Abstract
Trained African giant-pouched rats (Cricetomys gambianus) can detect Mycobacterium tuberculosis and show potential for the diagnosis of tuberculosis (TB). However, rats' ability to discriminate between clinical sputum containing other Mycobacterium spp. and nonmycobacterial species of the respiratory tract is unknown. It is also unknown whether nonmycobacterial species produce odor similar to M. tuberculosis and thereby cause the detection of smear-negative sputum. Sputum samples from 289 subjects were analyzed by smear microscopy, culture, and rats. Mycobacterium spp. were isolated on Lowenstein-Jensen medium, and nonmycobacterial species were isolated on four different media. The odor from nonmycobacterial species from smear- and M. tuberculosis culture-negative sputa detected by ≥2 rats (“rat positive”) was analyzed by gas chromatography-mass spectrometry and compared to the M. tuberculosis odor. Rats detected 45 of 56 confirmed cases of TB, 4 of 5 suspected cases of TB, and 63 of 228 TB-negative subjects (sensitivity, 80.4%; specificity, 72.4%; accuracy, 73.9%; positive predictive value, 41.7%; negative predictive value, 93.8%). A total of 37 (78.7%) of 47 mycobacterial isolates were M. tuberculosis complex, with 75.7% from rat-positive sputa. Ten isolates were nontuberculous mycobacteria, one was M. intracellulare, one was M. avium subsp. hominissuis, and eight were unidentified. Rat-positive sputa with Moraxella catarrhalis, Streptococcus pneumoniae, Staphylococcus spp., and Enterococcus spp. were associated with TB. Rhodococcus, Nocardia, Streptomyces, Staphylococcus, and Candida spp. from rat-positive sputa did not produce M. tuberculosis-specific volatiles (methyl nicotinate, methyl para-anisate, and ortho-phenylanisole). Prevalence of Mycobacterium-related Nocardia and Rhodococcus in smear-negative sputa did not equal that of smear-negative mycobacteria (44.7%), of which 28.6% were rat positive. These findings and the absence of M. tuberculosis-specific volatiles in nonmycobacterial species indicate that rats can be trained to specifically detect M. tuberculosis.
INTRODUCTION
Novel methods for rapid diagnosis of tuberculosis (TB) are urgently needed to complement smear microscopy, which has low sensitivity (20), and culture, which is slower and requires specialized laboratory conditions not available in resource-constrained settings. Trained African giant pouched rats (Cricetomys gambianus) possess profound potential for rapid detection of TB with higher sensitivity and specificity (38). An increase in TB case detection rate of 43 to 44% is achieved when Cricetomys rats are used as a second-line screening tool, after smear microscopy (18, 25). Rats have a highly developed sense of smell among mammalian species (22), which renders them trainable to specifically sense TB odor in sputum samples with a broad range of acid-fast bacilli (AFB) counts (see Materials and Methods). These rats also detect smear-negative sputa, which may contain few acid-fast bacilli missed by microscopy. Moreover, smear-negative results may persist in thorough reexamination of smears made after rat results.
In the present study, we investigated the extent to which TB detection rats can discriminate between clinical sputa with different Mycobacterium spp. (Mycobacterium tuberculosis) and nontuberculous mycobacteria (NTM) and other microorganisms of the respiratory tract (Nocardia spp., Rhodococcus spp., Streptomyces spp., Moraxella spp., Candida spp., and Streptococcus pneumoniae), which can also be found in sputum. We also investigated whether the nonmycobacterial species found in sputum produce odor compounds similar to M. tuberculosis odor, which could cause false detection of smear-negative sputum without M. tuberculosis.
MATERIALS AND METHODS
Specimens.
Sputa (n = 514) from 289 individuals presenting for TB diagnosis in six selected Tanzanian TB clinics (i.e., Dar Es Salaam = 5; Morogoro = 1) were analyzed from April to June 2009 (252 sputa from 161 individuals) and July 2010 (262 sputa from 128 individuals). Sputum aliquots (1 ml) for cultures were aseptically transferred into sterile screw-cap microtubes. The remaining volume (≥3 ml) in polypropylene sputum containers with lid was processed for TB detection by sniffer rats. Processing of sputa for rats included adding 5 ml of phosphate-buffered saline (PBS) to increase volume and avoid drying of sputum during inactivation. Sputum samples (≥8 ml, with PBS) were heat inactivated at 90°C for 30 min, cooled to room temperature, and stored at −20°C until later use in TB diagnosis by the rats at Sokoine University of Agriculture, and Anti-Persoonmijnen Ontmijnende Product Ontwikelling (SUA-APOPO) laboratory, Morogoro, Tanzania.
