Abstract
Investigations of Campylobacter jejuni and Campylobacter coli in samples of drinking water suspected of being at the origin of an outbreak very often lead to negative results. One of the reasons for this failure is the small volume of water typically used for detecting these pathogens (10 to 1,000 ml). The efficiencies of three microfilters and different elution procedures were determined using real-time quantitative PCR to propose a procedure allowing detection of Campylobacter in 20 liters of drinking water or low-turbidity water samples. The results showed that more than 80% of the bacteria inoculated in 1 liter of drinking water were retained on each microfilter. An elution with a solution containing 3% beef extract, 0.05 M glycine at pH 9, combined with direct extraction of the bacterial genomes retained on the cellulose ester microfilter, allowed recovery of 87.3% (±22% [standard deviation]) of Campylobacter per 1 liter of tap water. Recoveries obtained from 20-liter volumes of tap water spiked with a C. coli strain were 69.5% (±10.3%) and 78.5% (±15.1%) for 91 CFU and 36 CFU, respectively. Finally, tests performed on eight samples of 20 liters of groundwater collected from an alluvial well used for the production of drinking water revealed the presence of C. jejuni and C. coli genomes, whereas no bacteria were detected with the normative culture method in volumes ranging from 10 to 1,000 ml. In the absence of available epidemiological data and information on bacterial viability, these last results indicate only that the water resource is not protected from contamination by Campylobacter.
INTRODUCTION
Thermotolerant Campylobacter species are an important cause of gastroenteritis disease throughout the world (7, 13, 23). Campylobacter jejuni and Campylobacter coli are the main species implicated in human Campylobacter infections. These bacteria have been shown to be widespread in the natural aquatic environment (6, 11, 12, 25). Feces of contaminated poultry, cattle, or birds and discharge of sewage constitute sources of pollution for the natural aquatic environment (21, 29). Consumption of contaminated drinking water constitutes a major route of transmission of Campylobacter and has been implicated in numerous water outbreaks in various countries (14, 16, 17, 18). However, isolation of Campylobacter from samples of water suspected of being the source of an outbreak remains difficult and frequently produces negative results (2, 18). There are three reasons that could explain this low recovery.
First, the period between the onset of human infection and the sampling of water involved in the outbreak may be too long to recover the implicated bacteria (18, 30). This period can vary widely depending on (i) the incubation time of the infection, (ii) notification of several cases to the authorities in charge of organizing the investigation, and (iii) the lapse of time until suspected water are sampled and analyzed by specialized laboratories. The longer the interval between disease onset and sampling, the lower the probability of detecting the microorganism, especially if the contamination is an isolated event.
Second, the current methodology used to detect Campylobacter may be ineffective and lead to false-negative results for different reasons. Laboratories generally use the normative procedure ISO 17995 (20) for detecting this pathogen in water in the event of a waterborne outbreak. This method requires filtering water through a cellulose ester microfilter. The filter is then directly placed in a selective broth for enrichment, followed by subculture on selective agar plates. The enrichment procedure increases the recovery of damaged cells present in water (19) and could also increase the growth of any background microflora present, particularly in fecally contaminated water (1, 15), that could prevent the detection of Campylobacter. Moreover, the presence of viable but noncultivable (VBNC) forms, under stressful environmental conditions (2, 8, 10), may also produce a negative result. Moreover, cells initially present in water can be irreversibly damaged if sampling or storage is not appropriate, and these can also not be detected. Molecular methods, such as PCR, are an alternative to traditional culture methods. PCR could be sensitive, specific, and provide results in a few hours (5 h compared to 4 to 6 days when using the ISO 17995 method). Furthermore, PCR can detect Campylobacter genomes from viable, VBNC, and dead cells. The detection of DNA from these three forms could constitute an advantage during the investigation of an outbreak. Characterization and typing the sequences of the bacterial genomes detected in water samples and comparison with those detected in feces of infected humans can help confirm that this pathogen is involved in the waterborne outbreak. Several protocols for PCR assays to detect Campylobacter in drinking water, well water, and surface water have been described (6, 25, 26).
Third, the volume of water used for Campylobacter detection may not be sufficient. Typically, volumes of 10 ml to 1 liter of water are tested (20). Hänninen et al. (18) reported that these volumes were too small for detecting Campylobacter present in drinking water samples collected during a waterborne outbreak. In their study, 4- to 10-liter samples of tap water or groundwater suspected to be at the origin of waterborne outbreaks were used to detect C. jejuni and C. coli using a culture method.
Taking into account all these observations, the aim of the study was to develop a sensitive and rapid method for detecting C. jejuni and C. coli in 1 to 20 liters of drinking water or in low-turbidity waters. We first selected methods to concentrate bacteria present in the sample by using microfilters, typical of the ones that environmental water analysis laboratories may use, and then compared recovery rates using real-time quantitative PCR (qPCR) and culture methods. The selected method was then assessed for natural water collected from an alluvial well used for the production of drinking water.
MATERIALS AND METHODS
Campylobacter strains and preparation of inocula.
C. coli strain ATCC 33559 and C. jejuni strain NCTC 11168 purchased from the National Reference Center for Campylobacter and Helicobacter (Bordeaux, France) were used as controls in the study. Each strain was cultured under a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) in Preston broth (Preston base with 5% defibrinated horse blood and antibiotic supplement [Oxoid, Dardilly, France]). After 44 ± 4 h at 37 ± 1°C, enrichments of bacteria were streaked onto mCCDA plates (modified charcoal cefoperazone desoxycholate agar; Oxoid). Plates were incubated at 41.5 ± 1°C for 44 ± 4 h under a microaerobic atmosphere. Bacterial cells obtained on agar were suspended in sodium chloride-peptone water composed of 0.85% sodium chloride and 0.1% peptone in purified water (AES Chemunex, Bruz, France) and enumerated on mCCDA plates from 10-fold serial dilutions to obtain a stock solution of 106 to 107 CFU/ml. This solution was used to spike the different waters tested in this work.
