Abstract
Enterococcus faecalis NKR-4-1 isolated from pla-ra produces a novel two-peptide lantibiotic, termed enterocin W, comprising Wα and Wβ. The structure of enterocin W exhibited similarity with that of plantaricin W. The two peptides acted synergistically, and their order of binding to the cell membrane was important for their inhibitory activity.
TEXT
Bacteriocins produced by some lactic acid bacteria (LAB) are antimicrobial peptides active against bacteria that are closely related to the producer strains and against food-borne Gram-positive spoilage bacteria. Nisin A, a substance generally recognized as safe (GRAS), is currently the only LAB bacteriocin approved for use as a food preservative. The proposed classification by Cotter et al. (3) divides bacteriocins into two distinct classes, I (lantibiotics) and II (nonlantibiotics). Class I bacteriocins, to which nisin A belongs, are lanthionine-containing bacteriocins, including two-peptide lantibiotics (1, 6, 9, 13). Two-peptide lantibiotics share the following characteristics with nonlantibiotic two-peptide bacteriocins: their complete antimicrobial activity requires the synergistic action of the two peptides in equimolar concentrations, the individual peptides have little or no activity, and each peptide is encoded by adjacent open reading frames in the same operon (5).
In the present report, we describe the identification and characterization of a novel two-peptide lantibiotic, enterocin W, produced by Enterococcus faecalis NKR-4-1, which was isolated from pla-ra, a traditional Thai fermented fish. The culture supernatant in MRS medium at 30°C for 16 h was tested for antimicrobial activity by using the spot-on-lawn method (11), and it exhibited a narrow spectrum (Table 1). Enterocin W was purified from the culture supernatant by a three-step procedure consisting of hydrophobic interactions (Amberlite XAD-16 resin; Sigma-Aldrich, St. Louis, MO), cation-exchange chromatography (SP-Sepharose Fast Flow column; GE Healthcare, Uppsala, Sweden), and reverse-phase high-performance liquid chromatography (HPLC; Resource RPC 3-ml column; GE Healthcare) sequentially according to previously described procedures (11). The inhibitory spectra of purified enterocin W are presented in Table 1. For MIC and fractional inhibitory concentration (FIC) (2) determination, the maximum concentrations of enterocin Wα, enterocin Wβ, and the mixture were 254.8, 311.6, and 6.8 μM (mixed in equimolar concentrations), respectively. The combination of the two purified peptides exhibited a broader spectrum and greater antimicrobial activity than each of the individual peptides against almost all the indicator strains. Interestingly, the culture supernatant exhibited no inhibitory activity against Bacillus subtilis JCM 1465T. However, the purified mixture exhibited remarkable inhibitory activity against this strain and against other Bacillus strains (Table 1). These results indicate that medium components and/or organic acids produced by E. faecalis NKR-4-1 may influence the activity of enterocin W against several strains.
Table 1.
Antimicrobial spectra of the single and synergistic activities of enterocins Wα and Wβa
| Indicator strain | Activity in culture supernatant (AU/ml) | MIC (μM)b |
FICc | ||
|---|---|---|---|---|---|
| Enterocin Wα | Enterocin Wβ | Equimolar mixture | |||
| Bacillus coagulans JCM 2257T | 400 | 7.95 | 9.7 | 0.22 | 0.05 |
| B. circulans JCM 2504T | 0 | 31.8 | 77.9 | 1.70 | 0.07 |
| B. subtilis JCM 1465T | 0 | NA | NA | 0.85 | ND |
| Kocuria rhizophila NBRC 12708 | 0 | NA | NA | NA | ND |
| Listeria innocua ATCC 33090T | 0 | 7.95 | 311.6 | 1.70 | 0.22 |
| Pediococcus pentosaceus JCM 5885 | 1,600 | 31.6 | 77.9 | 1.70 | 0.07 |
| Enterococcus faecalis JCM 5803T | 100 | 15.9 | 19.7 | 0.21 | 0.02 |
| E. faecalis NKR-4-1 (producer strain) | 0 | NA | NA | NA | ND |
| Lactococcus lactis subsp. lactis JCM 7638 | 3,200 | 33.8 | 4.8 | 0.42 | 0.15 |
| L. lactis subsp. lactis ATCC 19435T | 200 | 3.97 | 36.7 | 0.11 | 0.03 |
| Lactobacillus sakei subsp. sakei JCM 1157T | 400 | 7.95 | 77.0 | 0.22 | 0.02 |
ATCC, American Type Culture Collection, Rockville, MD; JCM, Japan Collection of Microorganisms, Saitama, Japan; NBRC, NITE Biological Resource Center, Chiba, Japan. The data were confirmed by three independent experiments.