Population characteristics.
The age of sputum donors varied from <1 year to 86 years (mean ± the standard deviation, 32 ± 3 years). The gender ratio was 1.08 (150 males to 139 females). The subjects were classified into three TB diagnostic categories: (i) confirmed TB cases were individuals with two smear-positive (AFB+) sputa and/or positive mycobacterial (M. tuberculosis) culture; (ii) suspected TB cases were individuals with only one smear-positive culture-negative sample (M. tuberculosis); and (iii) non-TB cases (negative) were individuals with smear-negative and M. tuberculosis culture-negative sputum. Individuals with NTM isolates were classified in the non-TB category. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of M. tuberculosis detection by rats were determined as described elsewhere (13) using confirmed TB and non-TB subjects. Suspected TB cases were excluded in the subsequent evaluation. Microorganisms from smear-negative, M. tuberculosis culture-negative but rat-positive sputa were used to determine odor profiles and confounding impact of microbes to TB diagnosis by rats.
Isolation and identification of mycobacteria.
A total of 380 sputum samples from 289 subjects (one to two samples per subject) were cultured to isolate mycobacterial species. After processing, 252 sputum samples from 161 donors were inoculated on Lowenstein-Jensen medium (LJ) with pyruvate and LJ with glycerine, and 128 sputum samples (128 donors) were inoculated on LJ with glycerine only. Processing included decontamination with 4% sodium hydroxide added to sputum in 1:1 ratio, mixing well, and being left to stand for 45 min. The mixture was centrifuged at 3,000 × g for 20 min, the supernatant was decanted, and the sediment was neutralized with 14% potassium dihydrogen phosphate. Cultures were incubated at 37°C for a minimum of 8 weeks with weekly examination for growth (44). Isolates were stained using the Ziehl-Neelsen (ZN) method, and DNA was extracted from all AFB by the bead-beating method (35). Multiplex real-time PCR for genus Mycobacterium, M. tuberculosis sp. complex (MTC) and M. avium complex (MAC) was performed as previously described (26, 31). A conventional multiplex PCR for this genus was also performed (42). Isolates negative for PCR were identified by 16S rRNA gene sequencing (1). MTC isolates were subjected to multispacer sequence typing (MST) (11).
Nonmycobacterial respiratory tract microbes.
Chocolate agar, Sabouraud dextrose agar, buffered charcoal yeast extract agar, and paraffin agar were used to isolate nonmycobacterial respiratory tract microorganisms from 394 sputa (289 donors). Cultures were incubated at 37°C for 6 weeks, with isolates preliminarily identified by colony and cell morphology and by biochemical tests. Moraxella and Streptococcus spp. were subjected to PCR for M. catarrhalis and S. pneumoniae (15). Nocardia sp., Rhodococcus sp., and Streptomyces sp. were identified by growth characteristics in different media, including the opacification of Middlebrook 7H11 medium and the formation of chalky white colonies (12). Nocardia isolates were further subjected to specific PCR (5, 14). 16S rRNA gene sequencing using fD1 and rP2 universal primers (39) was used to identify Rhodococcus, Enterococcus, and Staphylococcus spp.
Determining microorganisms in rat-positive sputum.
