Abstract
In China, rubella vaccination was introduced into the national immunization program in 2008, and a rubella epidemic occurred in the same year. In order to know whether changes in the genotypic distribution of rubella viruses have occurred in the postvaccination era, we investigate in detail the epidemiological profile of rubella in China and estimate the evolutionary rate, molecular clock phylogeny, and demographic history of the predominant rubella virus genotypes circulating in China using Bayesian Markov chain Monte Carlo phylodynamic analyses. 1E was found to be the predominant rubella virus genotype since its initial isolation in China in 2001, and no genotypic shift has occurred since then. The results suggest that the global 1E genotype may have diverged in 1995 and that it has evolved at a mutation rate of 1.65 × 10−3 per site per year. The Chinese 1E rubella virus isolates were grouped into either cluster 1 or cluster 2, which likely originated in 1997 and 2006, respectively. Cluster 1 viruses were found in all provinces examined in this study and had a mutation rate of 1.90 × 10−3 per site per year. The effective number of infections remained constant until 2007, and along with the introduction of rubella vaccine into the national immunization program, although the circulation of cluster 1 viruses has not been interrupted, some viral lineages have disappeared, and the epidemic started a decline that led to a decrease in the effective population size. Cluster 2 viruses were found only in Hainan Province, likely because of importation.
INTRODUCTION
Rubella had been considered a relatively benign infection in children and adults. However, the rubella virus is a potent, highly infectious, and teratogenic agent. Infection in the first trimester of pregnancy can lead to fetal death or various birth defects, including deafness, cataracts, and heart disease, known as congenital rubella syndrome (CRS) (11, 16). At least 100,000 cases of CRS occur each year globally (24). An effective rubella and CRS surveillance program has not yet been established nationwide in China. Thus, there is a lack of information on the prevalence of congenital rubella-associated defects and disabilities. The estimated number of CRS cases in China in 2005 was at least 20,000 (17).
The Chinese immunization program has a long history, and a national immunization program first established in 1978 was a routine immunization schedule that included 4 basic vaccines against 6 contagious diseases (tuberculosis, diphtheria, neonatal tetanus, whooping cough, poliomyelitis, and measles). By 1988, 1990, and 1995, China achieved its universal childhood immunization goals of >85% coverage by province, county, and village, respectively. Hepatitis B vaccine was added into the routine immunization schedule in 2002, and in 2008, the national immunization schedule was expanded from 5 vaccines against 7 contagious diseases to 14 vaccines against 15 infectious diseases, and rubella vaccine was introduced into the national immunization schedule at this opportunity.
The Chinese immunization program is widely regarded to be one of the country's most successful public health endeavors under challenging conditions: (i) a very large population (estimated population, 1.336 billion in 2009; proportion of population aged <15 years, 18.5%; birth rate, 12.09‰; birth cohort, estimated at 16.46 million); (ii) high population density with an unbalanced distribution, average of 134 persons per square kilometer; eastern coastal area, >400 persons per square kilometer; central area, about 200 persons per square kilometer; western plateau area, <10 persons per square kilometer; and (iii) hard-to-reach populations in the mountainous area (32). Despite the challenging conditions, all Chinese children have the opportunity to receive vaccination against these infectious diseases free of charge, and vaccine-preventable diseases targeted by the expanded program on immunization are at low levels of incidence overall.
Rubella is one of the preventable contagious diseases, and with the first introduction of the rubella vaccine in 1969, a significant reduction in morbidity and mortality has occurred. The cycle of rubella epidemics has been broken; pandemics have been prevented, and the number of cases of CRS has been reduced (22). Rubella vaccination was instituted in the Chinese national immunization program in 2008. Prior to this, 2 types of rubella vaccine, including the imported RA27/3 and domestic BRDII vaccines, had been used in several large cities in eastern China since the 1990s (35).
The rubella virus is the sole member of the Rubivirus genus in the Togaviridae family and is a positive-sense RNA virus with a genome of 9,762 nucleotides (nt) which encodes 2 nonstructural polypeptides (P150 and P90) and 3 structural polypeptides (C, E2, and E1). A 739-nt region (nt 8731 to 9469, amino acids 159 to 404 within the E1 protein) within the E1 glycoprotein contains important functional domains, including a hemagglutination-inhibiting and -neutralizing epitope, and antigenic sites (5, 11) and has been designated to be the minimum acceptable sequence window for assigning genotypes by comparison with reference virus sequences (29). Thus far, 2 clades including 13 genotypes have been described: 9 genotypes (1B, 1C, 1D, 1E, 1F, 1G, 2A, 2B, and 2C) are recognized, and 4 genotypes (1a, 1h, 1i, and 1j) are provisional.
The results of molecular epidemiology studies of rubella viruses in China indicate that 5 (1a, 1E, 1F, 2A, and 2B) out of the 13 genotypes have been present in China since 1979, and cocirculation of these different genotypes has been observed (34). Before the introduction of the rubella vaccine into the national immunization program in 2008, rubella epidemics were common. According to the rubella surveillance data, rubella epidemics occurred approximately every 6 to 8 years in China. A shift in the predominant genotype from 1F and 2B to 1E coincided with the 2001 rubella epidemic, and 1E subsequently became the most common genotype (34).
In this study, we examined whether changes in the genotypic distribution of rubella viruses have occurred in the postvaccination era, particularly following the recent rubella epidemic peak. The epidemiological profile of rubella and the origin and evolution of the predominant rubella viruses circulating in China are described in detail. We sought to provide data that would contribute to our genetic knowledge base on these viruses and help to form a rational scientific basis for the prevention and control of rubella and CRS in China. We also used a Bayesian approach to estimate the evolutionary rates and the demographic history of rubella viruses in a series of isolates collected between 1995 and 2009.
MATERIALS AND METHODS
Rubella incidence data sources.