The highest concentrations of enterocins Wα, Wβ, and the equimolar mixture were 254.8 μM, 311.6 μM, and 3.2 μM (of each peptide), respectively. NA means no activity against indicator strains, even with the highest concentrations of the indicated peptides (>3.2 μM).
The FIC was calculated as FIC index = MICWα in combination/MICWα alone + MICWβ in combination/MICWβ alone, where Wα and Wβ were the two respective purified peptides tested. Synergy was defined as an FIC value of <0.5, whereas FIC values between 0.5 and 1.0 were considered to be the result of additive antimicrobial effects, and FIC values of >1 indicated antagonistic activity. ND, not determined.
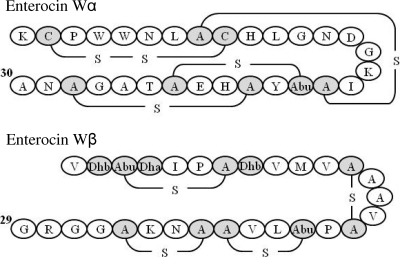
The molecular masses of purified enterocin Wα and enterocin Wβ determined by electrospray ionization–time of flight mass spectrometry were found to be 3,256.5 and 2,728.6 Da, respectively. The purified enterocin W peptides were subjected to N-terminal amino acid sequencing by automated Edman degradation after several chemical treatments. Enterocin Wα was treated by reduction and pyridylethylation (4, 8) and then sequenced. As a result, 21 amino acid residues of the peptide were obtained by 30 cycles of Edman degradation as follows: KCPWWNLXCHLGNDGKXXXYXHXXTAXXNA (X represents amino acid residues that could not be determined). Enterocin Wβ was treated by reduction under alkaline conditions to cleave the monosulfide bridges of (2S,6R)-lanthionine (Lan) and (2S,3S,6R)-3-methyllanthionine (MeLan) (10) and then sequenced. Consequently, 18 amino acid residues of the peptide were obtained by 29 cycles of Edman degradation as follows: VXXXIPXXVMVXAAVXPXLVXXNKXGGRG. In addition, treatment of enterocin Wβ with CNBr (7), which cleaves the C-terminal end of the methionine residue and converts it into a homoserine lactone (Hse), yielded a product with a molecular mass of 931.5 Da [putative amino acid sequence, V1-Dhb2-Abu3-Dha4-I5-P6-A7-Dhb8-V9-Hse10, where Abu is α-aminobutyric acid, Dha is 2,3-didehydroalanine, and Dhb is (Z)-2,3-didehydrobutyrine]. This result suggests that enterocin Wβ is divided into N-terminal and C-terminal regions by a methionine residue.
A part of the structural gene of enterocin Wα was obtained by short degenerated primers (pepA primers) with 11 to 14 bases (nested anchored rapid PCR [NAR-PCR]) on the basis of the obtained amino acid sequences (Table 2). Nested and anchored PCRs were performed using enterocin-specific primers (NKA primers) and vector-specific primers (M13 primers), according to a previously described protocol (11). The PCR products were purified and directly sequenced to confirm the sequences obtained. As a result, the DNA sequences of enterocins Wα and Wβ, including the respective putative N-terminal leader sequences, were obtained (Fig. 1). According to the DNA sequence obtained, the calculated molecular mass of enterocin Wα was 3,312.7 Da. The observed molecular mass was approximately 56 Da lower than the calculated mass, indicating that the cysteine, serine, and threonine residues in this peptide form a disulfide bridge (−2 Da) and 3 dehydrated residues (−18 × 3 = −54 Da) with or without monosulfide bridges. Likewise, the calculated molecular mass of enterocin Wβ was 2,855.4 Da. The molecular mass was approximately 126 to 127 Da lower than the calculated mass, indicating the occurrence of 7 dehydrations (−18 × 7 = −126 Da) of all the serine and threonine residues. Considering this difference and the peptide fragmentation pattern, enterocin Wβ was proven to have 3 dehydrated amino acids as well as 2 and 2 Lan and MeLan residues, respectively. The proposed primary structures of enterocins Wα and Wβ are shown in Fig. 2. The amino acid sequences of the prepeptides of enterocin W showed the highest identity with those of plantaricins Wα and Wβ (63.3 and 44.7%, respectively) (6).
Table 2.
