Abstract
A marine bacterium, designated strain MCTG13d, was isolated from a laboratory culture of the dinoflagellate Lingulodinium polyedrum CCAP1121/2 by enrichment with polycyclic aromatic hydrocarbons (PAHs) as the sole carbon source. Based on 16S rRNA gene sequence comparisons, the strain was most closely related to Porticoccus litoralis IMCC2115T (96.5%) and to members of the genera Microbulbifer (91.4 to 93.7%) and Marinimicrobium (90.4 to 92.0%). Phylogenetic trees showed that the strain clustered in a distinct phyletic line in the class Gammaproteobacteria for which P. litoralis is presently the sole cultured representative. The strain was strictly aerobic, rod shaped, Gram negative, and halophilic. Notably, it was able to utilize hydrocarbons as sole sources of carbon and energy, whereas sugars did not serve as growth substrates. The predominant isoprenoid quinone of strain MCTG13d was Q-8, and the dominant fatty acids were C16:1ω7c, C18:1ω7c, and C16:0. DNA G+C content for the isolate was 54.9 ± 0.42 mol%. Quantitative PCR primers targeting the 16S rRNA gene of this strain showed that this organism was common in other laboratory cultures of marine phytoplankton. On the basis of phenotypic and genotypic characteristics, strain MCTG13d represents a novel species of Porticoccus, for which the name Porticoccus hydrocarbonoclasticus sp. nov. is proposed. The discovery of this highly specialized hydrocarbon-degrading bacterium living in association with marine phytoplankton suggests that phytoplankton represent a previously unrecognized biotope of novel bacterial taxa that degrade hydrocarbons in the ocean.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are organic compounds composed entirely of fused benzene rings and found naturally occurring in coal, crude oil, and refined fossil fuels. Based on their poor water solubility, toxicity, persistence, and potential to bioaccumulate, these compounds are recognized as high-priority pollutants in the environment and are of significant concern for human health (1). PAHs can enter the marine environment from various sources. In semiclosed estuarine areas and in active tectonic zones, where burial of large amounts of organic matter takes place, polyarenes (43) can form in diagenetic processes at their fossilization (44). These areas act as point sources of pyrogenic PAH input into the marine environment (35), as evidenced by the presence of these chemicals in sediments at high concentrations where no apparent pollutant input occurs (34). The sea surface, particularly along inhabited coastal areas, is generally prone to high levels of PAH input, such as from atmospheric fallout, domestic and industrial effluents, land runoff, and oil spill contamination. Upon their entry into marine surface waters, these chemicals become subjected to various influences, both abiotic (e.g., photolysis and chemical oxidation) and biotic (e.g., microbial degradation), that determine their fate. Their ultimate removal, however, is carried out by PAH-degrading bacteria, whose activities are recognized as critical to the purging of the marine water column and sediment (19, 58). Since these microorganisms are the foundation of hydrocarbon remediation, a critical understanding of these processes is at the heart of designing successful bioremediation methods.
There is compelling evidence in the literature suggesting that phytoplankton play a significant role in the fate of PAHs and other organic pollutants upon their entry into marine surface waters. For example, field studies performed in the Baltic Sea have shown a high correlation between PAH removal and particulate organic carbon, particularly along the coast and during the warmer months corresponding to peaks in phytoplankton blooms (55, 56). In a study by Kowalewska (23), phytoplankton cells significantly contributed to the transport of PAHs from the upper layers of the Southern Baltic ecosystem to the sea floor by sedimentation. Although the hydrophobic nature of PAHs (log Kow = 3 to 8) greatly limits their solubility in seawater, this property would favor their adsorption to marine particulate matter, including phytoplankton and detritus (11, 27). Kowalewska (23) quantitatively demonstrated that different PAH compounds conferred different correlation coefficients for binding to phytoplankton cells. Similarly, Binark et al. (4) showed that cell surfaces of several marine microalgae were capable of adsorbing up to 14 different types of PAHs at relatively high concentrations. During periods of high productivity, which lead to larger vertical fluxes of particles, eutrophication of the euphotic zone would be expected to enhance the vertical transport of particle-associated pollutants (6, 31, 57). This has potentially profound implications for the natural purging of the marine water column and overall health of the ocean. There is also evidence to suggest that phytoplankton may be a biogenic source of PAHs. Studies conducted in the 1960s to 1980s showed that some microalgae possessed the ability to synthesize these compounds (3, 16) and translocate them into the algal cell wall (15, 16, 38, 59). More recently, two open-ocean sites were found to contain a suite of chlorinated aromatic compounds and, based on the global inventory and isomer distribution of this class of compounds, these chemicals were proposed to be likely produced in situ by marine microbes, such as phytoplankton (41).
Our understanding of the microbial diversity that contributes to the degradation of PAHs and other toxic pollutants in the ocean has progressed, but it remains far from complete. One area that has received little attention in this respect is the bacterial community associated with the cell surface layer, or phycosphere, of marine phytoplankton. Whether through biogenic synthesis or adsorption from the surrounding seawater, the cell surface of microalgal cells may act as a hot spot with which PAH-degrading bacteria can associate. Supporting this concept, we recently isolated two novel genera of bacteria from laboratory cultures of marine phytoplankton that exhibit a preference for utilizing PAHs as sole sources of carbon and energy (T. Gutierrez, D. H. Green, P. D. Nichols, W. B. Whitman, K. T. Semple, and M. D. Aitken, submitted for publication; T. Gutierrez, D. H. Green, W. B. Whitman, P. D. Nichols, K. T. Semple, and M. D. Aitken, submitted for publication). In the present study, we report on the isolation and characterization of another PAH-degrading bacterium, designated strain MCTG13d, from a nonaxenic laboratory culture of the marine dinoflagellate Lingulodinium polyedrum CCAP1121/2. Polyphasic analysis suggested that the strain belongs to the recently described genus Porticoccus and may be classed as a putative “specialist” PAH degrader based on its nutritional preference for PAH compounds as sole carbon and energy sources. Using newly designed PCR primers that target the 16S rRNA gene of strain MCTG13d, we identified this organism in other cultures of marine phytoplankton. The isolation of PAH-degrading bacteria from phytoplankton has only recently been discovered through our work and raises important questions about the function of these organisms in the ocean and their relationship with their microalgal hosts.
MATERIALS AND METHODS
Algal strains and inoculum preparation.
Nonaxenic laboratory cultures of dinoflagellates, diatoms, and coccolithophores were obtained from the Culture Collection of Algae and Protozoa (CCAP; Oban, Scotland) and from the Center for Culture of Marine Phytoplankton (CCMP; Maine). Table 1 lists the various strains and locations from where they were originally isolated. The strains were maintained in algal medium that is specific to their growth requirements and in temperature-controlled illuminated incubators, per the supplier's guidelines.
Table 1.