The number of all clinically diagnosed and laboratory-confirmed rubella and measles cases and the annual rubella incidence rates were taken directly from reports from the National Notifiable Disease Reporting System (NNDRS), which has been in place since the 1950s and covers all hospitals in mainland China. Hospitals reported cases by posting a card to the county Center for Disease Control and Prevention (CDC), and the data were collected monthly in the county CDCs and then submitted through the prefectural and provincial CDC and finally reached the China CDC. On the basis of the official publication of the China Ministry of Health Country Norm of Information Report and Management on Public Health Emergency Events, a rubella outbreak was defined to be 10 or more laboratory-confirmed cases occurring in the same school, kindergarten, village, or community within 1 week. Nationwide rubella information could be obtained starting from 2004, when rubella case reporting was introduced into the NNDRS, and it was used as the official source of data on the number of reported rubella cases since then.
Viruses.
All rubella virus strains were isolated from throat swab specimens and urine samples from patients with clinically suspected rubella within 7 days after rash onset. The samples were collected between 2008 and 2009 in 15 provinces (Anhui, Hainan, Henan, Jilin, Jiangxi, Liaoning, Inner Mongolia, Ningxia, Shandong, Sichuan, Fujian, Gansu, Guangdong, Shanxi, and Tianjin) which covered the east, central, and western regions of China. Clinical specimens were inoculated onto monolayers of African green monkey kidney (Vero) cells, Vero/SLAM cells, or rabbit kidney (RK13) cells, according to standard methods (35). Cells inoculated with clinical specimens were incubated at 35°C for 7 days. The culture supernatant was harvested and used to inoculate fresh cells for up to 2 additional passages. The presence of viral RNA was detected via reverse transcription-PCR (RT-PCR), which amplified a 185-nt fragment of the E1 coding region as previously described (35).
RT-PCR amplification and sequence determination.
RT-PCR was performed using a Titanium one-step RT-PCR kit (BD Bioscience, Palo Alto, CA) to amplify a 1,107-nt (nt 8656 to 9762) product containing the 739-nt WHO-recommended sequence window (nt 8731 to 9469) as previously described (3). The Titanium Taq DNA polymerase used in this kit contains a thermostable DNA polymerase and TaqStart antibody, which provides an integrated hot start for increased specificity and yield, and this powerful PCR feature made amplification of GC-rich templates possible by the addition of GC-Melt reagent, which destabilizes base pairing in GC-rich regions. The PCR products were purified using a QIAquick gel extraction kit (Qiagen, Valencia, CA), and the amplicons were bidirectionally sequenced using an ABI Prism 3100 genetic analyzer (Applied Biosystems, Hitachi, Japan).
Phylogenetic analysis.
The 739-nt E1 sequences of the rubella virus strains were aligned, and phylogenetic analysis using neighbor joining (NJ) and maximum likelihood (ML) was performed in order to gain further insights into molecular epidemiology using the MEGA (version 5.03) program (Sudhir Kumar, Arizona State University, Tempe, AZ) (25). Tamura-Nei (which takes into account unequal base frequencies, variable transition frequencies, equal transversion frequencies, and unequal nucleotide substitution rates among sites) and Kimura two-parameter (which takes into account the fact that transitions should occur more often than transversions at equal base frequencies) evolutionary models were selected for rubella virus phylogenetic analysis. The branch lengths of the NJ tree were determined from the topology of the trees and were obtained by majority-rule consensus among 1,000 bootstrap replicates, and bootstrap values greater than 80% were considered statistically significant for grouping. For ML trees, a heuristic search was performed with a subtree pruning-regrafting (SPR) branch-swapping algorithm.
Evolutionary analysis based on Bayesian MCMC method.
The evolutionary rate, the molecular clock phylogeny, and the demographic history of the 1E rubella virus global genotypes were coestimated using the Bayesian Markov chain Monte Carlo (MCMC) method in the BEAST (version 1.6.1) program (9), and the time of the most recent common ancestor (tMRCA) with a 95% highest posterior density (HPD) was estimated. A Bayesian skyline plot (BSP; a nonparametric piecewise-constant model) was used under both strict and relaxed (uncorrelated lognormal distributed [UCLD]) clock conditions to estimate the demographic history. BSP uses an MCMC method that allows estimates of effective population size over time with credibility intervals at every time depending on errors due to the phylogeny reconstruction and the stochastic nature of the coalescent process (10).
In order to reduce the computation load, sequences with high homogeneity and identical isolation years were deleted. Data were analyzed under both the Hasegawa-Kishino-Yano (HKY) and the general time reversible (GTR) nucleotide substitution models with a gamma distribution of among-site rate variation. Two different models of rate variation among branches were implemented in our analysis: the strict clock and the UCLD relaxed molecular clock. Both constant and exponential growth (EG) population size coalescences were used as tree priors. For each model, the MCMC chain was run for 30,000,000 steps and sampled every 1,000 steps. Uncertainty in the estimates was indicated by 95% HPD intervals. The parameter outputs generated by Bayesian MCMC runs and convergence on the basis of the effective sampling size (ESS) after a 10% burn-in were analyzed by the TRACER (version 1.5) program, and the trees were summarized in a target tree by the Tree Annotator program included in the BEAST package by choosing the tree with the maximum product of posterior probabilities after a 10% burn-in.
Nucleotide sequence accession numbers.
The nucleotide sequences of 53 representative viruses of the 118 Chinese rubella virus strains that were isolated in this study have been deposited in the GenBank database under accession numbers JF702819 to JF702871. An additional 35 sequences of rubella viruses from China collected from 2000 to 2007 were retrieved from the GenBank database and had accession numbers FJ875029, FJ875033, FJ875036 to FJ875044, FJ875047 to FJ875056, and FJ875058 to FJ875071.
RESULTS
Epidemiological profile of rubella in China.
