon_0426), with various degrees of homology to the few previously characterized fucose permeases.
In addition to determining the location of the genes of fucosylated oligosaccharide transport, how B. longum subsp. infantis metabolizes fucose remains enigmatic. An examination of the ATCC 15697 genome did not reveal the genes of the canonical fucose utilization pathway. An alternative explanation is that fucose is metabolized via a cryptic oxidative pathway. There is nascent evidence for the existence of an oxidative pathway, based on identification of previously characterized oxidative pathway genes (37). There are two gene clusters within the B. longum subsp. infantis ATCC 15697 genome that are putatively involved with oxidative catabolism of fucose. One is localized to the HMO cluster (Blon_2337-Blon_2340) and is immediately adjacent to the fucosidases encoded by Blon_2335 and Blon_2336. Of the six enzymes involved in the oxidative pathway, there is evidence for the existence of four of them in the ATCC 15697 genome (37). It is possible that some of these enzymes are multifunctional or have yet to be identified. Another possibility is that ATCC 15697 does not metabolize fucose but rather cleaves this moiety from oligosaccharides to afford access to a utilizable portion of the molecule.
Of interest, one ATCC 15967 α-fucosidase (Blon_0426) displayed considerably higher enzymatic efficiency toward CNP-fucose than its paralog (Blon_0248). This hints that these α-fucosidases are active on disparate substrates despite their sequence similarity. It is unknown what biological function, if any, is bolstered by Blon_0426's higher efficiency, detectable on CNP-fucose. However, based on MS analyses, Blon_0248 did exhibit greater activity on purified HMOs, indicating that this enzyme likely processes longer-chain fucosylated glycans. Curiously, it was inactive on smaller-chain oligosaccharides as tested by TLC. This contrasts with the HMO cluster enzymes (Blon_2335 and Blon_2336) that clearly are active on both these small-chain linkages and large-chain linkages as assayed by TLC and MS, respectively.
In addition, the crystal structure of Blon_2336 further demonstrates the structural conservation of the catalytic domain, in particular the active site of α-l-fucosidase, as well as the structural uniqueness of the GH29 family glycosyl hydrolases. That the active site component residues, including a catalytic nucleophile, are highly conserved can be hardly appreciated simply by sequence alignment due to generally low amino acid identities between these fucosidases. The slight positional variation of the catalytic acid/base and one of the most sequentially diversified regions after the catalytic residue, as described earlier, may be one structural determinant of their substrate specificities and catalytic efficiencies.
The Blon_2336 structure, together with other four α-l-fucosidase structures, also supports the implication that the catalytic acid/base residue of α-l-fucosidase of mammals, including humans, is an aspartate residue, based on an analysis of a large number of samples of α-l-fucosidase genes (9). The key residue substitution may lead to different substrate specificities and potentially different catalytic mechanisms. The physiologic or even pathological relationship between the α- l-fucosidases of humans and the α- l-fucosidases of enteric bacteria such as bifidobacteria remains an interesting area for further investigation.
Conclusions.
B. longum subsp. infantis ATCC 15697 utilizes several small-mass neutral oligosaccharides, some of which are fucosylated. Whereas previous MS glycoprofiles of neutral HMO consumption examined an end point in early stationary phase, this study revealed a temporal-dependent utilization profile. Specifically, among preferred HMOs, LNT is favored early, with LNH and F-LNH utilized fully though slightly later in the fermentation. DF-LNH is utilized at the same point as LNH and F-LNH, although it is consumed to a lesser degree. In order to utilize fucosylated HMOs, the ATCC 15697 chromosome encodes five fucosidase genes, four of which are GH29 α- l-fucosidases. Interestingly, the HMO cluster fucosidase genes provide the greatest evidence for involvement in HMO metabolism, as they are active on a wider assortment of HMO linkages and their expression is induced by this purified substrate.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the University of California Discovery Grant Program, the California Dairy Research Foundation, USDA NRI-CSREES award 2008-35200-18776, and National Institutes of Health NICHD awards R01HD059127, R01HD065122, and R01HD061923. D.A.S. was supported by a predoctoral training grant (NIH-NIGMS T32-GM08799). This work was also supported by National Institutes of Health grant GM074942 and by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, NIH, or DOE.