Types of water samples. (i) Spiked water.
Different types of drinking water were included in the test (Table 1). Tap water (TW) was collected in the laboratory by using sterile polyethylene bottles containing 20 mg/liter of sodium thiosulfate (Sigma, Saint-Quentin Fallavier, France). Bottled spring water (SW) and two types of bottled natural mineral water, containing low and high salt concentrations (MW1 and MW2), were also tested. Drinking water mixed with surface water was also tested using the method to simulate water contamination. Surface water used for this simulation was collected from the Moselle River. Tap water was mixed with about 10% (vol/vol) surface water (TWm1) or 50% (vol/vol) surface water (TWm2). All these waters samples were spiked with the appropriate quantity of bacteria from the stock solution.
Table 1.
pH, conductivity, and turbidity results for the tested waters
| Water sample | pH | Conductivity (mS/cm) | Turbidity (NFU) |
|---|---|---|---|
| Drinking waters | |||
| TW | 7.7 | 0.38 | 0.03 |
| SW | 7.2 | 0.53 | 0.03 |
| MW1 | 7.2 | 0.57 | 0.02 |
| MW2 | 7.2 | 2.73 | 0.21 |
| Tap waters contaminated with surface water | |||
| TWm1 | 7.6 | 0.32 | 0.44 |
| TWm2 | 7.5 | 0.4 | 1.91 |
(ii) Groundwater.
Eight samples of 20 liters of groundwater (GW) were collected from an alluvial aquifer used for the production of drinking water in a water supply near the greater Nancy area. All samples were collected in summer 2010, and the pH ranged from 7.02 to 7.57, conductivity ranged from 0.49 mS/cm to 0.56 mS/cm, and turbidity ranged from 0.05 NTU to 0.24 NTU (nephelometric turbidity units).
Microfilters.
Three 47-mm-diameter flat disc microfilters were selected for the study. Two 0.45-μm-pore-size filters were tested: (i) a cellulose ester filter (reference EZHAWG474; Millipore, Molsheim, France) and (ii) an electropositive, charge-modified, diatomaceous earth/cellulose filter (catalog number NM04701; Zetapor; Cuno, Cergy-Pontoise, France). A third electropositive flat filter not rated by pore size, containing a mixture of fiberglass and cellulose (catalog number NM04711; Zetaplus Virosorb 1-MDS; Cuno, Cergy-Pontoise, France), was also included in this work.
Concentration and elution of bacteria.
After filtration of 1 to 20 liters of the spiked water or groundwater samples through the microfilters, they were introduced into a 50-ml tube containing 3 ml of elution buffer, and the whole mixture was shaken for 10 min for eluting the bacteria retained or adsorbed on the filters. Three types of buffer were tested: (i) a solution of 1% beef extract with glycine (0.05 M) at pH 7, (ii) a solution of 3% beef extract with glycine (0.05 M) at pH 7, and (iii) a solution of 3% beef extract with glycine (0.05 M) at pH 9.
Campylobacter detection methods. (i) Detection of Campylobacter in groundwater by using the ISO method.
Concentration and isolation of Campylobacter organisms present in GW were carried out according to the ISO 17995 method. For each sample, volumes of 10, 100 and 1,000 ml were assessed in duplicate. After filtration of the test volume, one microfilter (a cellulose ester filter with 0.45-μm pore size; Millipore) was immersed in Preston broth, and the other microfilter was immersed in Bolton broth (Bolton enrichment broth with 5% defibrinated horse blood, Campylobacter growth supplement, and selective supplement from Oxoid). Broths were incubated at 37 ± 1°C for 44 ± 4 h under a microaerobic atmosphere and bacteria, present in broths, were streaked on mCCDA plates after the enrichment step. After incubation of the plates at 41.5 ± 1°C for 44 ± 4 h, typical Campylobacter colonies were streaked onto two Columbia blood agar plates (Oxoid) for confirmation. We checked for the absence of growth after incubation under an aerobic atmosphere at 41.5 ± 1°C for 21 ± 3 h, confirmed the presence of curved forms and cell mobility by phase-contrast microscopy (Olympus, BX51TF), and used a latex agglutination test to identify enteropathogenic Campylobacter spp. (Dryspot Campylobacter test kit; Oxoid).
(ii) Detection of Campylobacter in spiked water using the culture method.
Ten-fold serial dilutions of 100 μl of spiked water collected before and after filtration or in the elution buffer used for the elution step were carried out using sodium chloride-peptone water (AES Chemunex). A 100-μl aliquot of each dilution was spread over the surface of mCCDA agar supplemented with CCDA supplement (Oxoid) by using a sterile spreader. Plates were incubated at 41.5 ± 1°C for 44 ± 4 h under a microaerobic atmosphere.
(iii) Detection of Campylobacter using real-time qPCR.
Real-time qPCR was used to determine the recoveries of each type of microfilter or each condition tested on spiked water and also to detect Campylobacter in the natural aquatic environment (GW). Bacterial genomes were extracted prior to performing qPCR.
(a) DNA extraction.
After concentrating Campylobacter from waters samples using the microfilters, DNA extraction was performed on the entire volume (3 ml) of the tested elution buffer. Extraction was carried out by applying one thaw-freeze cycle (100°C for 10 min and −80°C for 10 min) in the presence of 2.5% (wt/vol) Chelex (Bio-Rad, Marnes la Coquette, France). After centrifugation for 10 min at 4,000 rpm, the supernatant was collected and mixed with 4 ml of a guanidine thiocyanate buffer provided by Qiagen and left for 10 min at room temperature. Then, 4 ml of ethanol was added and the mix was filtered through a silica column (Qiagen, Courtaboeuf, France). DNA bound on the column was eluted in 50 μl of sterile water by centrifugation for 1 min at 8,000 rpm. DNA was stored at −20°C until use.