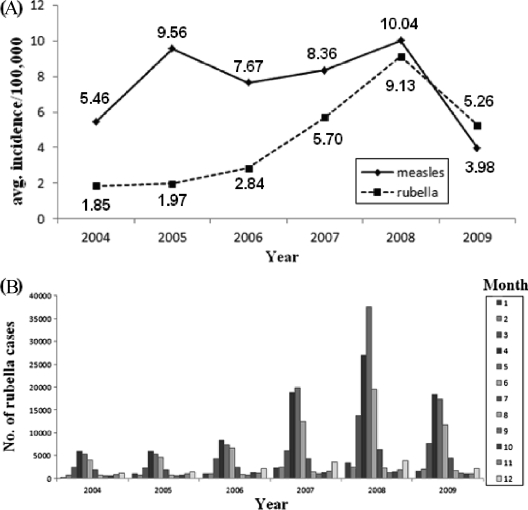
Since 2004, the average reported incidence rate of rubella cases increased from 1.85/100,000 (24,051 cases) to 9.13/100,000 (120,614 cases) in 2008 and then decreased to 5.26/100,000 (69,821 cases) in 2009 (Fig. 1a). Compared to measles, the incidence of rubella was lower from 2004 to 2008, while it was slightly higher in 2009 (measles incidence, 3.98/100,000). From 2004 to 2009, rubella cases were reported in all 12 months, and most of them were concentrated between March and June, but small peaks also occurred in December (Fig. 1b).
Fig 1.
Reported rubella cases in China by year of onset (A) and by month of onset (B), 2004 to 2009.
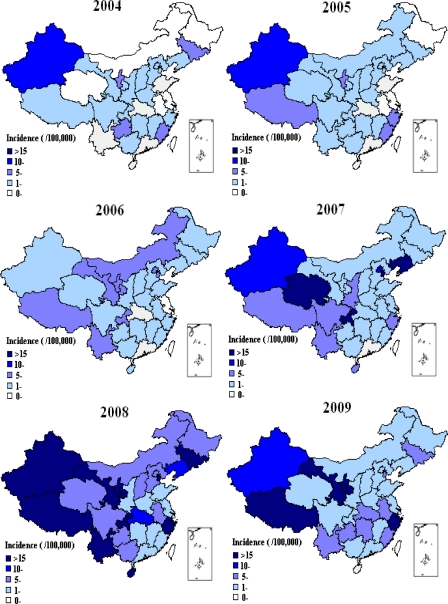
In 2004 and 2005, an incidence of rubella of >10/100,000 occurred only in Xinjiang Province (western China). The incidence of rubella in all 31 provinces was <10/100,000 in 2006. However, the number of rubella cases increased in 2007, with incidence rates of >10/100,000 in 5 of 31 provinces. Tianjin and Liaoning Provinces (eastern China) and Chongqing and Qinghai Provinces (western China) had incidence rates of >15/100,000. Among these, Liaoning had the highest incidence, at 48.90/100,000. The nationwide rubella epidemic continued and reached a peak in 2008. In 2008, 10/31 provinces had a rubella incidence of >15/100,000 (Tianjin, Zhejiang, and Hainan [eastern China], Jilin [central China], Chongqing, Yunnan, Tibet, Gansu, Ningxia, and Xinjiang [western China]). The epidemic was widespread throughout China, with the highest incidence being in Tianjin (49.98/100,000). In 2009, the epidemic subsided, with an incidence of >15/100,000 limited to 4 provinces, including Zhejiang (eastern China), Tibet, Gansu, and Ningxia (western China) Provinces, and with Tibet having the highest incidence (58.15/100,000) (Fig. 2).
Fig 2.
Geographic distribution of rubella cases in China, 2004 to 2009.
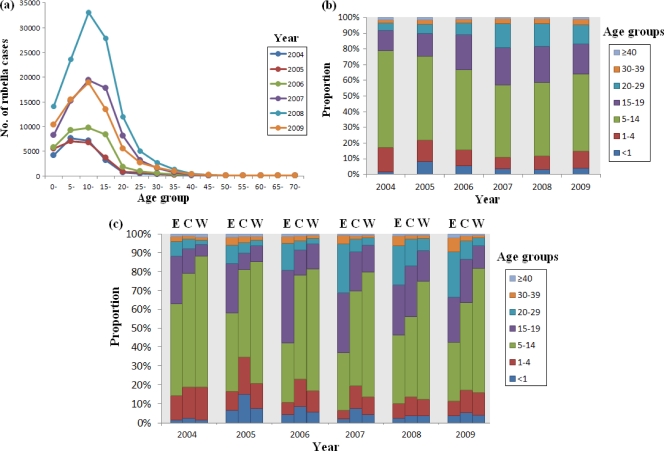
Between 2004 and 2009, the total number of reported rubella outbreaks was approximately 972, of which 931 (95.78%) occurred in schools. The reported rubella cases were concentrated in those under 15 years of age (Fig. 3a), with the proportions being 78.70%, 75.34%, 66.66%, 57.16%, 58.69%, and 64.05% for each year between 2004 and 2009, respectively (Fig. 3b). The proportion of reported rubella cases within the 15- to 39-year-old age group increased each year from 2004 (19.77%) to 2007 (42%), maintained a similar level in 2008 (40.51%), and then decreased to 34.71% in 2009 (Fig. 3b). The proportion of rubella cases in individuals aged <15 years among the eastern, central, and western regions of China was quite varied. Between 2004 and 2009, the proportions of rubella cases in individuals aged <15 years in eastern China were 62.84%, 58.07%, 42.04%, 37.02%, 46.40%, and 42.45%, respectively, for each of these years. These were relatively lower than those in central and western China, whereas the proportion of reported rubella cases within the 15- to 39-year-old age group in eastern China was higher than that in central and western China (35.80%, 40.27%, 56.63%, 62.07%, 52.51%, and 55.57%, respectively, for each of these years) (Fig. 3c).
Fig 3.