Oligonucleotide primers used to obtain the genes encoding enterocins Wα and Wβ
| Primer name | Sequence (5′-3′) |
|---|---|
| pepA.F1 | AARTGYCCNTGGTG |
| pepA.F2 | CAYYTNGGNAAYGA |
| NKA.F1 | AAAGTGTCCGTG |
| NKA.R1 | CACGGACACTTT |
| NKA.F2 | TGGAATCTTTC |
| NKA.R2 | GAAAGATTCCA |
| NKA.F3 | TGTCATTTAGG |
| NKA.R3 | CCTAAATGACA |
| EnA.F1 | TGTTGTTAACTTGATTTTTGGG |
| EnA.R1 | TAATTATGCATTACAACCT |
| EnB.F1 | TTGTGGGGATAAATAATGACT |
| EnB.R1 | AATGGTAGTATAGACTGCCAT |
| 1stMup13-f | TTAACTATGCGGCATCAGA |
| 1stMup13-r | TAATGTGAGTTAGCTCACTC |
| Mup13-f | AAGGCGATTAAGTTGGGTA |
| Mup13-r | GTATGTTGTGTGGAATTGTG |
| s-M13-f | GTAAAACGACGGCCAGT |
| s-M13-r | TTCACACAGGAAACAGG |
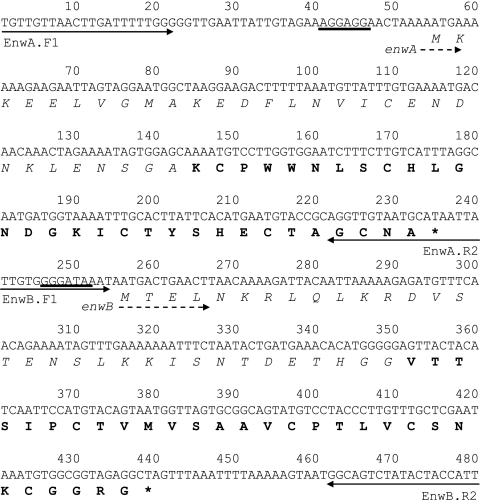
Fig 1.
Nucleotide sequence of the region containing the structural genes of enterocin W. Starting points of enterocin W genes (enwA and enwB) are indicated by dotted arrows. The leader peptides are indicated in italics. Putative ribosome binding sites and stop codons are indicated by underlining and asterisks, respectively. The sequences corresponding to the mature peptides are shown in bold.
Fig 2.
Proposed primary structures of enterocin W. Cysteine and unusual amino acids are indicated in gray.
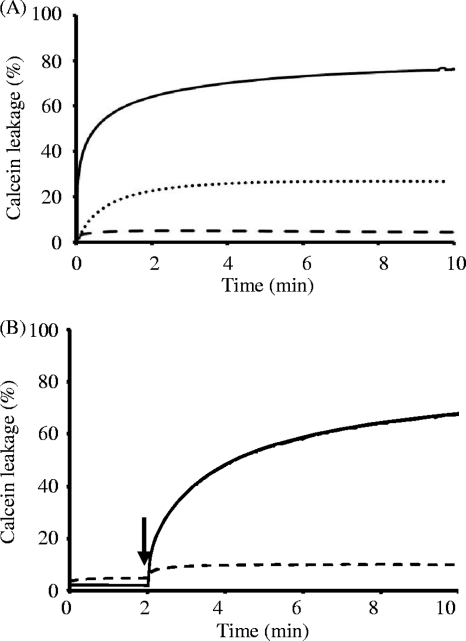
The modes of action of enterocin W peptides against egg yolk l-α-phosphatidylcholine–l-α-phosphatidyl-dl-glycerol (PC-PG; 1:1) liposomes, which mimic the general Gram-positive cell membrane, were examined by tryptophan fluorescence spectroscopy, light scattering, and the calcein leakage test according to previously reported methods (12). PC-PG liposomes were prepared according to previously described procedures (12) in which calcein fluorescent dye was entrapped for the leakage experiments. During tryptophan fluorescence spectroscopy in the presence of PC-PG liposomes, the emission maximum of enterocin Wα changed to a lower wavelength (blue shift) than that of the control (data not shown). Moreover, it was confirmed by the light scattering test that the addition of the peptides caused no morphological changes in the liposomes (data not shown). These results revealed that enterocin W was bound to the liposomes without their morphological changes. Calcein leakage from PC-PG liposomes induced by enterocin Wα or Wβ alone was scarcely detected (Fig. 3A), even at a high concentrations (>5 μM) (data not shown). Conversely, remarkable calcein leakage was detected at a low-concentration mixture of the two peptides (0.25 μM each). Moreover, remarkable calcein leakage was detected when the addition of enterocin Wα was followed by that of enterocin Wβ (0.25 μM each) (Fig. 3B). In contrast, calcein leakage was scarcely detected when the addition of enterocin Wα was preceded by that of enterocin Wβ (Fig. 3B). This result suggested that the order of peptide binding to the cell membrane appeared to be important for the inhibitory activity of enterocin W. In addition, nisin A (0.50 μM), which requires lipid II (docking molecules) for cell membrane binding, did not induce calcein release from the PC-PG liposomes (data not shown) (12). The results of calcein leakage suggested that enterocin W can act on the cell membrane even without docking molecules.