Phytoplankton strains that were examined in this study
| Algal straina | Origin |
|---|---|
| Dinoflagellates | |
| Lingulodinium polyedrum CCAP1121/2b,c | Loch Creran, Argyll, Scotland |
| Isochrysis sp. CCMP1324c | Mataiva, Tahiti, South Pacific Ocean |
| Heterosigma akashiwo CCMP1870c | Long Beach, CA, USA |
| Diatoms | |
| Pseudo-Nitzschia sp. CCAP1061/25c | Loch Creran, Argyll, Scotland |
| Skeletonema costatum CCAP1077/1Cc | North Sea (approximate location unknown) |
| Thalassiosira weissflogii CCMP1587c | Jakarta Harbor, Indonesia |
Dinoflagellate and diatom strains were maintained on f/2 and f/2+Si medium, respectively (15a, 15b).
Strain used for the enrichment and isolation of PAH-degrading bacteria.
Strains screened by qPCR using primers that were designed and optimized in this study for the targeted detection of strain MCTG13d.
Enrichment experiments and isolation.
To isolate PAH-degrading bacteria, enrichment cultures were prepared using acid-washed (0.1 N HCl), steam-sterilized glass test tubes (16 by 125 mm) fitted with screw caps lined with aluminum foil to prevent PAH loss via adsorption. Stock solutions (ca. 3,000 mg/liter) of phenanthrene, anthracene, fluorene, and pyrene dissolved in acetone were prepared. Four sets of 6 test tubes each were prepared containing one of each of the PAHs and 2.8 ml of ONR7a medium (9). Another set of 24 culture tubes was prepared in the same way with 2.8 ml of ZM/10 marine medium (10-fold dilution of 2216 marine medium). All 48 test tubes were then inoculated with 200 μl of an exponentially growing culture of L. polyedrum CCAP1121/2. Uninoculated controls (24 tubes), acid-killed controls (6 tubes), and tubes that were inoculated but without any added PAHs (6 tubes) were also prepared. All 84 test tubes were incubated in the dark with gentle shaking (100 rpm) at 21°C. PAH degradation was determined spectrophotometrically. The initial amounts of the PAHs in the respective tubes were 0.23 ± 0.01 mg for phenanthrene, 0.17 ± 0.03 mg for anthracene, 0.22 ± 0.0 mg for pyrene, and 0.29 ± 0.0 mg for fluorene.
To isolate PAH-degrading bacteria, 5-μl samples from each PAH incubation were taken at 2 and 4 weeks and streaked onto ONR7a and ZM/10 agar plates that were then sprayed with the respective PAH compound used in the enrichments (22). The plates were stored in closed plastic bags in the dark at room temperature. Colonies forming clearing zones were picked, purified, and stored frozen at −80°C with the addition of 20% (vol/vol) glycerol.
Laboratory cultures of the 6 phytoplankton strains examined in this study were independently enriched with pyruvate and phenanthrene for subsequent analysis of the target organism by quantitative PCR. Enrichments were conducted in 50-ml Erlenmeyer flasks containing 10 ml of algal medium used to maintain the strains (Table 1). When phenanthrene was used, about 10.5 mg/liter (final concentration) was added per flask. When pyruvate was used, it was added at 3,000 mg/liter per flask. Each flask was then inoculated with a 1-ml inoculum of a phytoplankton strain from cultures in mid- to late growth phase. The flasks were incubated in the dark with gentle shaking (100 rpm) at 21°C. After 6 days, biomass was collected from each incubation and whole DNA was extracted (51). At the time of preparing these enrichments, biomass samples of all 6 phytoplankton strains were stored at −20°C for whole DNA extractions; these were designated the “nonenrichments.” All DNA extracts were stored in the freezer for subsequent analysis for DNA and 16S rRNA gene quantification of the target bacterial isolate.
Measurement of PAH degradation.
A spectrophotometric method was used to quantify the amount of each PAH in the enrichments. For this, 3 test tubes from each PAH incubation were sacrificed at 2 and at 4 weeks for extraction with ethyl acetate (high-pressure liquid chromatography [HPLC] grade). This was performed by adding 2 ml of ethyl acetate to each tube and then vigorously shaking them by hand for 10 s and vortexing them for 30 s. When emulsions formed between the aqueous and nonaqueous phases, a few drops of saturated NaCl solution were added to separate the two phases. Aliquots of the nonaqueous top layer were diluted with ethyl acetate in quartz cuvettes for spectrophotometric analysis at 251 nm for phenanthrene, 356 nm for anthracene, 335 nm for pyrene, and 261 nm for fluorene. The A251, A356, A335, and A261 values were converted to concentrations of phenanthrene, anthracene, pyrene, and fluorene, respectively, using the molar absorptivity coefficients 6.3 × 104, 6.4 × 103, 4.83 × 104, and 2.27 × 104 liters mol−1 cm−1 (49).
Isolate characterization.
Phenotypic and biochemical characterization of strain MCTG13d was performed using cultures grown in ONR7a amended with acetate at 20°C in the dark, unless otherwise indicated. The various microbiological tests, including those for Gram stain reaction, oxidase and catalase, nitrate reduction, motility, phosphatase, gelatinase, and lipase, were performed according to standard methods (13, 28). Uninoculated medium and medium without an added carbon source were used as controls where appropriate. All incubations were performed in at least triplicate. Spectral and microscopic analysis (with Nile Blue) was used to determine whether the cells accumulate poly-β-hydroxybutyrate (PHB) granules (13). The Schaeffer-Fulton staining method was used for the determination of endospore formation. The formation of indigo from indole was tested on ONR7a agar medium amended with 2 mM indole, as previously described (10). Growth on high-nutrient medium was evaluated by streaking the cells onto R2A, ZM/10, and ZM/1 agar plates. To determine the organism's requirement for and tolerance to NaCl, the organism was inoculated into ONR7a liquid medium amended with acetate containing increasing concentrations of NaCl: 0.0, 1.0, 3.0, 6.0, 10.0, 15.0, and 20.0% (wt/vol). To determine the pH range for growth, ONR7a amended with acetate was prepared with the following buffers: 25 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.5 to 6.5; 25 mM TAPSO {3-[N-Tris(hydroxymethyl)methylamino]-2-hydroxypropanesulfonic acid}, pH 7.5; 25 mM Tris base-Tris HCl, pH 8.5 to 9.5. To determine the temperature range for growth, cultures of ONR7a amended with acetate were incubated at the following temperatures: 4, 10, 15, 20, 30, 37, and 45°C.
To determine whether different substrates could be used as sole sources of carbon, representative aliphatic and aromatic hydrocarbons, carbohydrates, and common metabolic intermediates were added at concentrations between 0.1 and 0.2% (wt/vol for solid substrates; vol/vol for liquid substrates) to ONR7a liquid medium. These tests were also performed using ONR7a agar medium. Due to their relative toxicity, some substrates were added at lower concentrations or added to the lids of plates. Substrates tested included glucose, fructose, mannitol, xylose, arabinose, pyruvate, acetate, methanol, salicylate, phthalate, hexane, pentane, decane, n-hexadecane, phenanthrene, anthracene, fluorene, and pyrene. Liquid cultures were incubated with gentle rotary shaking (100 rpm) in the dark at 21°C under aerobic conditions, and growth was monitored spectrophotometrically at 600 nm over 2 to 4 weeks. Uninoculated medium and medium without an added carbon source were used as controls. All incubations were performed in at least triplicate. Growth on the various carbon sources was also evaluated on agar plates which were incubated in closed plastic bags in the dark at 21°C, and the development of colonies was observed over a period of up to 6 weeks. Growth on methane was tested by inoculating ONR7a agar plates and placing them in a desiccator, after which 30% of the air atmosphere was replaced with methane. Anaerobic growth was tested using ONR7a agar plates that were incubated in a desiccator that was rendered oxygen free.