Age distribution of rubella cases in China, 2004 to 2009. (a) Age specificity of rubella cases in mainland China from 2004 to 2009, as reported by NNDRS; (b) proportion of rubella incidences in different age groups from 2004 to 2009 in mainland China, as reported by NNDRS; (c) rubella incidences in different age groups from 2004 to 2009 in eastern (E), central (C), and western regions (W) of China, as reported by NNDRS.
Phylogenetic analysis of rubella viruses.
A total of 118 rubella virus isolates were collected from throat swab or urine specimens in 15 of 31 provinces in China during the rubella epidemic between 2008 and 2009. The sequences of the 739-nt region within the E1 gene from virus isolates from the outbreaks were very similar (<0.54% difference) or identical, so 53 rubella viruses were selected (randomly selected on the basis of their genetic relationships) as representative viruses for phylogenetic analysis. All viruses were named according to the WHO systematic nomenclature for rubella viruses. The nomenclature system of strain naming includes epidemiological information that is essential for interpretation of the molecular data. Strains or sequences are designated either RVi, rubella virus isolate in cell culture, or RVs, rubella virus sequence derived from RNA extracted from clinical material. Other information to be included in the strain/sequence name include town/city of isolation, country, date of specimen collection by epidemic week and year, isolate number, genotype, and special designation for a sequence if it derived from CRS cases.
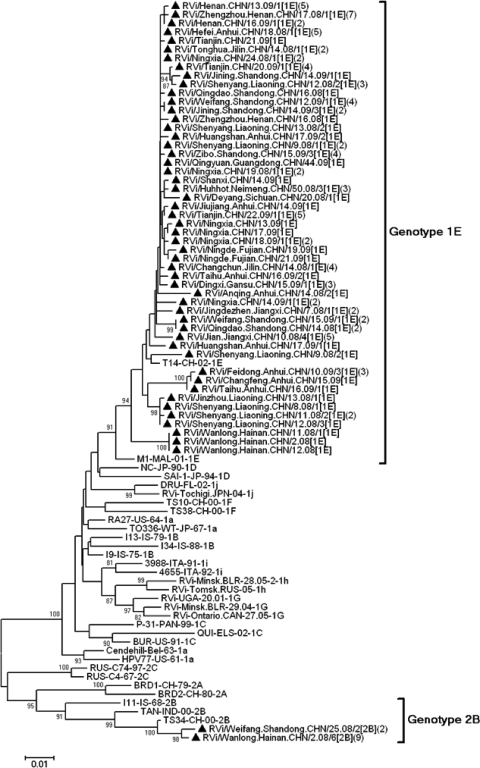
Both NJ and ML analyses with the Tamura-Nei and Kimura two-parameter models gave similar results. The 53 sequences of Chinese rubella viruses collected during 2008 and 2009 were divided into 2 genotypes, genotype 1E (51 strains, 96.2%) and genotype 2B (2 strains, 3.8%), indicating that 1E was predominant. These genotype assignments were supported by high bootstrap scores (Fig. 4).
Fig 4.
Phylogenetic analysis of sequences of 53 representative Chinese rubella viruses from 2008 to 2009 compared to the WHO reference sequences based on the WHO standard sequence window within the E1 gene (nt 8731 to 9469). Numbers in parentheses are the numbers of identical or similar sequences found in the same outbreak. The genotype 1E rubella viruses isolated during 2008 and 2009 are indicated by solid triangles.
Genotype 1E rubella viruses were found in all 15 provinces, and a 96.7 to 100% nucleotide sequence identity (99.1% to 100% amino acid sequence identity) was found among these viruses. Identical and similar sequences of genotype 1E rubella viruses collected during 2008 and 2009 were found in different provinces, including Guangdong, Liaoning (eastern China), Anhui, Jilin (central China), and Ningxia (western China) (Fig. 4); this indicates that highly similar or identical 1E rubella virus sequences circulated in the various provinces with no apparent geographic restriction.
Origin, evolutionary rate, molecular clock phylogeny, and demographic history.
The sequences of the 739-nt window within the E1 gene of the representative Chinese rubella virus isolates recovered during 2008 and 2009 (n = 51), from 2001 to 2007 (n = 35, from 13 provinces), and from 14 other countries (n = 19), including the Bahamas (33), Canada (33), Germany (33), Italy (33), United States (33), Suriname (33), France (26), Tunisia (26), Belarus (12), United Kingdom (14), Russia, Ukraine, Sudan, and Kazakhstan (Table 1), representing the global genotype 1E rubella virus population, were selected for divergence time and substitution rate estimation using the Bayesian MCMC method. Different models were used for data analysis, and it was found that UCLD with EG fit our data best, while the HKY and GTR nucleotide substitution models had no significant impact on the analysis (Table 2). The coefficients of variation of the estimated evolutionary rates among branches estimated by the HKY model and GTR model were 0.19 (95% HPD, 0.01 to 0.44) and 0.20 (95% HPD, 0.01 to 0.46), respectively, indicating that rate heterogeneity exists among the different branches. Analysis of the partial E1 fragment sequences showed that the substitution rate of the 1st and 2nd codon positions was significantly lower than that calculated for the 3rd codon position (mean relative substitution rates, 0.17 with a 95% HPD of 0.08 to 0.28 and 2.69 with a 95% HPD of 2.56 to 2.82, respectively).
Table 1.