(b) Real-time qPCR.
Campylobacter detection in DNA extracts was assessed using a fluorescence resonance energy transfer (FRET)-PCR assay targeting the gyrA gene as previously described by Ménard et al. (24) with minor modifications in the sequence of the probes and of the reverse primer used for C. jejuni (Table 2). The anchor and anchorJ probes were fluorescein labeled at the 5′ end, and the sensor and sensorJ probes were phosphorylated at the 3′ end. Each PCR mixture contained 1× LightCycler FastStart DNA Master HybProbe (Roche Diagnostics, Meylan, France), 3 mM MgCl2, 0.5 μM each primer, 0.2 μM each probe, and 5 μl of template DNA in a total volume of 25 μl. Amplification was done on a Rotor-Gene 6000 thermocycler using the following program: 10 min at 95°C, followed by 45 cycles of 6 s at 95°C, 30 s at 54°C, and 25 s at 72°C. Rotor-Gene software was used to analyze the parameters of each run and define the threshold cycle (CT) values. The CT value corresponds to the PCR cycle number at which the generated fluorescence crosses the threshold. It is inversely correlated to the logarithm of the quantity of genome copies.
Table 2.
Primers and probes used for detection of C. jejuni and C. coli by qPCR
| Species detected | Primer or probe | Sequence (5′→3′)a | Localization (nucleotide position)b |
|---|---|---|---|
| C. coli | Primers | ||
| F3-gyrA-CJ-CC | GTACTTTTGGTGTGATTATG | 986–1005 | |
| R4-gyrA-CJ-CC | TTATCTCTTTTAATTCATCGCG | 1429–1408 | |
| FRET probes | |||
| Sensor | Red 640-GTTCGTCTGATAATCACTGTTTTTCTATG-p | 1100–1072 | |
| Anchor | GCTCTTGCTCTTGCTTTTTGAAGTTCAA- F | 1133–1106 | |
| C. jejuni | Primers | ||
| F3-gyrA-CJ-CC | GTACTTTTGGTGTGATTATG | 986–1005 | |
| R4J-gyrA | TWATYTCTTTTAATTCATCGCG | 1429–1408 | |
| FRET probes | |||
| SensorJ | LC705 - GTTCTTCTAATAATAACTGTTTTTCTATG-p | 1100–1072 | |
| AnchorJ | GCTCTTGCTCTTGCCTTTTGAAGTTCAA- F | 1133–1106 |
p, phosphate; F, fluorescein.
The nucleotide positions of primers and probes were compared to the gyrA gene numbering system used for the Campylobacter jejuni NCTC 11168 reference strain.
Evaluation of efficiencies of methods using qPCR to detect bacterial genomes.
Evaluation of the efficiency of the detection method was performed on 3 ml of PBS (phosphate-buffered saline, pH 7.4). PBS was spiked with serial 10-fold dilutions of C. coli to obtain final concentrations ranging from 2.3 × 104 to 2.3 × 10−1 CFU/ml in PBS. A test on a theoretical concentration of 2.3 × 10−1 ml was conducted to confirm the limit of detection of the method used. The same was done for C. jejuni, to obtain final concentrations ranging from 8.3 × 103 to 8.3 × 10−1 CFU/ml in PBS.
To explore the impact of the beef extract present in the elution solution, 3 ml of elution buffer containing 3% beef extract with 0.05 M glycine was used. Buffer was spiked with serial 10-fold dilutions of C. coli to obtain final concentrations ranging from 3.9 × 105 to 3.9 × 100 CFU/ml in the buffer. The same was done for C. jejuni to obtain final concentrations ranging from 8.3 × 103 to 8.3 × 10−1 CFU/ml.
Each test was repeated three times. The slope (S) of the 10-fold dilution curve was used to assess the efficiency (E) of the procedure according to equation 1:
| (1) |
The correlation coefficient squared (r2), the sensitivity, and the limit of detection of the procedure were also determined.
Evaluation of recovery rates.
Different types of recovery were explored in the study.
(i) Retention recovery.
Retention recovery (RR) was defined as the microfilter's efficiency in removing, by size exclusion or adsorption, bacteria from the spiked test water. The RR was determined for the three microfilters tested with 1 liter of tap water contaminated with a known quantity of Campylobacter. Three-milliliter water samples were collected before and after filtration and were analyzed using a real-time qPCR assay after DNA extraction. From the CT values obtained, recoveries (as percentages) were calculated using equation 2:
| (2) |
where CT(i) and CT(f) are the CT values that correspond to the quantity of bacteria after DNA extraction from the water sample before [CT(i)] and after [CT(f)] filtration and S is the slope of the standard calibration curve. Retention recovery was also determined using the plate count method on mCCDA plates (RRC) and equation 3:
| (3) |
where Ci is the concentration of bacteria present in the spiked water before filtration and Cf is the concentration of bacteria present in the filtered water.
(ii) Global recovery.