The cell morphology and motility of the isolate were observed using transmission and scanning electron microscopy of cells that had been grown in ONR7a amended with fluorene as the sole carbon and energy source. For negative staining, bacterial suspensions were stained with 3% ammonium molybdate (pH 7.0) (18), prior to viewing with an LEO EM910 transmission electron microscope (TEM) operating at 80 kV. Digital images were acquired using a Gatan Orius SC1000 charge-coupled device (CCD) digital camera and 3.11.0 digital micrograph (Gatan, Inc., Pleasanton, CA). For preparing thin sections, glutaraldehyde fixation was followed by postfixation with 1% OsO4 before the samples were dehydrated gradually and embedded in Polybed 812. Sectioned samples (∼100 nm) were then stained with uranyl acetate and lead citrate prior to visualization with a JEOL 100CX TEM. For scanning electron microscopy, glutaraldehyde fixation was followed by postfixation with 2% OsO4. After critical-point drying, the cells were sputter coated with gold-palladium and observed using a Hitachi 4700 FEG scanning electron microscope (SEM).
The DNA G+C content was determined by HPLC (30). Fatty acid profiles, obtained following extraction of biomass and methylation of the total extractable lipids, were analyzed (32) from cells grown in ONR7a amended with fluorene and repeated for cells grown in ONR7a amended with n-hexadecane for up to 12 days at 21°C. Isoprenoid quinones were extracted from lyophilized cells, and the samples were purified and analyzed by the DSMZ Identification Service (Braunschweig, Germany).
16S rRNA sequencing and phylogenetic analysis.
Total genomic DNA was recovered using a Wizard genomic DNA purification kit (Promega, Madison, WI), according to the manufacturer's instructions. The 16S rRNA gene was amplified by PCR with primers 27f (54) and 1492r (24). The resulting product was then cloned into the plasmid PCR4-Topo using the Topo-TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). The insert was sequenced with primers M13f, M13r, 338f, and 338r (2); 907f (25); and 907r (54) at the University of North Carolina Genome Analysis Facility. Sequences were assembled using the program Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI). The consensus sequence was submitted to GenBank and checked for close relatives and phylogenetic affiliation using the BLASTN search program and RDP-II (8). The search results were used as a guide for tree construction. Additional related 16S rRNA sequences identified from the BLASTN and RDP-II search were retrieved from GenBank. The CLUSTAL_X program (50) was used to align the sequences and construct neighbor-joining trees with TREEVIEW (WIN32) version 1.5.2 (37). The trees were bootstrapped 1,000 times, and gaps in the alignment were ignored. To evaluate dendrogram topology and confirm phylogenetic affiliation, sequences were imported and automatically aligned in the ARB-SILVA SSURef 94 database (40) and manually refined taking into account the secondary structural information of the rRNA molecule. Tree reconstruction was performed using the neighbor-joining, maximum parsimony, and maximum likelihood methods and applying various filters.
Quantitative PCR primer design.
16S rRNA-targeted primers for quantitative real-time PCR (qPCR) were developed to quantify the novel MCTG13d isolate. The Probe Design and Probe Match tools of the ARB software (40) were used to design primers that were specific for this sequence. The primers were named 13d_469F (5′-ACT GTC AGC CCT GAC GTT-3′) and 13d_637R (5′-CTA GTC AGA CAG TTC TGG-3′). Primer specificity was confirmed with the Probe Check tool of RDP-II. The 13d_469F primer was complementary to five unclassified Gammaproteobacteria, four of which formed a phylogenetic cluster with strain MCTG13d with a bootstrap value of 88% (1,000 replications). The 13d_637R primer was complementary to 103 unclassified Gammaproteobacteria, one uncultured Dasnia strain, one uncultured Pseudomonas strain, and one unclassified alphaproteobacterium. The optimal annealing temperature of the primer set (56°C) was determined using an Eppendorf (Hauppauge, NY) Mastercycler gradient thermal cycler. The template for construction of a standard curve for quantitative PCR was a plasmid containing a representative sequence which had been linearized using PstI (New England BioLabs, Ipswich, MA) and purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA). The limit of quantification for the target strain using these primers was five gene copies per reaction. When threshold cycle (CT) values beyond the highest value in the linear range of the standard curve (CT range, 12.2 to 34.0) were measured, the gene was considered detected but below the quantification limit of the assay. The amplification efficiency of the primers (39) was determined to be 1.75. Further support for the specificity of the newly designed primers for strain MCTG13d was evaluated by performing qPCRs using DNA from nontarget organisms, including Escherichia coli, Pseudomonas putida, and other marine strains isolated from some of the phytoplankton species examined in this study (T. Gutierrez, D. H. Green, P. D. Nichols, W. B. Whitman, K. T. Semple, and M. D. Aitken, submitted for publication; T. Gutierrez, D. H. Green, W. B. Whitman, P. D. Nichols, K. T. Semple, and M. D. Aitken, submitted for publication). The CT values measured from these reactions (>36.0) were outside the limit of detection.
Quantitative detection of the isolate in phytoplankton cultures.
DNA extracted from the six nonaxenic laboratory cultures of phytoplankton examined in this study (enriched and nonenriched) was quantified using a NanoDrop ND-3300 fluorospectrometer (Thermo, Waltham, MA) and the Quant-iT Picogreen double-stranded DNA (dsDNA) kit (Invitrogen, Carlsbad, CA). Measurements were performed on triplicate DNA extracts. Identification of the target strain, MCTG13d, in these extracts was determined by qPCR as described previously (45).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain MCTG13d was deposited with GenBank under accession number JN088732.
RESULTS
Enrichments and strain isolation.
Enrichments were performed with individual PAH compounds rather than a PAH mixture because of the possibility that competitive inhibition might occur (47). Degradation of each of the four PAHs was measured during incubation of nonaxenic Lingulodinium polyedrum CCAP1121/2 with the four different PAH compounds. For the enrichments utilizing ZM/10 medium, the amounts of phenanthrene, anthracene, and pyrene degraded after 4 weeks were 31.9% ± 5.1%, 40.1% ± 10.0%, and 10.1% ± 2.1% of the total initial amount added, respectively. No degradation of fluorene was measured in these enrichments. For enrichments utilizing ONR7a medium, the amounts of phenanthrene and fluorene degraded after 4 weeks were 16.1% ± 3.5% and 23.7% ± 2.2%, respectively. No degradation of anthracene or pyrene was detected in incubations using ONR7a medium. Uninoculated controls showed no significant loss of PAHs.