Processed sputa (n = 514) in polypropylene containers were analyzed by a group of 10 rats selected from 22 qualified rats, which consistently detected more than 80% of known smear-positive sputa (TB positive). Detection procedures described by Weetjens et al. (38; http://www.youtube.com/watch?v=KoRvdyuHxdE) (32) were used. Briefly, rats were rewarded with food if they paused for at least 5 s to sniff TB-positive sputum confirmed by smear-microscopy and/or culture. Smear-positive sputa consisted of various AFB counts: 1 to 9 AFB, 1+, 2+ to 3+, whereby 1 to 9 AFB refers to 1 to 9 AFB per 100 microscopy fields; 1+ is 10 to 99 AFB per 100 fields; 2+ is 1 to 10 AFB per field, and 3+ is more than 10 AFB per field (8). Rats were not rewarded on pausing at TB-negative samples. On average, one rat analyzed the set of 70 samples at a rate of 8 min per session. Each rat analyzed the 70 sputa twice (thus two sessions = 16 min). A sample was considered TB positive if a minimum of two rats gave a positive signal on it. Therefore, consensus results (two rats × two sessions each) of 70 samples were obtained in 32 min. The training session was conducted by two teams of trainers handling five rats each, in 95 min. This duration includes time for changing rats, cleaning/wiping of the training cage floor with 70% ethanol to remove odor residues between rat sessions, and changing of metal bars containing the 70 sputa. Detection of negative sputa by two rats, based on microscopy and/or culture, was indicative of M. tuberculosis (rat positive) and the sputum was subjected to thorough investigation.
Odor analysis by GC-MS.
Volatile compounds of the microorganisms from AFB smear-negative, M. tuberculosis culture-negative, and rat-positive sputa were identified using gas chromatography-mass spectrometry (GC-MS). Analyses were carried out on an Agilent 7890A GC system connected to an Agilent 5975C inert mass detector fitted with an HP-5MS fused silica capillary column (30 m, 0.25 mm [inner diameter], 0.25-μm film; J&W Scientific). The conditions were as follows: inlet pressure, 77.1 kPa, He 23.3 ml min−1; injection volume, 2 μl; transfer line, 300°C; and electron energy, 70 eV. The GC program was set as follows: 5 min at 50°C, increasing with 5°C min−1 to 320°C, operated in the splitless mode (60-s valve time); the He carrier gas flow was 1.2 ml min−1.
Briefly, selected bacteria and yeast isolates were grown on suitable media, and headspace samples were collected for 24 h using a closed-loop stripping apparatus as described previously (28), fitted with an activated charcoal filter (Chromtech; precision charcoal filter, 5 mg). The collected volatiles were eluted from the filter for GC-MS analysis using 30 μl of dichloromethane (Suprasolv; Merck, Germany). Compounds were identified by comparison of GC-MS retention indices with those of mass spectral libraries and comparison with synthetic reference compounds. Retention indices I were determined from a homologous series of n-alkanes (C8 to C35) (36).
Statistical analysis.
A Fisher exact test was used to determine whether the distribution of rat-positive and -negative sputa with M. tuberculosis was different from that of sputa with NTM and nonmycobacterial species, with a P value of <0.05 for statistical significance.
RESULTS
Detection of TB in sputum by rats.
There were 56 confirmed TB cases based on smear microscopy (n = 19) and culture (n = 37), 228 TB-negative cases, and 5 suspected TB cases. Rats detected 45 (true positive) of the 56 confirmed TB cases and 63 (false positive) of the 228 negative subjects. Four (80%) of the five suspected TB cases with one AFB-positive sputum were detected by rats (rat positive). The sensitivity and specificity were 80.4 and 72.4%, respectively. The PPV was 41.7%, and the NPV was 93.8%, with an accuracy for TB diagnosis of 73.9%.
Mycobacterium isolation.
Mycobacterium spp. were isolated from 47 of 289 patients (16.3%). Thirty-seven isolates from 37 patients were MTC (78.7%) based on specific multiplex PCRs for the Mycobacterium genus and MST analyses. Details of the genotypic analyses (MST) of MTC isolates will be reported elsewhere (G. F. Mgode et al., unpublished data). The majority of MTC (75.7%) were from rat-positive sputa (Table 1 and Fig. 1).
Table 1.
Mycobacterium spp. from smear-positive and smear-negative sputum samples (n = 47) tested by trained Cricetomys gambianus ratsa
| Mycobacterial designation | No. of samples |
Distribution (%) | Rat-positive samples |
Rat-negative samples |
Detection (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Smear+ | Smear− | Total | Smear+ | Smear− | Total | Smear+ | Smear− | |||
| M. tuberculosis | 37 | 25 | 12 | 78.7 | 28 | 25 | 3 | 9 | 0 | 9 | 75.7 |
| NTM | 8 | 1 | 7 | 17.0 | 4 | 1 | 3 | 4 | 0 | 4 | 50.0 |
| M. avium subsp. hominissuis | 1 | 0 | 1 | 2.1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0 |
| M. intracellulare | 1 | 0 | 1 | 2.1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0 |
Smear positive, smear+; smear negative, smear−. The combined rat-positive sputum samples (detection) with NTM and M. avium subsp. hominissuis and M. intracellulare is 40%.