Representative genotype 1E strains from countries other than China used in this study
| Virus isolate | Isolation site, yr | GenBank accession no. | Reference |
|---|---|---|---|
| RVs/Caen.FRA/23.95/1E | France, 1995 | FN546967 | 16 |
| FRI-BAH97 | Bahamas, 1997 | AY326359 | 15 |
| DES/MB-CAN97 | Manitoba, Canada, 1997 | AY326358 | 15 |
| G432-GER99 | Germany, 1999 | AF551761 | 15 |
| 6423/PV-ITALY-1997 | Pavia, Italy, 1997 | AY161374 | 15 |
| CAS/FL-USA97 | Florida, USA, 1997 | AY326356 | 15 |
| S633-SUR98 | Suriname, 1998 | AY326363 | 15 |
| CAB/NY-USA00 | New York, USA, 2000 | AY326355 | 15 |
| RVs/Angers.FRA/36.03[1E] | France, 2003 | FN547016 | 16 |
| RVs/TUN/7.03[1E] | Tunisia, 2003 | FN547014 | 16 |
| RVi/Minsk.BLR/24.04/1[1E] | Minsk, Belarus, 2004 | AM258954 | 17 |
| Rvi/Deweim.SDN/24.05[1E]CRS | Sudan, 2005 | FJ775000 | |
| RVi/Minsk.BLR/18.05/2[1E] | Minsk, Belarus, 2005 | AM258955 | 17 |
| RVs/London.GBR/08.05[1E]CRS | London, UK, 2005 | EF210051 | 18 |
| RVs/Lille.FRA/25.05[1E] | France, 2005 | FN547019 | 16 |
| Bar4-108.RUS/06 | Barnaul City, Russia, 2006 | EF421978 | |
| RVs/Chernivtcsi.UKR/13.07[1E] | Ukraine, 2007 | FJ711683 | |
| RVi/Vladimir.RUS/9.08[1E] | Russia, 2008 | FJ711681 | |
| RVi/Almaty.KAZ/13.06[1E] | Kazakhstan, 2006 | FJ711684 |
Table 2.
Origin and evolutionary rate inferred from the Bayesian MCMC method on the 739-nt window within the E1 gene
| Parameter | Mean value of parameter (95% HPD) determined by: |
|
|---|---|---|
| HKY + UCLD + EG | GTR + UCLD + EG | |
| tMRCA (calendar yr) | ||
| Genotype 1E | 1995.2 (1995.1–1995.4) | 1995.2 (1995.1–1995.2) |
| Cluster 1 | 1997.5 (1997.1–1998.5) | 1997.5 (1997.2–1998.4) |
| Cluster 2 | 2006.3 (2006.2–2006.12) | 2006.3 (2006.2–2006.12) |
| Evolutionary rate (10−3 substitutions/site/yr)a | ||
| Mean | 1.65 × 10−3 (1.32 × 10−3–2.00 × 10−3) | 1.66 × 10−3 (1.32 × 10−3–2.01 × 10−3) |
| Cluster 1 evolutionary rate | 1.90× 10−3 (1.44 × 10−3–2.40 × 10−3) | 1.95× 10−3 (1.45 × 10−3–2.48 × 10−3) |
| Coefficient of variation | 0.19 (0.01–0.44) | 0.20 (0.01–0.46) |
Rate of molecular evolution given as numbers of nucleotide substitutions per site per year.
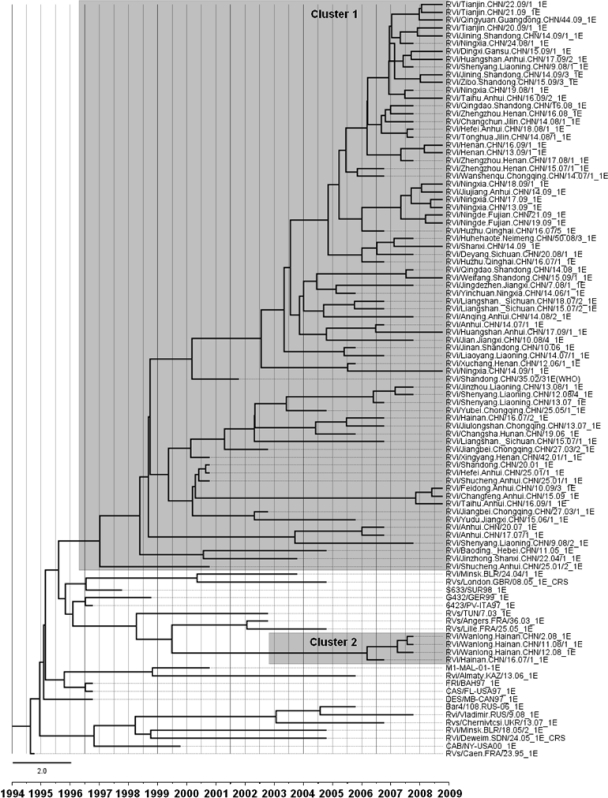
Chinese rubella viruses were distributed in multiple different clusters within genotype 1E, and the sequences of 1E rubella viruses circulating in 2008 and 2009 interdigitated with the sequences from 2001 to 2007. In the molecular clock phylogenetic tree (Fig. 5), the Chinese strains belonged to either cluster 1 or cluster 2, composed of isolates from 2001 to 2009 and 2007 to 2008, respectively. Cluster 1 was unique to China. Most of the viruses (81/86) belonged to cluster 1 and were present in all 19 provinces. Some viruses in cluster 1 (such as isolates from Shandong, Anhui, and Henan Provinces) were isolated in 2001, and 9 years have passed since the last isolation. In contrast, recent isolates from these provinces are located in different lineages, indicating that the older strains may have been eliminated. Five viruses (5/86) from Hainan Province belonged to cluster 2. Although most Chinese genotype 1E rubella viruses had a distinct cluster (cluster 1), some (cluster 2) still group with viruses isolated from the United Kingdom, Belarus, Suriname, Italy, Germany, the United States, Bahamas, Russia, Canada, France, Tunisia, Ukraine, and Kazakhstan recovered between 1997 and 2008 (Fig. 5).
Fig 5.
MCMC tree of the 739-nt region of the E1 sequences of genotype 1E rubella viruses throughout the world visualized in FigTree. The width of a branch reflects the evolutionary rate of individual sequences and their reconstructed ancestors. Genotype 1E rubella viruses from China recovered from 2001 to 2009 segregated into 2 clusters (clusters 1 and 2). Chinese genotype 1E viruses were named according to WHO standard nomenclature.