Agar plates that were streaked from these enrichments yielded several isolates that formed clearing zones on PAH-sprayed agar plates. Forty-eight colonial morphotypes were picked and transferred to obtain pure cultures. One isolate developed large pale yellow-green colonies surrounded by clearing zones on ONR7a agar plates sprayed with different PAHs. This isolate did not yield any growth on ONR7a medium without any added carbon source. The isolate was designated strain MCTG13d and selected for further study.
Phenotypic and biochemical characterization.
An evaluation of different medium types revealed that ONR7a amended with a carbon source (e.g., acetate, fluorene, phenanthrene, or n-hexadecane) satisfied the specific nutritional requirements of this fastidious strain. Other media that are often used to grow marine strains, such as R2A and marine medium 2216 (Difco), did not support growth of strain MCTG13d. On ONR7a agar plates amended with acetate, colonies were small (0.5 to 2 mm in diameter), round, and nonpigmented after 2 weeks. On ONR7a agar sprayed with a PAH compound (e.g., phenanthrene, anthracene, fluorene, or pyrene), the colonies that formed were larger (4 to 7 mm in diameter) with rough surfaces, were pigmented green-yellow, were slightly raised with undulate margins, and displayed discernible clearing zones, which was evidence of PAH degradation. The strain displayed a narrow nutritional spectrum with a preference for assimilating hydrocarbon substrates. The following served as sole carbon sources for growth: fluorene, anthracene, pyrene, phenanthrene, n-hexadecane, indole, and acetate. No growth was observed on the following as sole sources of carbon: mannitol, fructose, glucose, xylose, arabinose, decane, hexane, pentane, pyruvate, methanol, and methane.
Cells of strain MCTG13d were short to long, non-spore-forming slightly bent rods, 1.0 to 2.0 by 0.5 to 0.6 μm in average size (Fig. 1). They stained Gram negative, contained intracellular inclusion bodies and few surface blebs, and were motile by means of a single polar monotrichous flagellum (Fig. 1). Cells stained with Nile Blue and viewed under the epifluorescence microscope revealed the presence of numerous intracellular fluorescent granules (size range, 0.2 to 0.5 μm). Spectral analysis, however, indicated that these were not poly-β-hydroxybutyrate granules. Strain MCTG13d was obligately aerobic and both catalase and oxidase positive. The strain grew at temperatures ranging from 10 to 37°C (optimal, 15°C) and at pH values ranging from 6.5 to 9.0 (optimal pH, 8.0). The strain was negative for lipase (Tween 80) and the hydrolysis of agar and gelatin. The strain was positive for phosphatase activity. The strain exhibited slight halotolerance, since it grew in medium containing NaCl concentrations up to 6%, although growth was markedly reduced in the absence of NaCl. No growth was measured at 10% NaCl.
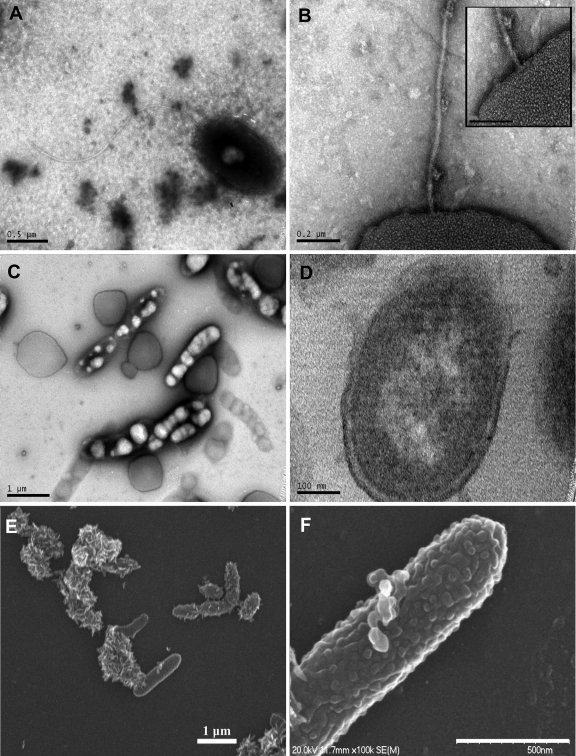
Fig 1.
Transmission (negative staining, A to D) and scanning (E and F) electron micrographs of strain MCTG13d. Cells contain a single polar monotrichous flagellum (A) attached to the cell membrane and apparently unsheathed (B and inset). Many cells were found to be packed with inclusion bodies (C) that fluoresced when viewed under the epifluorescence microscope (not shown). Thin sections show a cell envelope and cell membrane that are typical of Gram-negative bacteria (D). Cells picked from colonies growing on ONR7a agar with fluorene show cells among crystals of fluorene (E). Blebs exuding from the cell surface were observed on some cells (F).
Table 2 shows the fatty acid (derived from the total extractable lipid) profiles of strain MCTG13d and its closest relative, Porticoccus litoralis IMCC2115T. The dominant fatty acids for strain MCTG13d were C16:1ω7c, C18:1ω7c, and C16:0, with these fatty acids accounting for 88.8% of total fatty acids. The following fatty acids were detected as minor components (1 to 5%): C17:1ω8c, C18:1ω9c, and C18:0, accounting for 8.1% of total fatty acids. The predominant isoprenoid quinone for strain MCTG13d was Q-8. The DNA G+C content for the isolate was 54.9 ± 0.4 mol%.
Table 2.
Cellular fatty acid composition of strain MCTG13d and its closest relative Porticoccus litoralis IMCC2115Tb
| Fatty acid | % of total fatty acids in strain: |
|
|---|---|---|
| MCTG13dT | IMCC2115T | |
| C11:0 3-OH | — | 1.4 |
| Iso-C15:0 | — | 3.7 |
| Anteiso-C15:0 | — | 67.6 |
| C16:0 | 27.8 | 6.9 |
| C16:1ω7c | 31.5 | — |
| Iso-C17:0 | — | 3.9 |
| Anteiso-C17:0 | — | 14.4 |
| C17:1ω8c | 4.0 | 1.8a |
| C18:0 | 1.3 | 1.3 |
| C18:1ω7c | 29.5 | — |
| C18:1ω9c | 2.8 | — |
| Iso-C19:0 | — | 1.2 |
| Anteiso-C19:0 | — | 3.2 |
Double bond position not specified in the work of Oh et al. (36).
Lipid extraction of biomass followed by methylation of the total extractable lipids was used for strain MCTG13d, with whole-cell methylation protocols used for strain IMCC2115T. The latter also recovers lipopolysaccharide-derived OH fatty acids. Values are percentages of the total fatty acids; components that represented <1% in all strains were not included; —, fatty acid not detected. Data for strain IMCC2115T were from the work of Oh et al. (36).
Phylogenetic analysis.