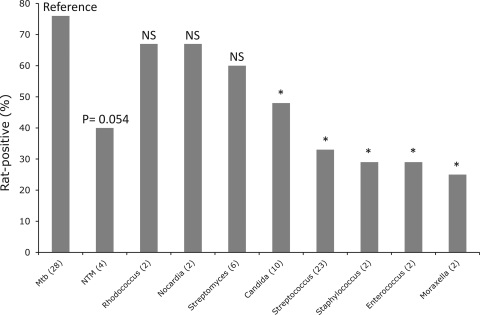
Fig 1.
Rat-positive (%) sputum samples with different individual microorganisms. Number of isolates of each species in detected sputum is indicated in brackets. Statistically significant differences (P < 0.05, Fisher exact test) between rat-positive sputa with M. tuberculosis (reference) and rat-positive sputa with nonmycobacterial species are indicated by an asterisk. Rat-positive samples not significantly different from sputa with M. tuberculosis are indicated by “NS”.
Ten mycobacterial isolates were NTM, of which two were M. intracellulare and M. avium subsp. hominissuis. Eight NTM (17%) of 47 mycobacterial isolates were not identified to species level. Four of the eight NTM were from rat-positive sputa, of which one was smear-positive. The detection trend for sputa with NTM (including M. avium subsp. hominissuis and M. intracellulare) was marginally different from detection of sputa with M. tuberculosis (P = 0.054, Fisher exact test) (Fig. 1). Isolates identified as M. avium subsp. hominissuis and M. intracellulare were from rat-negative sputa (Table 1). Nine of the ten NTM were from smear-negative sputa corroborating previous reports showing increasing occurrence of NTM in clinical samples (7, 10). Overall, 21 (44.7%) of all mycobacterial isolates (n = 47) were from smear-negative sputa, revealing that a significant proportion of smear-negative sputa contained mycobacterial species, which were probably the cause of detection of these sputa by rats. Indeed, 6 (28.6%) of the 21 mycobacterial isolates from smear-negative sputa were rat positive, indicating an increased detection rate for smear-negative TB by >28%.
Isolation and species distribution of opportunistic pathogens of the respiratory tract.
Of the four media used, paraffin agar improved the isolation success of species, which included Nocardia sp., Streptomyces sp., Candida sp., and one NTM. These were identified by their characteristic colony morphologies and pigmentation in this medium. Rhodococcus spp. were isolated on chocolate agar and buffered charcoal yeast extract agar, whereas Moraxella spp., Streptococcus sp., and Enterococcus spp. were isolated on chocolate agar. Yeast species were isolated on all four media used.
Streptococcus spp. were the most abundant among the respiratory tract nonmycobacterial isolates (n = 69). Thirteen isolates from rat-positive sputa were identified as S. pneumoniae by specific PCR (15), and the remaining streptococcal isolates were assigned to S. pneumoniae based on colony and cell morphology, which were similar to 13 isolates identified by PCR. Other microorganisms from sputum are shown in Table 2. Two of the three Nocardia isolates were identified as N. farcinica (5, 14), of the N. asteroides complex. Rhodococcus spp., Enterococcus spp., Staphylococcus succinus, and other Staphylococcus spp. were identified by 16S rRNA gene sequence (39). Candida spp. were identified by Gram stain. Nonmycobacterial microorganisms co-occurred in some individuals. For example, Streptococcus spp. co-occurred with Candida spp. (n = 5) and with M. catarrhalis (n = 3), Streptomyces spp. (n = 3), Rhodococcus spp. (n = 1), and Nocardia spp. (n = 1). Co-occurrence was also found in Nocardia spp. and Streptomyces spp. (n = 2), Candida spp., and M. catarrhalis (n = 4). Nonmycobacterial species also co-occurred with M. tuberculosis (Table 2 and Fig. 3). Rat-positive sputa with Staphylococcus and Enterococcus spp. were also either smear positive or mycobacterial culture positive. The rats' detection of sputum containing mycobacterial and nonmycobacterial species is presented in Tables 1 and 2 and Fig. 1 to 3.