Different estimated evolutionary rates for different branches were observed, and the mean rate was 1.65 × 10−3 substitutions per site per year (HKY) or 1.66 × 10−3 substitutions per site per year (GTR). Cluster 1 viruses within the 1E genotype had a faster estimated evolutionary rate of 1.90 × 10−3 per site per year (HKY) or 1.95 × 10−3 per site per year (GTR). The most recent common ancestor of global genotype 1E rubella virus may be traced back to February 1995 (range, January 1995 to April 1995). The tMRCA estimates for Chinese clusters were dated to May 1997 (cluster 1) and March 2006 (cluster 2).
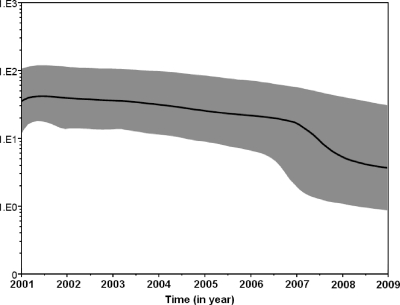
Both the strict and relaxed clock models were implemented using the Bayesian skyline model for population growth, with the second being preferred. The BSP (Fig. 6) showed that the effective number of infections remained constant from the time of the root until about 4 years ago (2001 to 2007), when the epidemic started a decline that led to a decrease in the effective population size, although a rubella epidemic occurred in the epidemiology in 2008.
Fig 6.
Bayesian skyline plot obtained by analyzing the 73 1E genotype rubella virus sequences sampled at different times. The ordinate is the number of effective infections at the indicated time; the abscissa is time (in years). The thick solid line represents the median, and the gray area represents the 95% HPD of the number of effective infections at time estimates.
DISCUSSION
The rubella epidemiological data collected in this study indicate that the high prevalence of rubella in China alternated between different regions and that the endemicity continuously occurred in areas covering eastern, central, and western portions of the country. Overall, the incidence of rubella in western China was significantly higher than that in other regions.
Reported rubella cases were concentrated in those under 15 years of age, most of whom were primary and middle school students. However, in eastern China, the proportion of rubella cases in those less than 15 years of age decreased significantly, and the proportion within the 15- to 39-year-old age group increased. This may be due to the introduction of the rubella vaccine into some provinces in eastern China, such as Shandong, since the 1990s. The immunization strategy involved 2 doses of vaccine, including the first dose for infants at between 8 and 18 months of age and the second dose for 7- or 12-year-olds (30). If routine vaccine coverage in children is not maintained, immunization of children could alter transmission dynamics and potentially lead to an increase in susceptibility in older age groups (27). A shift in risk to older age groups has already occurred in Brazil and Costa Rica (4). This is of concern because older age groups include women of childbearing age, thus raising the potential for an increase in the incidence of CRS.
In 2008, the rubella vaccine was introduced into the national immunization program in China. Measles-rubella vaccine (MR) was introduced for 8-month-old infants, and measles-rubella-mumps vaccine (MMR) was given to infants at 18 to 24 months of age. However, in some circumstances, when rubella vaccines were not available, they were replaced by measles vaccines in order to achieve the measles elimination goal initiated by WHO (19). Furthermore, seroepidemiological surveys of Chinese females in the cities of Shenzhen (20), Beijing, and Chongqing (18) revealed that only about 80% were immunized against rubella, which is too low to provide herd immunity to the population. Considering the shift in the ages at risk and the seroepidemiological data, the development of a routine rubella vaccination program should be a priority, and both children and women of childbearing age should be included.
In 2005, the WHO Regional Committee of the Western Pacific Region (WPR) formally declared regional measles elimination a goal with a target date set for 2012 (1). Measles and rubella are similar diseases, both characterized by a rash that may be difficult to differentiate clinically (35). In addition, rubella epidemics usually occur in the spring or early summer, similar to measles, peaking between April and May (13). Therefore, during the measles elimination campaign in China, particularly in 2009, when the incidence of rubella was higher than that of measles, measles control might have been hampered because large numbers of suspected measles cases, which were in reality rubella cases, may have appeared. Therefore, it is crucial to eliminate rubella during measles eradication campaigns. However, countries wishing to control and eliminate rubella must not only maintain high vaccine coverage but also be supported by high-quality surveillance, including molecular epidemiological studies, in order to obtain information about the circulation of indigenous viruses and the importation of new strains from other parts of the world (29).
The data presented in this study demonstrate that 1E rubella virus has been the predominant virus genotype since 2001 and that only 2 provinces had incidences of the genotype 2B virus in 2008. Therefore, the genotype 1E rubella virus is most likely to cause a nationwide epidemic. From 2001 to 2007, multiple transmission chains of genotype 1E rubella viruses were found in different parts of China (34). Some transmission chains of 1E faded, while other similar sequences continued to circulate in various provinces between 2008 and 2009. This might be related to the large-scale use of rubella vaccine during the nationwide immunization program in 2008, which may have interrupted some transmission chains. Some genotype 1E strains still survive and circulate.
Chinese genotype 1E rubella virus isolates were grouped into 2 clusters. Cluster 1 isolates were collected from 2001 to 2009, revealing the long-term circulation of this cluster in China since its emergence in 1997 (based on Bayesian MCMC analysis). Cluster 2 consisted of isolates from Hainan Province from between 2007 and 2008 (and may have emerged in 2006, based on Bayesian MCMC analysis), suggesting that it may recently have been introduced into Hainan Province.
Cluster 1 strains within the 1E genotype predominate in China and have not been found elsewhere, although sequences of many other 1E strains from other countries are available for comparison. This indicates that cluster 1 is unique to China. These viruses may have arisen from mutations and random genetic drifts that conferred a selective advantage to this lineage following its emergence in 1997.