Global recovery (GR) was defined as the efficiency of the concentration procedure, including filtration of water through the tested microfilter and also the elution step, to recover the bacteria in spiked tap water. GR values were determined for the three microfilters tested on 1 liter of tap water spiked with a known quantity of Campylobacter cells. After concentrating the bacteria, each microfilter was introduced into a 50-ml tube containing 3 ml of elution solution composed of glycine (0.05 M) and 1% beef extract at pH 7 and vortexed for 10 min. Before DNA extraction, the microfilter was removed. For the cellulose ester microfilter, two other buffers containing glycine (0.05 M) and 3% beef extract at pH 7 and pH 9 were also tested. The quantity of genomes obtained in the 3 ml of buffer solution after the removal of the microfilter was compared to the quantity of genomes of the bacteria present in the solution used for water contamination from a final volume of 3 ml in the same elution buffer. This was done to avoid bias in the calculation of the GR due to the chemical composition of the buffer. From the CT values, GRs (as percentages) were calculated using equation 4:
| (4) |
where CT(0) is the value corresponding to the quantity of genomes of the bacteria in solution used to contaminate the water sample, CT(x) is the value corresponding to the quantity of genome obtained in the 3 ml of buffer after the concentration procedure, and S is the slope of the standard qPCR curve for calibration.
(iii) Global direct recovery.
To determine the global direct recovery (GdR) rate, DNA was extracted from 3 ml of elution buffer in the presence of the tested microfilters. GdR was measured using the same principle and equation as for the GR (equation 4).
Statistical analysis.
For all data, values are expressed as means (± standard deviations). Analysis of variance (ANOVA) was used to compare the differences among recovery rates (GR and GdR) obtained for tap water spiked with Campylobacter cells under various conditions. Post hoc analyses using Bonferroni's test were used for pairwise multiple comparisons. All the statistical analyses were performed using Statistica (StatSoft Inc.).
RESULTS
Efficiency of the genome detection method using qPCR.
After filtration of 1 to 20 liters of drinking water using a 47-mm flat microfilter, bacteria were successfully recovered in a volume of 3 ml of elution buffer. To achieve high sensitivity, we first evaluated and validated the method described above to extract DNA on the entire volume of elution buffer. This evaluation was done with PBS and with a buffer containing beef extract.
The standard curves obtained from qPCR on the serial 10-fold dilutions of C. coli in 3 ml of PBS resulted in a slope of −3.791 (±0.01) with a correlation coefficient (r2) of 0.998 (±0.002) (Table 3). According to equation 1, the efficiency of the procedure was estimated at 83.7%. The limit of detection was determined to be 2.3 × 100 CFU/ml. For C. jejuni, an efficiency of 80.7% was calculated from a slope of −3.890 (±0.029). The r2 value was 0.990 (±0.011), and the limit of detection of the procedure for this strain was 8.3 × 100 CFU/ml.
Table 3.
Slopes, correlation coefficients, and limits of detection of the qPCR assays used to detect C. coli and C. jejuni genomes present in 3 ml of buffer
| Species | Medium | Mean slope (± SD) | Mean r2 (± SD) | Limit of detection (CFU/ml) |
|---|---|---|---|---|
| C. coli | PBS | −3.791 (± 0.01) | 0.998 (±0.002) | 2.3 × 100 |
| Elution buffer | −3.939 (±0.143) | 0.996 (±0.004) | 3.9 × 101 | |
| C. jejuni | PBS | −3.890 (±0.029) | 0.990 (±0.011) | 8.3 × 100 |
| Elution buffer | −3.71 (±0.608) | 0.995 (±0.000) | 8.3 × 100 |
With 3 ml of elution buffer containing 3% beef extract, the slope for C. coli was −3.939 (±0.143) with an r2 coefficient of 0.996 (±0.004), which corresponds to an efficiency of 79.5%. The limit of detection was 3.9 × 101 CFU/ml. For C. jejuni the slope of the curve was −3.71 (±0.608), with an r2 of 0.995 (±0.000), which corresponds to an efficiency of 86.2%. The limit of detection of the procedure was 8.3 × 100 CFU/ml.
Recovery rates for the three tested microfilters.
RRPCR and RRC were evaluated with C. coli for each type of filter in 1 liter of tap water spiked with 1.3 × 105 CFU or 3 × 107 CFU, respectively. RRPCR values were 90.3% (±4.1%), 96% (±3.5%), and 83.8% (±7.9%) for cellulose ester, Zetapor, and Zetaplus Virosorb 1-MDS microfilters, respectively. RRc values were 90.2% (±5.7%), 89.6% (±4.4%), and 89.7% (±4%) for cellulose ester, Zetapor, and Zetaplus Virosorb 1-MDS microfilters, respectively.
GRs, calculated using equation 4, were assessed in 1 liter of tap water spiked with 340 CFU of C. coli after elution using buffer containing 0.05 M glycine and 1% beef extract at pH 7. GR values of 12.2% (±4.9%) and 19.2% (±3.5%) were obtained for the cellulose ester and Zetapor microfilters, respectively. The Virosorb 1-MDS filter showed the lowest recoveries (<1%). For the cellulose ester filter, the percentages of bacteria recovered after elution, estimated using real-time qPCR, were similar to those estimated using the culture method (17.3% ± 4.9%). Taking into account these results, we chose to use the cellulose ester microfilter in subsequent experiments.
Improved recoveries with cellulose ester microfilters.
Samples of tap water (1 liter) spiked with 66 to 340 CFU of C. coli were filtered through cellulose ester microfilters, and bacteria were eluted with different elution buffers. GR and GdR rates were determined in triplicate for each experimental condition using the real-time qPCR assay after extracting the bacterial genomes. The results are presented in Table 4. We found that for GR, increasing the concentration of beef extract to 3% and the pH to 9 significantly enhanced the GR to 85.6% ± 3.6% (versus 12.2% ± 4.9%) (P < 0.05). As with the GR, the GdR was also enhanced when a buffer at pH 7 containing 1% or 3% beef extract was used: 55.4% (±6.2%) and 69.3% (±12.9%), respectively (P < 0.05). However, similar recoveries were obtained when a buffer containing 3% beef extract at pH 9 was employed, with or without direct extraction (85.6% ± 3.6% versus 87.3% ± 22%). Under these latter conditions, tests performed on 1 liter of tap water spiked with 196 CFU of C. jejuni showed similar results for GR (71.6% ± 28.5%) and GdR (70.3% ± 19.8%). Given these results and with the aim of minimizing handling of the microfilters, we decided to use the direct extraction procedure with the buffer containing 3% beef extract at pH 9.