Table 2.
Respiratory tract microbes from smear-positive and smear-negative sputum samples tested by rats
| Speciesa | No. of samples |
Rat-positive samples |
Rat-negative samples |
Smear− detection (%)b | Significance (P)c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Smear+ | Smear− | Total | Smear+ | Smear− | Total | Smear− | Smear− | |||
| Moraxella catarrhalis | 8 | 1 | 7 | 2 | 1 | 1 | 6 | 0 | 6 | 12.5 | 0.011 |
| Rhodococcus sp. | 3 | 0 | 3 | 2 | 0 | 2 | 1 | 0 | 1 | 66.7* | NS |
| Nocardia farcinica | 3 | 0 | 3 | 2 | 0 | 2 | 1 | 0 | 1 | 66.7* | NS |
| Streptomyces sp. | 10 | 2 | 8 | 6 | 2 | 4 | 4 | 0 | 4 | 40.0* | NS |
| Candida sp. | 21 | 3 | 18 | 10 | 3 | 7 | 11 | 0 | 11 | 33.3* | 0.045 |
| Streptococcus sp. | 69 | 14 | 55 | 23 | 13 | 10 | 46 | 1 | 45 | 14.5 | 3.85e-5 |
| Enterococcus sp. | 7 | 2 | 5 | 2 | 2 | 0 | 5 | 0 | 5 | 0.0 | 0.025 |
| Staphylococcus sp. | 7 | 2 | 5 | 2 | 2 | 0 | 5 | 0 | 5 | 0.0 | 0.025 |
| Unidentified | 11 | 4 | 7 | 6 | 4 | 2 | 5 | 0 | 5 | 18.2 | NS |
| Total (%) | 139 | 28 (20.1) | 111 (79.9) | 55 (39.6) | 27 (49.1) | 28 (50.9) | 84 (60.4) | 1 (1.2) | 83 (98.8) | ||
Two Streptococcus pneumoniae isolates from smear-negative M. tuberculosis culture-positive sputum not detected by rats are not presented in this table.
*, Frequently detected microorganisms from TB-negative, rat-positive sputa had their volatile compounds analyzed by GC-MS and compared to volatile compounds of M. tuberculosis (see Table 3).
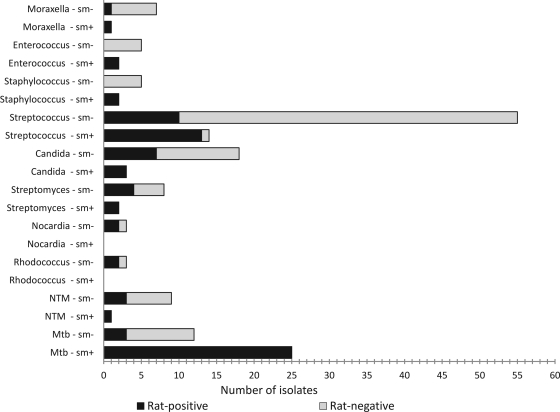
Fig 3.
Patterns of rat positive and rat-negative in smear-positive (sm+) and smear negative (sm−) sputum samples with Mycobacterium and nonmycobacterial microorganisms. Smear-positive sputa with M. tuberculosis have a higher frequency of being rat positive than smear negative (100 versus 28%, respectively).
A comparison of rat-positive (detected) and rat-negative (undetected) sputa with M. tuberculosis (Table 1) versus sputa with nonmycobacterial species (Table 2) revealed that detection of sputa with M. catarrhalis, S. pneumoniae, Candida spp., Enterococcus spp., Staphylococcus succinus, and other Staphylococcus spp. was significantly different from M. tuberculosis (P < 0.05) (Table 2, Fig. 1). Thus, nonmycobacterial species did not cause detection of sputum by rats compared to M. tuberculosis. The distribution of rat-positive and rat-negative sputa with Rhodococcus spp., Nocardia spp., Streptomyces spp., and few unknown microorganisms was not significantly different from that of M. tuberculosis. However, these species were not as abundant in detected sputa as M. tuberculosis (Tables 1 and 2 and Fig. 1) and have low prevalence (1%).