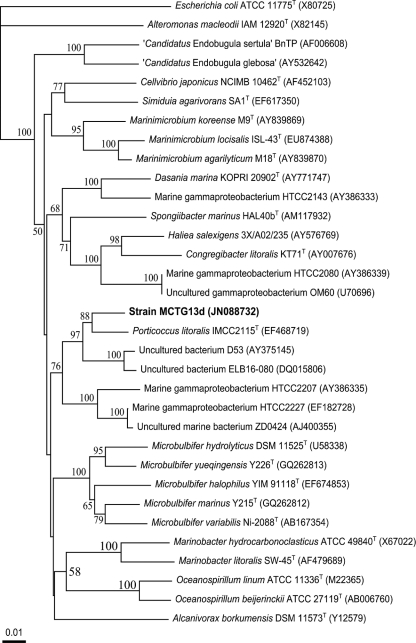
An almost complete sequence of the 16S rRNA gene (1,503 bp) was obtained for strain MCTG13d. From a BLASTN analysis, strain MCTG13d was most closely related to Porticoccus litoralis IMCC2115T (96.5% sequence identity), which originated from coastal surface seawater in the Yellow Sea, South Korea. The next closest cultivated relatives included members of the Microbulbifer (91.4 to 93.7%) and Marinimicrobium (90.4 to 92.0%) genera. These and other related sequences were used to construct the neighbor-joining tree (Fig. 2). The affiliation of strain MCTG13d with the genus Porticoccus was supported by a moderate bootstrap value of 85%. Using the RDP-II Classifier tool (52) with a confidence threshold of 80% indicated the novel strain to be an unclassified member of the family Alteromonadaceae. Further analysis revealed that strain MCTG13d was distinctly grouped within a clade of mainly uncultivated bacterial clones that lies adjacent to the OM60 clade, represented by strain HTCC2080 (7), and the SAR92 clade, represented by strain HTCC2207 (46). The only cultured representative within this clade is Porticoccus litoralis IMCC2115T. To clarify the phylogenetic position of strain MCTG13d, tree construction was performed with the neighbor-joining, maximum parsimony, and maximum likelihood methods. In all cases, the position of strain MCTG13d was distinct within this clade, which comprised Porticoccus litoralis IMCC2115T as the only cultured organism, and a few hundred uncultured clones that included the representative bacterial clones D53 (60) and ELB16-080 (14).
Fig 2.
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences (>1,200 bp), showing the relationships between strain MCTG13d and representatives of related taxa. Bootstrap values (expressed as percentages of 1,000 replications) of >50% are shown at each node. GenBank accession numbers are shown in parentheses. Bar, 1 substitution per 100 nucleotide positions. The maximum parsimony and maximum likelihood trees showed essentially the same topology (data not shown).
Quantitative detection of strain MCTG13d in phytoplankton cultures.
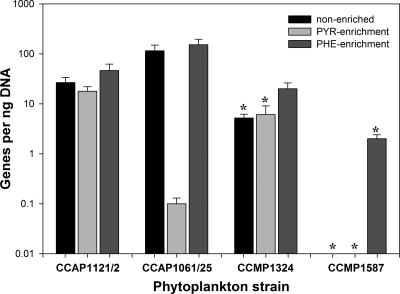
Primers used for qPCR that targeted the 16S rRNA gene of strain MCTG13d were developed to determine whether various nonaxenic laboratory cultures of phytoplankton (Table 1) harbor this novel PAH degrader. Four phytoplankton strains (CCAP1121/2, CCAP1061/25, CCMP1324, and CCMP1587), out of six that were analyzed, contained MCTG13d and possibly other Porticoccus-related bacteria (Fig. 3). However, based on the quantification limit for the qPCR assay, the abundance in only three of these strains (i.e., CCAP1121/2, CCAP1061/25, and CCMP1324) could be reliably measured. As expected, strain MCTG13d was abundant in L. polyedrum CCAP1121/2, from which it was isolated. Except for strain CCMP1061/25, the 16S rRNA gene copy number of MCTG13d was higher for the other three phytoplankton strains after they had been incubated on phenanthrene. For CCMP1061/25, the abundances of MCTG13d genes were similar for cultures incubated with and without phenanthrene. MCTG13d was below the quantification limit in CCMP1324 incubated with and without pyruvate, and in CCMP1587, CCMP1870, and CCMP1077/1C under all three incubation conditions tested (not shown for the latter two).
Fig 3.
Abundance of strain MCTG13d 16S rRNA genes in phytoplankton species enriched with pyruvate (PYR-enrichment), phenanthrene (PHE-enrichment), or no added carbon source (non-enriched). Bars are the average and standard deviation of triplicate qPCRs measuring the abundance of MCTG13d-specific 16S rRNA genes per ng DNA. Phytoplankton strains: Lingulodinium polyedrum CCAP1121/2; pseudo-nitzschia CCAP1061/25; Isochrysis sp. CCMP1324; Thalassiosira weissflogii CCMP1587. Asterisks represent values that were below the quantification limit (<5 gene copies per reaction) of the assay, which varied based on template DNA concentration per reaction.
DISCUSSION
A novel bacterium, strain MCTG13d, was isolated from a laboratory culture of the marine dinoflagellate L. polyedrum CCAP1121/2 which displays a nutritional preference for utilizing hydrocarbons as growth substrates. Strain MCTG13d is a member of a phylogenetic clade that lies adjacent to the OM60 and SAR92 clades. Except for P. litoralis IMCC2115T, this clade is almost entirely represented by uncultivated bacteria. Strain MCTG13d is a strictly aerobic, rod-shaped bacterium that stains Gram negative and is a halophile.
Physiology and ecology.
Recently, two novel genera of PAH-degrading bacteria were isolated from marine phytoplankton, Polycyclovorans algicola TG408T (T. Gutierrez, D. H. Green, P. D. Nichols, W. B. Whitman, K. T. Semple, and M. D. Aitken, submitted for publication) and Algiphilus aromaticivorans DG1253T (T. Gutierrez, D. H. Green, W. B. Whitman, P. D. Nichols, K. T. Semple, and M. D. Aitken, submitted for publication), for which the latter is proposed to represent a novel family. An investigation of their metabolic versatility revealed that, like strain MCTG13d, these organisms all share the ability to utilize hydrocarbons over other naturally occurring organic substrates as sole sources of carbon and energy. Hence, these organisms may be classed as specialist hydrocarbon degraders. It is not presently clear why these PAH degraders have not been identified in previous studies. It may be inferred that they have hitherto eluded cultivation because they occupy a specific biotope in the ocean (i.e., the phycosphere of phytoplankton) which has been poorly investigated in this respect. In addition, these organisms confer a narrow nutritional spectrum, with an apparent exclusive requirement for hydrocarbons as growth substrates, and are therefore difficult to isolate and cultivate. The application of targeted approaches for the cultivation of these organisms from phytoplankton, as performed in this and our earlier work, has revealed novel genera of specialist hydrocarbon degraders. To our knowledge, PAH-degrading bacteria specifically associated with phytoplankton have not been previously investigated.