Table 4.
Global recovery and global direct recovery rates for 1-liter samples of tap water spiked with Campylobacter coli based on use of three different elution buffers
| Microfilter tested | Elution buffer | Mean GR (%) (± SD) | Mean GdR (%) (± SD) |
|---|---|---|---|
| Cellulose ester | 1% beef extract, pH 7 | 12.2 (±4.9) | 55.4 (±6.2) |
| 3% beef extract, pH 7 | 27.4 (±8.7) | 69.3 (±12.9) | |
| 3% beef extract, pH 9 | 85.6 (±3.6) | 87.3 (±22) |
Impact on GdR of the physico-chemical composition of the water.
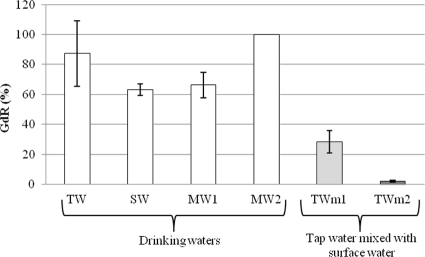
The impacts of the physico-chemical composition of the different waters tested on the efficiency of the method were assessed with 1-liter volumes spiked with 88 to 1,720 CFU of C. coli. GdR values were determined with the elution buffer containing 3% beef extract and 0.05 M glycine at pH 9. Each test was done in triplicate. Results obtained are presented in Fig. 1. For the various drinking waters, GdR values varied between 63.1% ± 3.9% and 99.99% ± 0.00% for mineral water with a high salt concentration (MW2). Simulated contamination of tap water with surface water caused a decrease in the efficiency of the method to 2.02% (±0.8%) for tap water mixed with 50% surface water (TWm2).
Fig 1.
GdR rates observed in different drinking waters and in tap water samples mixed with surface water spiked with C. coli, based on use of elution buffer containing 3% beef extract, 0.05 M glycine, at pH 9. Error bars indicate the standard deviations of the means.
Global direct recovery for the method with 1 and 20 liters of tap water.
The GdR for the method using the cellulose ester microfilter was assessed on 1 and 20 liters of tap water spiked with different quantities of C. coli from serial 10-fold dilutions of the solution stock (n = 2). The mean GdR values from 1 liter of tap water spiked with 4.3 × 104, 4.3 × 103, 4.3 × 102, and 4.3 × 101 CFU were 85.3% (±20.8%), 79.7% (±10.5%), 66.5% (±30.7%), and 83.5% (±23.4%), respectively. It was possible, for only one assay, to measure a recovery of 4.6% when a quantity of 4.3 × 100 CFU was tested. In 20 liters of tap water spiked with 9.1 × 103, 9.1 × 102, and 9.1 × 101 CFU, GdR values were, respectively, 15.1% (±0.8%), 27.5% (±0.1%), and 69.5% (±10.3%). The lowest quantity detectable in this trial was 9.1 × 101 CFU. This quantity was detected in the two trials conducted. To confirm the recovery for the lowest quantity of bacteria spiked in 20 liters, a supplementary trial was organized in triplicate with 3.6 × 101 CFU. The mean GdR was 78.5% (±15.1%).
Sensitivity of the method.
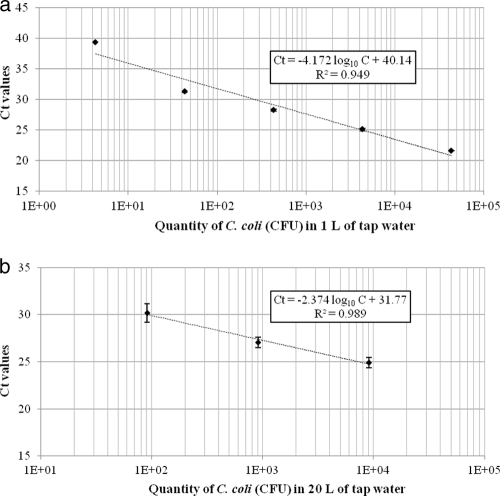
Sensitivity of the method was assessed on both 1 liter and 20 liters of tap water spiked with C. coli. Samples of water were spiked with different quantities of C. coli and filtered through an ester cellulose microfilter. Bacteria were recovered with the buffer containing 3% beef extract, 0.05 M glycine at pH 9, and the detection was performed by real-time qPCR. For 1 liter of tap water (Fig. 2a), the slope obtained from serial 10-fold dilutions ranging from 4.3 × 104 to 4.3 × 100 CFU was −4.172, and the r2 value was 0.949. According to equation 1, the efficiency was estimated to be 74%. The limit of detection was considered to be 43 CFU (n = 2). For 20 liters, a slope of −2.374 and a correlation coefficient of 0.989 were obtained with water spiked with 9.1 × 103 to 9.1 × 101 CFU (Fig. 2b). Given these data and those for supplementary on water spiked with 3.6 × 101 CFU (see above), the detection limit was considered to be between 3.6 × 101 CFU and 9.1 × 101 CFU.
Fig 2.