Odor analysis.
An odor analysis was performed on selected isolates from M. tuberculosis smear-negative, culture-negative, rat-positive sputa, namely, Rhodococcus sp., Candida sp., and Staphylococcus sp. isolates, as well as reference strains of Nocardia spp. (N. asteroides and N. africana) and Streptomyces spp. (S. coelicolor, S. griseoflavus, and S. antibioticus). Table 3 lists the compounds repeatedly found in these strains. Methyl nicotinate, methyl para-anisate, ortho-phenylanisole, and methyl phenylacetate were predominant in M. tuberculosis, as reported by Syhre and Chambers (33). A wide variety of compounds occurring in M. tuberculosis strains may serve as basis for the odor detection. We identified volatiles shared by M. tuberculosis, Nocardia spp., Streptomyces spp., and Rhodococcus sp., for example, 2-phenylethanol or 2-hydroxy-3-pentanone (Table 3), which are also produced by other microbial species (28, 29, 37, 41) and hence cannot be regarded as specific markers for M. tuberculosis. For example, aciphyllene, which is a known sesquiterpene from the endophytic fungus Muscodor albus (3), is a typical volatile compound of Nocardia spp. Candidate TB markers of M. tuberculosis were not found in nonmycobacterial species, which had a distribution of rat-positive and -negative sputa similar to sputa with M. tuberculosis (P > 0.05) (Table 2).
Table 3.
Volatile compounds of isolates from sputum samples and reference M. tuberculosis, Nocardia spp., and Streptomyces spp.
| Compounda | Microorganism tested (no. of isolates) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mycobacterium tuberculosis (35) | Rhodococcus isolate (2) | Staphylococcus isolate (2) | Candida isolate (2) | Nocardia asteroides (4) | Nocardia africana (6) | Streptomyces coelicolor (3) | Streptomyces antibioticus (3) | Streptomyces griseoflavus (4) | |
| Dimethyl disulfide | X | ||||||||
| Dimethyl trisulfide | X | X | X | X | X | X | |||
| Dimethyl tetrasulfide | X | X | X | ||||||
| Methyl methanethiosulfonate | X | ||||||||
| 2,3-Dimethyl-5-isopentylpyrazine | X | X | X | ||||||
| Unknown pyrazine | X | X | X | ||||||
| Camphor | X | X | |||||||
| Linalyl acetate | X | ||||||||
| Isobornyl acetate | X | ||||||||
| Aciphyllene | X | X | |||||||
| Unknown diterpenoid | X | X | |||||||
| 2-Hydroxy-3-butanone | X | X | X | ||||||
| 2-Hydroxy-3-pentanone | X | X | X | X | |||||
| 2,5-Dimethylthiopene | X | X | |||||||
| 1-Hexanol | X | X | |||||||
| 1-Octanol | X | ||||||||
| 4-Methyl-2-pentanone | X | X | |||||||
| 4-Methylpent-3-en-2-one | X | ||||||||
| γ-Methylbutyrolactone | X | X | |||||||
| 2-Phenylethanol | X | X | X | ||||||
| Ethyl phenylacetate | X | X | |||||||
| Methyl phenylacetate* | X | ||||||||
| Methyl nicotinate* | X | ||||||||
| Methyl para-anisate* | X | ||||||||
| ortho-Phenylanisole* | X | ||||||||
*, See Syhre and Chambers (33).