Strain MCTG13d displays a narrow nutritional spectrum, utilizing hydrocarbons, such as PAHs, and a few other compounds as growth substrates. Notably, it is unable to grow on sugars, which is a key feature that distinguishes it from its closest relatives Porticoccus, Microbulbifer, and Marinimicrobium, all of which are capable of utilizing sugars as growth substrates. Since MCTG13d was able to grow in medium containing NaCl concentrations up to 6% (wt/vol), this strain is therefore a slightly halotolerant and slightly halophilic bacterium and can be considered a marine strain (26). As it was unable to grow in typical marine medium, such as that amended with yeast extract and/or peptone, MCTG13d can be classed as a fastidious specialist hydrocarbon degrader, with a physiology acclimated for life under oligotrophic conditions in the ocean. Further work, however, will be needed to better understand how this organism responds to changes in its environment.
As far as we know, bacteria that specialize in degrading hydrocarbons have been isolated only from the marine environment (19). These hydrocarbon specialists are found distributed throughout the world's oceans and play a significant global role in the natural attenuation and mineralization of these compounds (12, 21, 58). In oil-impacted environments, they are strongly selected for and successively increase in numbers from nearly undetectable levels to levels constituting up to 70 to 90% of the total bacterial population (17, 29). With respect to specialist PAH degraders in the ocean, our knowledge of these organisms has hitherto been limited to members that comprise the genera Neptunomonas and Cycloclasticus. The discovery of novel PAH degraders living associated with phytoplankton brings to light the possible role of algal-bacterial associations in the natural purging of sites contaminated with hydrocarbons in the ocean. It is not clear to us at present what type of association these PAH-degrading bacteria might share with their algal hosts, though we can postulate that the degradative capacity of these bacteria is likely to benefit the algae by reducing the chemical toxicity of PAHs at the phycosphere.
Using newly designed qPCR primers targeting the 16S rRNA gene of MCTG13d, we confirmed the laboratory cultures of CCAP1121/2, CCAP1061/25, and CCMP1324 to harbor this novel PAH-degrading bacterium. For CCMP1587, the gene copy number of MCTG13d was just below the quantifiable limit. It is noteworthy that although a value for gene copy number was measured for this algal strain, albeit below the detection limit, this was from an incubation of the algal culture on phenanthrene—one of MCTG13d's preferred growth substrates. This result could suggest that this bacterium is associated with CCMP1587 at very low cell numbers and that a longer incubation time on phenanthrene may have yielded sufficiently higher cell numbers (equating to higher gene copies) that may then have been quantifiable by our qPCR assay. Since the maintenance of these phytoplankton cultures in the laboratory has not involved amendment with hydrocarbons, the association of MCTG13d with these algae cannot be explained by laboratory enrichment. Considering that laboratory cultures of phytoplankton represent a snapshot of the bacterial community associated with the algal cells at the time of their isolation (20) and that many of these cultures have been maintained for years and even decades in the laboratory, MCTG13d can be considered to be a member of the indigenous bacterial flora associated with CCAP1121/2, CCAP1061/25, and CCMP1324 in which this bacterium was quantifiably detected.
The enrichments of L. polyedrum CCAP1121/2 incubated on the four different PAHs revealed that this algal strain harbors a diverse community of associated PAH-degrading bacteria. Photosynthetically enhanced biodegradation of toxic aromatic pollutants has been demonstrated using artificial algal-bacterial consortia (5, 33, 42, 53). However, there is little known about these processes in natural algal-bacterial assemblages. In a recent report by Teira et al. (48) investigating the influence of PAH concentrations and seasonal variations on the abundance of Cycloclasticus in seawater samples taken from the middle Ria de Vigo (Spain), a correlation between periods of phytoplankton activity and an increase in the numbers of Cycloclasticus and PAH degradation was demonstrated. The generation of oxygen by the algae has been postulated as a mechanism to enhance the bacterial degradation of the hydrocarbons in these studies. However, a critical assessment to explain the role of the algae in the degradation process has not previously been made.
Interestingly, strain MCTG13d and the other novel PAH degraders that we have identified from phytoplankton are not well represented in the current pool of 16S rRNA sequence data that are currently available (i.e., in the GenBank and RDP databases). Based on the information that we already know about these organisms—i.e., association with phytoplankton and growth under nutrient-limited conditions—it may be inferred that their existence in the marine environment is confined to a life associated with certain species of phytoplankton and in low abundance reminiscent of background populations of oligotrophic planktonic bacteria. Events that can lead to the eutrophication of its surrounding environment (e.g., an oil spill) might be expected to negatively impact this organism, at least to possibly negate its ability to undergo cell division. More work, though, will be needed to more fully understand their association with their eukaryotic hosts, as well as their function and ecology in the wider context of oil degradation in the ocean. The demonstration that laboratory cultures of phytoplankton are a source of novel PAH-degrading bacteria should encourage more detailed study of these organisms and their occurrence with different species of phytoplankton comprising the different lineages, as well as to investigate the diversity of PAH degraders associated with natural algal-bacterial communities in the ocean.
Taxonomy.
Phylogenetic analysis revealed that strain MCTG13d belongs to a distinct phyletic line in the class Gammaproteobacteria which is represented by a clade of mainly uncultivated bacteria. Hitherto, this clade has been defined by just one cultivated type strain, Porticoccus litoralis IMCC2115T (36). Based on 16S rRNA analysis, IMCC2115T was found to be the closest relative to MCTG13d (96.5% sequence identity). Hence, phylogenetic inference did not strongly support the assignment of strain MCTG13d into a separate genus. Strain MCTG13d, therefore, warranted further characterization in order to determine its affiliation or assignment as a new bacterial taxon. As shown in Table 2, there are clear differences between the fatty acid profiles of strain MCTG13d and its closest relative, P. litoralis IMCC2115T. These and other phenotypic and biochemical differences that can be used to delineate strain MCTG13d from IMCC2115T are shown in Table 3. Several fatty acids were found in strain IMCC2115T that were absent in MCTG13d. Notably, two of the three major fatty acids of strain MCTG13d (i.e., C16:1ω7c and C18:1ω7c) were not present in P. litoralis IMCC2115T. Other features distinguishing between the two strains include differences in cell shape, their differential capacity to utilize sugars, their NaCl requirement for growth, and a difference in their DNA G+C content.
Table 3.
Key phenotypic characteristics that differentiate strain MCTG13d from its closest relative, Porticoccus litoralis IMCC2115T (36)b
| Characteristic | Strain 1 | Strain 2 |
|---|---|---|
| Cell morphology | Rod | Coccus |
| Flagellation | Single polar flagellum | No flagella |
| Optimal pH for growth | 7.5–8.5 | 7.0–8.0 |
| pH range for growth | 6.5–9.0 | 5.0–11.0 |
| Temp range for growth (°C) | 10–37 | 15–42 |
| Optimal growth temp (°C) | 15 | 20–25 |
| Maximum NaCl concn (%) at which growth occurs | 6.0 | 5.0 |
| NaCl range for growth (%) | 0.0–6.0 | 1.5–5.0 |
| Nitrate | + | − |
| Catalase activity | + | − |
| Utilization of | ||
| Mannitol | − | + |
| Fructose | − | + |
| Glucose | − | + |
| Arabinose | − | + |
| Indole production | + | − |
| DNA G+C (mol%) | 54.9 | 47.8 |
| Major fatty acids (% of total) | C16:1ω7c (31.5), C18:1ω7c (29.5), C16:0 (27.8) | ai-C15:0a (67.6), ai-C17:0a (14.4) |
The abbreviation “ai” denotes anteiso methyl branching.