(a) Standard curve obtained by filtration of 1 liter of tap water spiked with different quantities of C. coli through an ester cellulose microfilter. Elution of bacteria was performed with a buffer containing 3% beef extract, 0.05 M glycine, at pH 9 (n = 2). (b) Standard curve obtained by filtration of 20 liters of tap water spiked with different quantities of C. coli through an ester cellulose microfilter. Elution of bacteria was performed with a buffer containing 3% beef extract, 0.05 M glycine, at pH 9 (n = 2).
Validation of the method on natural groundwater samples.
The method combining cellulose ester microfilter and direct DNA extraction of the bacteria retained or adsorbed on the filter was tested with 20 liters of groundwater from an alluvial aquifer that is used for the production of drinking water. Of the eight samples collected in summer 2010, thermotolerant Campylobacter spp. were not detected using the standard method (ISO 17995 [2005]). Genomes of C. coli and C. jejuni were detected by real-time qPCR in four of the samples. C. coli were detected in three samples, C. jejuni was detected in two samples, and both species were detected in one sample.
DISCUSSION
The aim of this study was to develop a methodology to rapidly detect C. jejuni and C. coli in 20-liter samples of water by combining a simple concentration technique and real-time qPCR. Using a cellulose ester microfilter with 0.45-μm pore size, we showed that Campylobacter genomes could be detected in samples of 20 liters of groundwater from an alluvial aquifer. In a volume of this size, recoveries ranging from 69.5% (±10.3%) to 78.5 (±15.1%) were obtained for a C. coli strain when tap water was spiked with bacteria concentrations ranging from 9.1 × 101 CFU to 3.6 × 101 CFU, respectively. To our knowledge, this is the first time that use of this type of microfilter in combination with a molecular method, such as qPCR, has been reported. In fact, other authors (26) or the standard technical procedure (5) have even discouraged the use of this combination. Oyofo and Rollins (26) reported binding of DNA to the ester cellulose microfilter, which would impact the detection.
Typically, volumes of 10 to 1,000 ml of water are employed to detect Campylobacter in water (6, 20, 22). While these volumes are sufficient for detecting these bacteria in contaminated natural aquatic environments, such as surface waters, they are not satisfactory for analyzing drinking water involved in a waterborne outbreak, especially if the contamination is an isolated event. Various authors have already recommended the use of greater volumes for analysis. Hänninen et al. (18) described the detection of Campylobacter strains in samples of tap or groundwater suspected to be at the origin of waterborne outbreaks by analyzing 4- to 10-liter samples using a cellulose ester filter and a culture method. Analysis of water volumes greater than 1 liter is typically carried out to detect other pathogens that have different morphological characteristics, such as viruses and parasites. For example, standard procedures (3, 4) for detection of enteroviruses or Cryptosporidium spp. oocysts recommend testing between 10 and 1,000 liters of water, depending on water quality. For bacteria, various authors have evaluated the efficiencies of flat filters or cartridges for filtering large volumes of water. Polaczyk et al. (28) showed recoveries for Salmonella enterica of 12% to 29% for 104 CFU seeded in 20 liters of tap water, using an electropositive microfilter and various elution buffers. With electronegative filters, such as glass fiber filters, Block and Rolland (9) and Payment et al. (27) reported recoveries higher than 50% for Salmonella enterica serovar Typhimurium and Legionella pneumophila, respectively, seeded in volumes of tap water of ≥15 liters and treated with acid (pH 3.5) to enhance electrostatic attraction.
In this study, we demonstrated that the cellulose ester microfilter retained more than 90% of the C. coli inoculated in 1 liter of tap water when we used either a culture method or the qPCR method. The absence of complete retention can be attributed to the size and morphology of Campylobacter, which can be lower than the microfilter pore size. The use of membranes with a 0.22-μm pore size may improve retention of bacteria. However, with this type of membrane it is not possible to filter more than 2 liters of tap water (data not shown). Free bacterial genomes in the solution used for spiking the test water, which cannot be retained on the filter and consequently would not be detected after filtration, would not lead to complete retention. However, the similar values of RRs measured with the culture and qPCR methods (RRPCR and RRC) seem to exclude this possibility.
The use of an electropositive microfilter (Zetapor) with the same porosity as the cellulose ester microfilter did not improve retention efficiency. However, (i) the high recovery rates obtained with the electropositive membrane Zetaplus Virosorb 1-MDS, with a porosity of >1 μm (RRPCR and RRC, >80%) and, inversely, (ii) the very low GdR value obtained with this filter (<1%), even with an elution buffer containing 1% beef extract at pH 7, confirmed that there are electrostatic interactions between the surface of the bacteria and the microfilter. This type of interaction has been reported for other bacteria, such as S. enterica by Polaczyk et al. (28) with the Zetaplus Virosorb 1-MDS electropositive filter when used to detect these bacteria in tap water.
In this study, we showed that the efficiency of the procedure to concentrate bacteria depends on the elution step. The GR values obtained with the cellulose ester microfilter were enhanced with the use of an alkaline elution buffer (pH 9) containing 3% beef extract. These results can be explained by the change in the charge of the membrane, which becomes negative, thereby promoting repulsion with the negative charge of the bacteria. Other interactions, such as hydrophobic interactions, may be involved in the adsorption process. Proteins present in the beef extract buffer used to elute the bacteria may compete with bacterial wall surface proteins. Hydrophobic interactions between bacteria and membrane or cartridge filters have been reported by other researchers (28). The elution of Campylobacter retained on a filter is not realized when these bacteria are screened in drinking water using the conventional culture method (20). Filtration membranes are directly immersed in culture broth, which limits inactivation due to compounds present in the elution buffer. The use of a buffer at pH 9 has an effect on the viability of bacteria. At this pH, we showed that Campylobacter could no longer be detected using culture methods (data not shown), while DNA of Campylobacter was still detected by qPCR methods, which cannot differentiate between genomes from viable, VBNC, or dead bacteria.