Various methods based on Bayesian statistics that allow evolutionary rates, molecular phylogeny, and population dynamics to be coestimated in a single analysis starting from a nucleotide sequence alignment have recently been developed. The phylogeny and times of divergence of the rubella virus lineages were inferred using partial E1 fragment sequences sampled at different times and a relaxed molecular clock model, which incorporates the time-dependent nature of the evolutionary process by assuming independent rates in different branches, rather than a strict clock.
Recently developed methods based on coalescent theory for inferring the demographic history of a population on the basis of the gene sequences of a sample have allowed the reconstruction of the history of epidemics due to highly variable RNA viruses (6, 21). BSP, a nonparametric piecewise-constant model of population size, can fit several models, which solved the problem that coalescent methods usually require the assumption of a demographic model that is a mathematical description of the changes in effective population size, and information about the demographic behavior of a study was seldom obtained in advance. The BSP approach makes it possible to reconstruct novel and complex demographic scenarios (10). The greater sensitivity of the nonparametric BSP method showed that the rubella virus population size remained constant until 2007, when a decline in the effective number of infections occurred with the introduction of rubella vaccine into the national immunization program in 2008.
In vitro, RNA virus nucleotide misincorporation rates per site ranged from 10−3 to 10−6, due to the intrinsic error rates of the RNA polymerase and the lack of proofreading (2, 8). Genetic mutations are the basis of RNA virus evolution, as they allow the virus population to rapidly adapt to new environments and escape host antiviral responses (7). As an RNA virus, rubella virus has the potential to continually mutate, so close monitoring of the genetic variations of wild-type rubella virus strains is necessary. In this study, we estimated that the mean mutation rate of genotype 1E rubella viruses was 1.65 × 10−3 substitutions per site per year, based on the 739-nt window of the E1 gene, which is lower than the rates of the measles virus (0.78 × 10−2) (15), mumps viruses (1.86 × 10−2) (7), and coxsackievirus group A16 (0.91 × 10−2) (31). The effects of strong negative selection can be seen in the 16-fold lower evolutionary rate in the 1st and 2nd codon positions of the partial E1 genome sequence, frequently causing amino acid changes, than in the 3rd codon position, rarely resulting in amino acid substitution. The evolution rate of rubella virus is relatively low and may be due to the high degree of conservation in both nucleotide and amino acid sequences of rubella virus (11). Although the 739-nt region within the E1 glycoprotein for diagnostic applications contains important functional domains, including a hemagglutination-inhibiting and -neutralizing epitope, and antigenic sites, negative or positive selection pressures may differ considerably across a viral genome, and further studies on other genes or the whole genome are required to confirm this finding.
To our best knowledge, genotype 1E viruses were first identified in 1995 in France (26) and then in 1997 in the United States, Canada, the Caribbean, and Italy (23, 33). In addition to North America and Europe, 1E viruses have now been isolated in South America, Africa, and Asia (28, 35). Global genotype 1E viruses investigated in this study share the recent common ancestor that originated in 1995, the same year of the first isolation in France, suggesting that genotype 1E viruses first appeared in Europe. However, analysis of additional sequences is needed to determine the origin and evolution of genotype 1E rubella viruses with greater accuracy.
A shift in the predominant genotype from genotypes 1F and 2B to genotype 1E was found with the 2001 rubella epidemic in China. Subsequently, 1E rubella viruses continually circulated in China for more than 9 years. Epidemiological data show that a rubella epidemic occurred in 2008. The transmission dynamics of endemic viruses may change due to vaccine introduction in 2007, and these may be contained or even eliminated. Ongoing molecular epidemiological surveillance of circulating rubella viruses is necessary.
Because humans are the only reservoir of rubella virus, the transmission of rubella virus is influenced by various factors relating to the amount and virulence of the viruses, the number of unimmunized hosts, and their interaction. So, it can be postulated that the more recent decrease in the effective number of the rubella virus infections (those actually transmitted) might be due to a decrease in the virus population that is related to routine vaccination with rubella vaccine in the national immunization program and the decrease in the immunity gap related to susceptibility to the infection.
In conclusion, after the first isolation of 1E genotype rubella virus (cluster 1) in 2001 in China, it continuously circulated throughout the epidemic of 2008, but unlike the last rubella outbreak in 2001, no genotypic shift occurred. In addition, although the circulation of the virus has not been interrupted following the introduction of the rubella vaccine into the national immunization program in 2008, some lineages within the 1E genotype disappeared. Cluster 2 viruses within the 1E genotype were found only in Hainan Province, which is most likely the result of importation from other regions due to high rates of migration and tourism. The most recent common ancestor of global genotype 1E rubella virus may be traced back to February 1995 (range, January 1995 to April 1995). The tMRCA estimates for Chinese clusters were dated to May 1997 (cluster 1) and March 2006 (cluster 2). Estimated by the BSP method, the effective number of infections remained constant until 2007. Although a rubella epidemic occurred in the epidemiology in 2008, the epidemic started a decline that led to a decrease in the effective population size with the introduction of rubella vaccine into the national immunization program in the same year.
ACKNOWLEDGMENTS
This work is supported by the National Natural Science Foundation of China (project no. 81101244), the National Infectious Diseases Surveillance Program (2012ZX10004201, 2008ZX10004-008, 2009ZX10004-201, and 2009ZX10004-202), WHO measles regional reference laboratory funding (WPCHN1002802), and State Key Laboratory for Molecular Virology and Genetic Engineering funding.
We thank WHO headquarters and WPRO for the technical and financial support. We thank all the provincial and prefectural measles and rubella laboratory staff and the epidemiologists in China for providing clinical specimens, isolates, and epidemiologic data. We also acknowledge all of the laboratories that isolated the viruses used in this study, and we thank anonymous reviewers for comments that improved the manuscript.