Strains: 1, strain MCTG13d; 2, Porticoccus litoralis IMCC2115T. +, positive; −, negative.
Based also on 16S rRNA identity, members belonging to the genera Microbulbifer and Marinimicrobium are the next closest relatives to MCTG13d (90.4 to 93.7% sequence identity). However, important phenotypic and biochemical properties can be used to distinguish MCTG13d from these other genera. For example, one of the major fatty acids in MCTG13d was C16:1ω7c, which is either absent or found in low abundance (<5.4%) in members of the genus Microbulbifer. All members of Microbulbifer contain iso-C15:0 (5.7 to 43.5%), iso-C17:1 (6.5 to 23.0%), C15:0 (0.7 to 6.4%), C17:0 (0.2 to 9.7%), iso-C17:0 (0.9 to 13.5), and C19:0 cyclo (0.5 to 5.3), whereas these acids were not identified in strain MCTG13d. Members belonging to the genus Marinimicrobium contain cyclo C19:0ω8c as a major fatty acid, whereas this acid was not identified in strain MCTG13d. Except for Microbulbifer marinus and Microbulbifer yueqingensis, all members of Microbulbifer, as well as all members of Marinimicrobium, require NaCl for growth, whereas MCTG13d can grow in the absence of NaCl. Unlike MCTG13d, all members of these genera are able to grow on sugars as well as on nutrient-rich marine medium. The optimum growth temperature for members of Microbulbifer is ≥25°C, whereas that for MCTG13d was found to be considerably less at 15°C. Unlike MCTG13d, all species of Microbulbifer are unable to produce indigo.
Based on polyphasic analysis, strain MCTG13d is distinct from any previous reported bacterial isolate and represents a new species of the genus Porticoccus, for which we propose the name Porticoccus hydrocarbonoclasticus.
Description of Porticoccus hydrocarbonoclasticus sp. nov.
Porticoccus hydrocarbonoclasticus (hy′dro.car.bo.no.clas′ti.cus. M.L. part. adj. hydrocarbonoclastic, hydrocarbon dismantling). Cells are Gram-negative motile rods (1.0 to 2.0 by 0.5 to 0.6 μm) with a single polar flagellum. Colonies are pale yellow-green, slightly raised with undulate margins on defined (ONR7a) medium amended with a hydrocarbon substrate. Oxidase and catalase positive. Lipase and gelatinase negative. Phosphatase activity is positive. Reduction of nitrate to nitrite is positive. Many cells harbored intracellular granules that fluoresced after staining with Nile Blue, although these were not accumulation of PHB. Cells do not form endospores, and some formed blebs at the cell surface. The cells are strictly aerobic. They exhibit a narrow substrate spectrum, showing a preference for utilizing hydrocarbons, including phenanthrene, anthracene, pyrene, fluorene, and n-hexadecane, as sole or principal sources of carbon and energy. Methane and methanol are not utilized as a sole carbon source. Growth is observed on acetate. Sugars are not utilized. The strain does not require Na+ or other special nutrition for growth. Growth occurs at 10 to 37°C (optimum, 15°C) and at pH 6.5 to 9.0 (optimum, 7.5 to 8.5). Growth is detected at 0 to 6% NaCl. The major fatty acids are C16:1ω7c, C18:1ω7c, and C16:0. The predominant isoprenoid quinone is Q-8. The DNA G+C content of the type species is 54.9 mol%. Habitat is certain species of marine diatoms and dinoflagellates. The type and only species is Porticoccus hydrocarbonoclasticus. The type strain is MCTG13dT (=ATCC BAA-2274).
ACKNOWLEDGMENTS
This research was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the 7th European Community Framework Programme. Partial support was also provided through the U.S. National Institute of Environmental Health Sciences, grant 5 P42ES005948.
We thank Wallace Ambrose (UNC's Analytical and Nanofabrication Laboratory) for providing valuable assistance with the preparation of samples for electron microscopy and in the capture of electron micrographs. We also thank Danny Holdsworth, who managed the CSIRO gas chromatography (GC) and GC-mass spectrometry (GC-MS) facility. We also especially thank two independent, anonymous reviewers for making invaluable comments about the manuscript.
Footnotes
Published ahead of print 2 December 2011
REFERENCES
- 1. Agency for Toxic Substances and Disease Registry 2007. CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry, Atlanta, GA: http://www.atsdr.cdc.gov/SPL/index.html [Google Scholar]
- 2. Amann RI, et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andelman JB, Suess MJ. 1970. Polynuclear aromatic hydrocarbons in the water environment. Bull. World Health Organ. 43:479–508 [PMC free article] [PubMed] [Google Scholar]
- 4. Binark N, Guven KC, Gezgin T, Unlu S. 2000. Oil pollution of marine algae. Bull. Environ. Contamin. Toxicol. 64:866–872 [DOI] [PubMed] [Google Scholar]
- 5. Borde X, et al. 2003. Synergistic relationships in algal-bacterial microcosms for the treatment of aromatic pollutants. Bioresour. Technol. 86:293–300 [DOI] [PubMed] [Google Scholar]
- 6. Buesseler KO. 1998. The decoupling of production and particulate export in the surface ocean. Global Biogeochem. Cycles 12:297–310 [Google Scholar]
- 7. Cho JC, et al. 2007. Polyphyletic photosynthetic reaction centre genes in oligotrophic marine Gammaproteobacteria. Environ. Microbiol. 9:1456–1463 [DOI] [PubMed] [Google Scholar]
- 8. Cole JR, et al. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169–D172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyksterhouse SE, Gray JP, Herwig RP, Cano LJ, Staley JT. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116–123 [DOI] [PubMed] [Google Scholar]
- 10. Ensley BD, et al. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in biosynthesis of indigo. Science 222:167–169 [DOI] [PubMed] [Google Scholar]
- 11. Evans KM, Gill RA, Robotham PWJ. 1990. The PAH and organic content of sediment particle size fractions. Water Air Soil Pollut. 51:13–31 [Google Scholar]
- 12. Geiselbrecht A, Hedlund BP, Tichi MA, Staley JT. 1998. Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl. Environ. Microbiol. 64:4703–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 14. Glatz RE, Lepp PW, Ward BB, Francis CA. 2006. Planktonic microbial community composition across steep physical/chemical gradients in permanently ice-covered Lake Bonney, Antarctica. Geobiology 4:53–67 [Google Scholar]
- 15. Gol'man LP, Mikhaseva MF, Reznikov VM. 1973. Infrared spectra of lignin preparations of pteridophytes and seaweeds. Dokl. Akad. Nauk BSSR 17:1031–1033 [Google Scholar]
- 15a. Guillard RR, Ryther JH. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can. J. Microbiol. 8:229–239 [DOI] [PubMed] [Google Scholar]
- 15b. Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60 In Smith WL, Chanley MH. (ed), Culture of marine invertebrate animals. Plenum Press, New York, NY [Google Scholar]
- 16. Gunnison D, Alexander M. 1975. Basis for the resistance of several algae to microbial decomposition. Appl. Microbiol. 29:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harayama S, Kasai Y, Hara A. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205–214 [DOI] [PubMed] [Google Scholar]