To minimize the impact of the elution step on the overall efficiency of the process, we chose to directly extract DNA from the Campylobacter organisms retained on the filter before carrying out the qPCR. This procedure has already been used for these bacteria by Oyofo and Rollins (26). A protocol by which the membrane is completely immersed in 3 ml of buffer followed by a specific thaw-freeze cycle (100°C/−80°C) and treatment with guanidine thiocyanate was tested for DNA extraction efficiency. However, this protocol recovered DNA in a final volume of 7 ml. With the aim of obtaining DNA in a total volume of less than 100 μl, we chose to concentrate and purify DNA using a silica column. Given the efficiencies of the procedure obtained with PBS buffer and various quantities of C. coli and C. jejuni (83.7% and 80.7%, respectively), we are able to validate the extraction step. We also verified that the use of a solution containing proteins (3% beef extract) did not alter the efficiency of the process (79.5% and 86.2%). This was important to verify, because we decided to use this concentration of beef extract to perform direct extractions. A buffer containing 3% beef extract at pH 9 was chosen in order to minimize adsorption of naked DNA onto the cellulose ester filter by electrostatic interactions. Adsorption of naked DNA on cellulose ester filters has been reported by Oyofo and Rollins (26).
Efficiencies of (i) the method for concentrating bacteria when using a cellulose ester filter, (ii) DNA extraction, and (iii) the qPCR assay revealed a significant robustness when different types of drinking water (with different physical and chemical characteristics) were evaluated. GdR values of 99.9% were obtained on bottled mineral water containing high levels of salt. Testing this type of water showed that the salts present in the water did not alter any of the three steps of the method. High recovery with this particular water confirmed the presence of electrostatic interactions in the process of retention. In other studies, salt has even been added to water samples to enhance adsorption of bacteria on filters (9, 27). The test performed on tap water spiked with different quantities of bacteria showed that our procedure was sensitive enough to detect 4.3 × 101 bacteria for all replicates, with a GdR of 83.5% (±23.4%). Increasing the volume to 20 liters did not greatly affect the GdR values for 3.6 × 101 to 9.1 × 101 CFU spiked in 20 liters (GdR of 69.5% ± 10.3% to 78.5% ± 15.1%).
The simulation of drinking water contamination by adding up to 50% (vol/vol) river water showed that Campylobacter strains can be detected in spiked water. However, the overall performance of the method was greatly altered (GdR, 2.02% ± 0.8%). Environmental compounds present in water may be retained on the microfilter along with bacteria and may induce decreases in the efficiency of the elution step. Moreover, these compounds may also interfere with elution and DNA extraction or alter the amplification efficiency, causing a reduction in recovery. With a greater volume of mixed water, filtration would not be possible, because the filter would become clogged. However, for this proportion of contamination, analysis of 1 liter of water would be sufficient to detect the bacteria present in the water.
Detection of C. jejuni and C. coli in 20 liters of groundwater from an alluvial aquifer confirmed that the procedure developed in this study is effective for both species when tested in volumes of this size. C. jejuni or C. coli was detected using our method in four of the eight samples collected in summer of 2010, although no thermotolerant Campylobacter organisms were detected in 1 liter when the standard culture method was used (20). Moreover, neither Escherichia coli nor enterococci were detected by normative procedures (data not shown). Although the qPCR gave positive results in groundwater, it was not possible to draw conclusions concerning the viability of the bacteria and thus assess the health risks. As discussed above, the qPCR method cannot distinguish between viable and nonviable forms or viable but noncultivable forms. In this case, where no epidemiological data have been reported and in the absence of any fecal indicator or information on the bacterial viability, it is not possible to assess the risk. These results only indicate that the water resource is not protected from contamination by Campylobacter. Inversely, when epidemiological data are available, this method can provide rapid results to the authorities in charge of an investigation and to water supply managers. The comparison of genomic sequences detected in water with sequences detected in stools of infected patients can confirm that the strains found in the water are indeed the cause of a waterborne outbreak.
Conclusions.
In this study, we showed that it is possible to detect Campylobacter strains in 20-liter samples of water by using a cellulose ester filter to concentrate the sample and a molecular method to detect these bacteria. In this volume of water, high recoveries were measured, even for low concentrations of bacteria seeded in water. Among all the microfilters tested in this study, the cellulose ester filter was simple and inexpensive to use. This filter can be used to test large volumes of tap water without clogging. Moreover, these filters are typically employed in laboratories that control the microbiological quality of drinking water, which involves screening for bacteria used as fecal indicators. Compared to the normative method, the method developed in this study has the advantages of being quick (6 h for samples of 20 liters) and can detect Campylobacter in water samples with or without high levels of background bacteria. The application of this method during an investigation of a waterborne outbreak, in parallel with use of the normative method, should improve the identification of the pathogen involved. Finally, this method can also be used to characterize the protection of a water aquifer used for the production of drinking water against microbial pollutions. Given the quantitative aspect of real-time qPCR and the recovery rates measured in this study, the concentrations of these pathogens in such waters can be determined.
ACKNOWLEDGMENT
We thank the National Reference Center for Campylobacter and Helicobacter (Bordeaux, France) for providing the Campylobacter strains.