We report no conflicts of interest.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Anonymous 2009. Progress toward the 2012 measles elimination goal—Western Pacific Region, 1990-2008. MMWR Morb. Mortal. Wkly. Rep. 58:669–673 [PubMed] [Google Scholar]
- 2. Batschelet E, Domingo E, Weissmann C. 1976. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene 1:27–32 [DOI] [PubMed] [Google Scholar]
- 3. Caidi H, et al. 2008. Phylogenetic analysis of rubella viruses found in Morocco, Uganda, Cote d'Ivoire and South Africa from 2001 to 2007. J. Clin. Virol. 42:86–90 [DOI] [PubMed] [Google Scholar]
- 4. Castillo-Solorzano C, et al. 2003. New horizons in the control of rubella and prevention of congenital rubella syndrome in the Americas. J. Infect. Dis. 187(Suppl. 1):S146–S152 [DOI] [PubMed] [Google Scholar]
- 5. Chen MH, Icenogle JP. 2007. Molecular virology of rubella virus, p 1–18 In Banatvala J, Peckham C. (ed), Rubella virus. Elsevier, Oxford, United Kingdom [Google Scholar]
- 6. Cottam EM, et al. 2006. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J. Virol. 80:11274–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cui A, Myers R, Xu W, Jin L. 2009. Analysis of the genetic variability of the mumps SH gene in viruses circulating in the UK between 1996 and 2005. Infect. Genet. Evol. 9:71–80 [DOI] [PubMed] [Google Scholar]
- 8. Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192 [DOI] [PubMed] [Google Scholar]
- 11. Frey TK. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hubschen JM, et al. 2007. Co-circulation of multiple rubella virus strains in Belarus forming novel genetic groups within clade 1. J. Gen. Virol. 88:1960–1966 [DOI] [PubMed] [Google Scholar]
- 13. Ji Y, et al. 2010. Genetic characterization of wild-type measles viruses isolated in China, 2006-2007. Virol. J. 7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin L, Thomas B. 2007. Application of molecular and serological assays to case based investigations of rubella and congenital rubella syndrome. J. Med. Virol. 79:1017–1024 [DOI] [PubMed] [Google Scholar]
- 15. Kuhne M, Brown DW, Jin L. 2006. Genetic variability of measles virus in acute and persistent infections. Infect. Genet. Evol. 6:269–276 [DOI] [PubMed] [Google Scholar]
- 16. Lee JY, Bowden DS. 2000. Rubella virus replication and links to teratogenicity. Clin. Microbiol. Rev. 13:571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Hu JY, Tao LN, Zhang JG. 2005. Epidemiological characteristics and immunoprophylactic strategy of congenital rubella syndrome. Shanghai J. Prev. Med. 17:72–74 [Google Scholar]
- 18. Long QJ, et al. 2007. A survey on rubella antibody level of women at childbearing age in Beijing and Chongqing. J. Vaccine Immun. 12:144–149 [Google Scholar]
- 19. Ma J, Hao LX, Luo HM. 2010. Progress on rubella control and immunization strategies. Zhongguo Yi Miao He Mian Yi 16:69–71 (In Chinese.) [PubMed] [Google Scholar]
- 20. Mou J, et al. 2010. Seroprevalence of rubella in female migrant factory workers in Shenzhen, China. Vaccine 28:7844–7851 [DOI] [PubMed] [Google Scholar]
- 21. Pybus OG, Rambaut A, Harvey PH. 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155:1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reef SE, Cochi SL. 2006. The evidence for the elimination of rubella and congenital rubella syndrome in the United States: a public health achievement. Clin. Infect. Dis. 43(Suppl. 3):S123–S125 [DOI] [PubMed] [Google Scholar]
- 23. Reef SE, et al. 2002. The changing epidemiology of rubella in the 1990s: on the verge of elimination and new challenges for control and prevention. JAMA 287:464–472 [DOI] [PubMed] [Google Scholar]
- 24. Robertson SE, Featherstone DA, Gacic-Dobo M, Hersh BS. 2003. Rubella and congenital rubella syndrome: global update. Rev. Panam. Salud Publica 14:306–315 [DOI] [PubMed] [Google Scholar]
- 25. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 26. Vauloup-Fellous C, et al. 2010. Phylogenetic analysis of rubella viruses involved in congenital rubella infections in France between 1995 and 2009. J. Clin. Microbiol. 48:2530–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vynnycky E, Gay NJ, Cutts FT. 2003. The predicted impact of private sector MMR vaccination on the burden of congenital rubella syndrome. Vaccine 21:2708–2719 [DOI] [PubMed] [Google Scholar]
- 28. WHO 2005. Distribution of final diagnosis measles/rubella suspected cases MESS, 2004 (N = 12,666). WHO, Geneva, Switzerland [Google Scholar]
- 29. WHO 2006. Global distribution of measles and rubella genotypes to update. Wkly. Epidemiol. Rec. 81:474–479 [PubMed] [Google Scholar]
- 30. Xu Q, et al. 2005. Analysis on the changing of age patterns among rubella patients after rubella vaccine immunization for children in Shandong Province, China. Zhonghua Liu Xing Bing Xue Za Zhi 26:861–863 (In Chinese.) [PubMed] [Google Scholar]
- 31. Zhang Y, et al. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. J. Clin. Microbiol. 48:619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, et al. 2010. Type 2 vaccine-derived poliovirus from patients with acute flaccid paralysis in China: current immunization strategy effectively prevented its sustained transmission. J. Infect. Dis. 202:1780–1788 [DOI] [PubMed] [Google Scholar]
- 33. Zheng DP, et al. 2003. Global distribution of rubella virus genotypes. Emerg. Infect. Dis. 9:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu Z, et al. 2010. Rubella virus genotypes in the People's Republic of China between 1979 and 2007: a shift in endemic viruses during the 2001 rubella epidemic. J. Clin. Microbiol. 48:1775–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu Z, et al. 2007. Comparison of four methods using throat swabs to confirm rubella virus infection. J. Clin. Microbiol. 45:2847–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]