- 18. Hayat MA, Miller SE. 1990. Negative staining. McGraw-Hill Publishing Co, New York, NY [Google Scholar]
- 19. Head IM, Jones DM, Roling WFM. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173–182 [DOI] [PubMed] [Google Scholar]
- 20. Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN. 2005. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71:3483–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kasai Y, Kishira H, Harayama S. 2002. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl. Environ. Microbiol. 68:5625–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kiyohara H, Nagao K, Yana K. 1982. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl. Environ. Microbiol. 43:454–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalewska G. 1999. Phytoplankton—the main factor responsible for transport of polynuclear aromatic hydrocarbons from water to sediments in the Southern Baltic ecosystem. ICES J. Mar. Sci. 56:219–222 [Google Scholar]
- 24. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid sequencing techniques in bacterial systematics. John Wiley & Sons, New York, NY [Google Scholar]
- 25. Lane DJ, et al. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82:6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsen H. 1986. Halophilic and halotolerant microorganisms—an overview and historical perspective. FEMS Microbiol. Rev. 39:3–7 [Google Scholar]
- 27. Lee RF, Gardner WS, Anderson JW, Blayblack JF, Barwell-Clarke J. 1978. Fate of polycyclic aromatic hydrocarbons in controlled ecosystem enclosures. Environ. Sci. 17:282–286 [Google Scholar]
- 28. MacFaddin JF. 2000. Biochemical tests for identification of medical bacteria, 3rd ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Maruyama A, et al. 2003. Dynamics of microbial populations and strong selection for Cycloclasticus pugetii following the Nakhodka oil spill. Microb. Ecol. 46:442–453 [DOI] [PubMed] [Google Scholar]
- 30. Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167 [Google Scholar]
- 31. Millard ES, Halfon E, Minns CK, Charlton CC. 1993. Effect of primary productivity and vertical mixing on PCB dynamics in planktonic model ecosystems. Environ. Toxicol. Chem. 12:931–946 [Google Scholar]
- 32. Miller MR, Bridle AR, Nichols PD, Carter CG. 2008. Increased elongase and desaturase gene expression with stearidonic acid enriched diet does not enhance long-chain (n-3) content of seawater Atlantic salmon (Salmo salar L.). Nutrition 138:2179–2185 [DOI] [PubMed] [Google Scholar]
- 33. Muñoz R, Guieysse B, Mattiasson B. 2003. Phenanthrene biodegradation by an algal-bacterial consortium in two-phase partitioning bioreactors. Appl. Microbiol. Biotechnol. 61:261–267 [DOI] [PubMed] [Google Scholar]
- 34. Nemirovskaya IA. 2004. Uglevodorody v okeane (sneg–led–voda–vzves'–donnye osadki). Nauch. Mir, Moscow, Russia [Google Scholar]
- 35. Nemirovskaya IA. 2007. Hydrocarbons in the bottom sediments of the Northern Divina Estuary. Water Resource 34:699–706 [Google Scholar]
- 36. Oh H-M, Kim H, Kim K-M, Min G-S, Cho J-C. 2010. Porticoccus litoralis gen. nov., sp. nov., a gammaproteobacterium isolated from the Yellow Sea. Int. J. Syst. Evol. Microbiol. 60:727–732 [DOI] [PubMed] [Google Scholar]
- 37. Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 38. Pastuska G. 1961. Die Kieselgelschicht-Chromatographie von Phenolen und Phenolcarbensiuren. I. Z. Anal. Chem. 179:355–358 [Google Scholar]
- 39. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pruesse E, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Repeta DJ, Hartman NT, John S, Jones AD, Goericke R. 2004. Structure elucidation and characterization of polychlorinated biphenyl carboxylic acids as major constituents of chromophoric dissolved organic matter in seawater. Environ. Sci. Technol. 38:5373–5378 [DOI] [PubMed] [Google Scholar]
- 42. Safanova ET, Dmitrieva IA, Kvitko KV. 1999. The interaction of algae with alcanotrophic bacteria in black oil decomposition. Res. Conserv. Recycl. 27:193–201 [Google Scholar]
- 43. Scott LT, et al. 1999. Geodesic polyarenes with exposed concave surfaces. Pure Appl. Chem. 71:209–219 [Google Scholar]
- 44. Simoneit BRT. 1985. Hydrothermal petroleum: genesis, migration, and deposition in Guaymas Basin, Gulf of California. Can. J. Earth Sci. 22:1919–1929 [Google Scholar]
- 45. Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. 2006. Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol. 8:1736–1745 [DOI] [PubMed] [Google Scholar]
- 46. Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. 2007. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 73:2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stringfellow WT, Aitken MD. 1995. Competitive metabolism of naphthalene, methylnaphthalenes, and fluorene by phenanthrene-degrading pseudomonads. Appl. Environ. Microbiol. 61:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teira E, et al. 2007. Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ. Microbiol. 9:2551–2562 [DOI] [PubMed] [Google Scholar]
- 49. Thomas O, Burgess C. 2007. Techniques and instrumentation in analytical chemistry, vol 27 UV-visible spectrophotometry of water and wastewater. Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 50. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL_X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tillett D, Neilan BA. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251–258 [Google Scholar]
- 52. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warshawsky D, LaDow K, Schneider J. 2007. Enhanced degradation of benzo[a]pyrene by Mycobacterium sp. in conjunction with green alga. Chemos 69:500–506 [DOI] [PubMed] [Google Scholar]
- 54. Wilmotte A, Van der Auwera G, De Wachter R. 1993. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF') strain PCC7518, and phylogenetic analysis. FEMS Microbiol. Lett. 317:96–100 [DOI] [PubMed] [Google Scholar]
- 55. Witt G. 1995. Polycyclic aromatic hydrocarbons in water and sediment of the Baltic Sea. Mar. Pollut. Bull. 31:237–248 [DOI] [PubMed] [Google Scholar]
- 56. Witt G. 2002. Occurrence and transport of polycyclic aromatic hydrocarbons in the water bodies of the Baltic Sea. Mar. Chem. 79:49–66 [Google Scholar]
- 57. Wong CS, et al. 1995. Accumulation, inventory, and diagenesis of chlorinated hydrocarbons in Lake Ontario sediments. Environ. Sci. Technol. 29:2661–2672 [DOI] [PubMed] [Google Scholar]
- 58. Yakimov MM, Timmis KN, Golyshin PN. 2007. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18:257–266 [DOI] [PubMed] [Google Scholar]
- 59. Zelibor JL, Romankiw L, Hatcher PG, Colwell RR. 1988. Comparative analysis of the chemical composition of mixed and pure cultures of green algae and their decomposed residues by 13C nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 54:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zeng R, Zhao J, Zhang R, Lin N. 2005. Bacterial community in sediment from the Western Pacific ‘Warm Pool’ and its relationship to environment. Sci. China Ser. D Earth Sci. 48:282–290 [Google Scholar]