Footnotes
Published ahead of print 2 December 2011
REFERENCES
- 1. Abulreesh HH, Paget TA, Goulder R. 2005. Recovery of thermophilic campylobacters from pond water and sediment and the problem of interference by background bacteria in enrichment culture. Water Res. 39:2877–2882 [DOI] [PubMed] [Google Scholar]
- 2. Abulreesh HH, Paget TA, Goulder R. 2006. Campylobacter in waterfowl and aquatic environments: incidence and methods of detection. Environ. Sci. Technol. 40:7122–7131 [DOI] [PubMed] [Google Scholar]
- 3. AFNOR 1996. Testing water: detection of enterovirus. Method by concentration on glass wool and detection by cell culture. XP T90-451. Association Française de Normalisation, Saint-Denis La Plaine, France [Google Scholar]
- 4. AFNOR 2001. Qualité de l'eau: recherche et dénombrement d'oocystes de Cryptosporidium et de kystes de Giardia. Méthode de concentration et de dénombrement. NF T90-455. Association Française de Normalisation, Saint-Denis La Plaine, France [Google Scholar]
- 5. AFNOR 2006. Water quality: detection and quantification of Legionella and/or L. pneumophila by concentration and gene amplification by polymerase chain reaction (PCR). XP T90-471. Association Française de Normalisation, Saint-Denis La Plaine, France [Google Scholar]
- 6. Ahmed W, Sawant S, Huygens F, Goonetilleke A, Gardner T. 2009. Prevalence and occurrence of zoonotic bacterial pathogens in surface waters determined by quantitative PCR. Water Res. 43:4918–4928 [DOI] [PubMed] [Google Scholar]
- 7. Allos BM. 2001. Campylobacter jejuni infections: update of emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 8. Baffone W, et al. 2006. Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int. J. Food Microbiol. 107:83–91 [DOI] [PubMed] [Google Scholar]
- 9. Block JC, Rolland D. 1979. Method for Salmonella concentration from water at pH 3.5, using micro-fiber glass filters. Appl. Environ. Microbiol. 38:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cappelier JM, Magras C, Jouve JL, Federighi M. 1999. Recovery of viable but non-culturable Campylobacter jejuni cells in two animal models. Food Microbiol. 16:375–383 [Google Scholar]
- 11. Carter AM, Pacha RE, Clark GW, Williams EA. 1987. Seasonal occurrence of Campylobacter spp. in surface waters and their correlation with standard indicator bacteria. Appl. Environ. Microbiol. 53:523–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Denis M, et al. 2009. Description and sources of contamination by Campylobacter spp. of river water destined for human consumption in Brittany, France. Pathol. Biol. 59:256–263 [DOI] [PubMed] [Google Scholar]
- 13. European Food Safety Authority 2011. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2009. EFSA J. 9:2090 [Google Scholar]
- 14. Evans MR, Ribeiro CD, Salmon RL. 2003. Hazards of healthy living: bottled water and salad vegetables as risk factors for Campylobacter infection. Emerg. Infect. Dis. 9:1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fricker CR. 1987. The isolation of salmonellas and campylobacters. J. Appl. Bacteriol. 63:99–116 [DOI] [PubMed] [Google Scholar]
- 16. Friedman CR, Neimann J, Wegener HC, Tauxe RV. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121–138 In Nachamkin I, Blaser MJ. (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 17. Gillespie IA, et al. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8:937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hänninen ML, et al. 2003. Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl. Environ. Microbiol. 69:1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Humphrey TJ. 1989. An appraisal of efficacy of preenrichment for the isolation of Campylobacter jejuni from food and water. J. Appl. Bacteriol. 66:119–126 [DOI] [PubMed] [Google Scholar]
- 20. International Organization for Standardisation 2005. Water quality: detection and enumeration of thermotolerant Campylobacter species. ISO 17995. International Organization for Standardisation, Geneva, Switzerland [Google Scholar]
- 21. Jones K. 2001. Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90:68S–79S [DOI] [PubMed] [Google Scholar]
- 22. Khan IUH, et al. 2009. A methods comparison for the isolation and detection of thermophilic Campylobacter in agricultural watersheds. J. Microbiol. Methods 79:307–313 [DOI] [PubMed] [Google Scholar]
- 23. Levin RE. 2007. Campylobacter jejuni: a review of its characteristics, pathogenicity, ecology, distribution, subspecies characterization and molecular methods of detection. Food Biotechnol. 21:271–347 [Google Scholar]
- 24. Ménard A, Dachet F, Prouzet-Mauleon V, Oleastro M, Mégraud F. 2005. Development of a real-time fluorescence resonance energy transfer PCR to identify the main pathogenic Campylobacter spp. Clin. Microbiol. Infect. 11:281–287 [DOI] [PubMed] [Google Scholar]
- 25. Moore J, Caldwell P, Millar B. 2001. Molecular detection of Campylobacter spp. in drinking, recreational and environmental water supplies. Int. J. Hyg. Environ. Health 204:185–189 [DOI] [PubMed] [Google Scholar]
- 26. Oyofo BA, Rollins DM. 1993. Efficacy of filter types for detecting Campylobacter jejuni and Campylobacter coli in environmental water samples by polymerase chain reaction. Appl. Environ. Microbiol. 59:4090–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Payment P, Berube A, Perreault D, Armon R, Trudel M. 1989. Concentration of Giardia lamblia cysts, Legionella pneumophila, Clostridium perfringens, human enteric viruses, and coliphages from large volumes of drinking water, using a single filtration. Can. J. Microbiol. 35:932–935 [DOI] [PubMed] [Google Scholar]
- 28. Polaczyk AL, Roberts JM, Hill VR. 2007. Evaluation of 1MDS electropositive microfilters for simultaneous recovery of multiple microbe classes from tap water. J. Microbiol. Methods 68:260–266 [DOI] [PubMed] [Google Scholar]
- 29. Skelly C, Weinstein P. 2003. Pathogen survival trajectories: an eco-environmental approach to the modeling of human campylobacteriosis ecology. Environ. Health Perspect. 111:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. World Health Organization 2003. Assessing microbial safety of drinking water: improving approaches and methods. IWA Publishing, London, United Kingdom [Google Scholar]