Fig 3.
Calcein leakage from PC-PG liposomes induced by enterocin W peptides. (A) Enterocins Wα and Wβ and their mixture were tested for the leakage of calcein entrapped in PC-PG (1:1) liposomes. The solid, dashed, and broken lines show the traces of leakage when the mixture (0.25 μM each), enterocin Wα (2 μM), or enterocin Wβ (2 μM) was added to 50 μM liposomes, respectively. (B) The orders of action of enterocin W peptides were examined. The solid and broken lines show the traces of leakage of calcein when the addition of enterocin Wα and Wβ, respectively, was followed by that of the other peptide (enterocin Wβ or Wα) (final concentrations were 0.25 μM each) after 2 min as indicated by the arrow. The lipid concentration of the PC-PG liposomes was 50 μM. As controls, 0% and 100% leakage were obtained by the addition of buffer and 0.1% Triton X-100, respectively. The data were confirmed by three independent experiments.
In conclusion, E. faecalis NKR-4-1 produces a novel two-peptide lantibiotic. FIC indices demonstrated synergistic effects against almost all the indicator strains when enterocins Wα and Wβ were combined. In addition, enterocin W peptides were demonstrated to have high heat stability (data not shown). These characteristics make these peptides appropriate for further applications. The short degenerated primers used in the new approach, NAR-PCR, theoretically produce larger numbers of amplified products, including specific and nonspecific fragments. Nested PCR using the amplified fragments as templates enabled us to obtain specific fragments. NAR-PCR enables structural genes to be determined more simply and rapidly, even when the amino acid and DNA sequences of the target proteins or peptides are short.
Nucleotide sequence accession number.
The DNA sequence described in the present study has been deposited in DDBJ under the accession number AB600897.
ACKNOWLEDGMENTS
This work was supported by the Core University Program between Yamaguchi University and Kasetsart University and a scientific cooperation program in the Development of Thermotolerant Microbial Resources and Their Applications of the Japan Society for the Promotion of Science (JSPS) and National Research Council of Thailand (NRCT). It was also supported in part by a Grant-in-Aid for Scientific Research from JSPS, the Research Grant for Young Investigators of Faculty of Agriculture, Kyushu University, and the Kato Memorial Bioscience Foundation.
Footnotes
Published ahead of print 2 December 2011
REFERENCES
- 1. Booth MC, et al. 1996. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 21:1175–1184 [DOI] [PubMed] [Google Scholar]
- 2. Cain CC, et al. 2003. Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 47:2113–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 4. Friedman M, Krull LH, Cavins JF. 1970. The chromatographic determination of cystine and cysteine residues in proteins as S-β-(4-pyridylethyl) cysteine. J. Biol. Chem. 245:3868–3871 [PubMed] [Google Scholar]
- 5. Garneau S, Martin NI, Vederas JC. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577–592 [DOI] [PubMed] [Google Scholar]
- 6. Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. 2001. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology 147:643–651 [DOI] [PubMed] [Google Scholar]
- 7. Kaiser R, Metzka L. 1999. Enhancement of cyanogen bromide cleavage yields for methionyl-serine and methionyl-threonine peptide bonds. Anal. Biochem. 266:1–8 [DOI] [PubMed] [Google Scholar]
- 8. Katsumi A, Tuley EA, Bodo I, Sadler JE. 2000. Localization of disulfide bonds in the cystine knot domain of human von Willebrand factor. J. Biol. Chem. 275:25585–25594 [DOI] [PubMed] [Google Scholar]
- 9. Martin NI, et al. 2004. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43:3049–3056 [DOI] [PubMed] [Google Scholar]
- 10. Meyer HE, et al. 1994. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal. Biochem. 223:185–190 [DOI] [PubMed] [Google Scholar]
- 11. Sawa N, et al. 2010. Identification and characterization of novel multiple bacteriocins produced by Leuconostoc pseudomesenteroides QU 15. J. Appl. Microbiol. 109:282–291 [DOI] [PubMed] [Google Scholar]
- 12. Yoneyama F, et al. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 53:3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yonezawa H, Kuramitsu HK. 2005. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]