Footnotes
Published ahead of print 2 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Asakuma S, et al. 2011. Physiology of the consumption of human milk oligosaccharides by infant-gut associated bifidobacteria. J. Biol. Chem. 286:34583–34592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashida H, et al. 2009. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017 [DOI] [PubMed] [Google Scholar]
- 3. Cohen SX, et al. 2004. Towards complete validated models in the next generation of ARP/wARP. Acta Crystallogr. D Biol. Crystallogr. 60:2222–2229 [DOI] [PubMed] [Google Scholar]
- 4. Collaborative Computation Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 5. Emsley P, Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 6. Ficko-Blean E, Boraston AB. 2006. The interaction of a carbohydrate-binding module from a Clostridium perfringens N-acetyl-beta-hexosaminidase with its carbohydrate receptor. J. Biol. Chem. 281:37748–37757 [DOI] [PubMed] [Google Scholar]
- 7. Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gyorgy P, Jeanloz RW, von Nicolai H, Zilliken F. 1974. Undialyzable growth factors for Lactobacillus bifidus var. pennsylvanicus. Protective effect of sialic acid bound to glycoproteins and oligosaccharides against bacterial degradation. Eur. J. Biochem. 43:29–33 [DOI] [PubMed] [Google Scholar]
- 9. Intra J, Perotti ME, Pavesi G, Horner D. 2007. Comparative and phylogenetic analysis of alpha-L-fucosidase genes. Gene 392:34–46 [DOI] [PubMed] [Google Scholar]
- 10. Katayama T, Fujita K, Yamamoto K. 2005. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J. Biosci. Bioeng. 99:457–465 [DOI] [PubMed] [Google Scholar]
- 11. Katayama T, et al. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J. Bacteriol. 186:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleerebezem M, Vaughan EE. 2009. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 63:269–290 [DOI] [PubMed] [Google Scholar]
- 13. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 14. Lammerts van Bueren A, et al. 2010. Analysis of the reaction coordinate of alpha-L-fucosidases: a combined structural and quantum mechanical approach. J. Am. Chem. Soc. 132:1804–1806 [DOI] [PubMed] [Google Scholar]
- 15. Larson G, Falk P, Hoskins LC. 1988. Degradation of human intestinal glycosphingolipids by extracellular glycosidases from mucin-degrading bacteria of the human fecal flora. J. Biol. Chem. 263:10790–10798 [PubMed] [Google Scholar]
- 16. LoCascio RG, et al. 2009. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb. Biotechnol. 2:333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. LoCascio RG, et al. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 55:8914–8919 [DOI] [PubMed] [Google Scholar]
- 18. Marcobal A, et al. 2010. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 58:5334–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markowitz VM, et al. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344–D348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution. From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62:859–866 [DOI] [PubMed] [Google Scholar]
- 21. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 22. Nagae M, et al. 2007. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J. Biol. Chem. 282:18497–18509 [DOI] [PubMed] [Google Scholar]
- 23. Newstead SL, Watson JN, Bennet AJ, Taylor G. 2005. Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. Acta Crystallogr. D Biol. Crystallogr. 61:1483–1491 [DOI] [PubMed] [Google Scholar]
- 24. Ninonuevo M, et al. 2005. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis 26:3641–3649 [DOI] [PubMed] [Google Scholar]
- 25. Ninonuevo MR, et al. 2006. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54:7471–7480 [DOI] [PubMed] [Google Scholar]
- 26. Ninonuevo MR, et al. 2007. Methods for the quantitation of human milk oligosaccharides in bacterial fermentation by mass spectrometry. Anal. Biochem. 361:15–23 [DOI] [PubMed] [Google Scholar]
- 27. Parche S, et al. 2006. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188:1260–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schneider TR, Sheldrick GM. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58:1772–1779 [DOI] [PubMed] [Google Scholar]
- 29. Sela DA, et al. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sela DA, et al. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J. Biol. Chem. 286:11909–11918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sulzenbacher G, et al. 2004. Crystal structure of Thermotoga maritima alpha-L-fucosidase. Insights into the catalytic mechanism and the molecular basis for fucosidosis. J. Biol. Chem. 279:13119–13128 [DOI] [PubMed] [Google Scholar]
- 33. Ventura M, et al. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wada J, et al. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol. 74:3996–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 72:4497–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie Y, Liu J, Zhang J, Hedrick JL, Lebrilla CB. 2004. Method for the comparative glycomic analyses of O-linked, mucin-type oligosaccharides. Anal. Chem. 76:5186–5197 [DOI] [PubMed] [Google Scholar]
- 37. Yew WS, et al. 2006. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry 45:14582–14597 [DOI] [PubMed] [Google Scholar]
- 38. Zhang G, Mills DA, Block DE. 2009. Development of chemically defined media supporting high-cell-density growth of lactococci, enterococci, and streptococci. Appl. Environ. Microbiol. 75:1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.