DISCUSSION
Our study reveals that trained African giant pouched rats (C. gambianus) target M. tuberculosis in sputa and not other microorganisms of the respiratory tract. M. tuberculosis was the most frequently detected species among the microorganisms isolated from sputa of suspected cases (Fig. 2). Most of the rat-positive sputa containing opportunistic pulmonary pathogens also contained M. tuberculosis as confirmed by either smear microscopy or culture. Rats' detection of smear- and culture-negative sputa containing other pulmonary pathogens, such as M. catarrhalis, S. pneumoniae, Candida spp., Enterococcus spp., Staphylococcus succinus, and other Staphylococcus spp. appears to be due to M. tuberculosis, which could be below the detection thresholds of microscopy and culture (19). The statistically significant difference found in the distribution of rat-positive and -negative sputa with M. tuberculosis and nonmycobacterial species (P < 0.05) indicates that trained rats do not false-detect sputa with these microorganisms as M. tuberculosis-positive samples (Tables 1 and 2). Sputa with these species alone, excluding those with Rhodococcus spp., Nocardia spp., and Streptomyces spp. were less frequently detected (Table 2, Fig. 2). Sputa containing Enterococcus spp. and Staphylococcus spp. were also detected in the presence of M. tuberculosis. The detection of sputa with Rhodococcus spp., Nocardia spp., and Streptomyces was not significantly different from that of sputa with M. tuberculosis (P > 0.05), which suggests that sputa with these mycobacteria-related species could be false-detected by rats as samples with M. tuberculosis. However, analysis of odor compounds revealed that these species do not produce candidate odor markers for TB diagnosis produced by M. tuberculosis (Table 3). This can be taken as evidence for the presence of M. tuberculosis bacilli in sputa with these species, which were not detected by culture or microscopy.
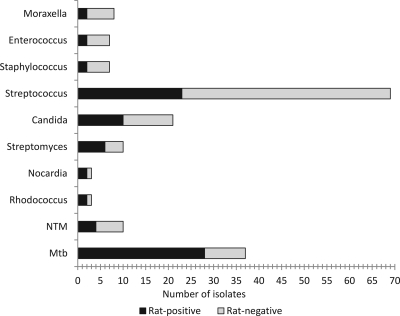
Fig 2.
Overall rat-positive and rat-negative sputum samples with different microorganisms. NTM, nontuberculous mycobacteria.
The prevalence of mycobacterium-related Nocardia spp. and Rhodococcus spp. in Tanzania was lower (1%) compared to 4 to 5% reported in other sub-Saharan countries (16). Hence, these species cannot account for the high proportion of smear-negative, rat-positive sputa, of which 28.6% were TB cases detected by rats and culture. Smear-negative, culture-positive Mycobacterium spp. contributed to 44.7% of the total mycobacterial isolates. This enabled evaluation of the causes of detection of smear-negative sputa and extent of detection of sputa with different mycobacterial species. However, the present study used randomly chosen sputa based on sample volume rather than patients' symptoms, such as bronchopneumonia, which increases the isolation rate of Nocardia spp. (23). Although the prevalence of Nocardia sp. in the present study could be an underestimate, the low prevalence of Rhodococcus sp. cannot be adequately discussed since there are no other data from Tanzania regarding this pathogen in humans. The prevalence of Streptomyces spp. and Candida spp. was higher (7.3 and 7.8%, respectively) but also cannot account for the proportion of smear-negative, rat-positive sputa.
The detection of sputa with NTM, as well as smear-negative, M. tuberculosis culture-negative sputa, could be due to low M. tuberculosis abundance, loss (death) of the few M. tuberculosis during decontamination and neutralization, or co-occurrence and competition between fast-growing NTM and slow-growing M. tuberculosis in culture. The prevalence of NTM is increasing (7, 10), especially in smear-negative TB/HIV coinfected patients associated with a low M. tuberculosis load in the sputum (9). Dormant M. tuberculosis bacilli also cause culture negativity (21). Further studies targeting isolation of dormant M. tuberculosis, for example, by incorporation of resuscitation promoting factors (rpf) in the medium (21), are needed to further determine the rate of detection of M. tuberculosis by rats in smear-negative samples and the extent of detection of sputa with a high proportion of NTM. The geneXpert MTB/RIF for M. tuberculosis (4) could assist in revealing M. tuberculosis in smear- and culture-negative, rat-positive sputa and provide deeper insight into the false-positive rate of the rats.
Odor analysis from selected nonmycobacterial isolates from smear-negative, rat-positive sputa and reference Nocardia spp. and Streptomyces spp. (Table 3) showed that these microorganisms produced volatile compounds different from those reported for M. tuberculosis (33). Previous studies on volatile compounds of related nonmycobacterial organisms such as R. fascians, S. epidermidis, and Streptomyces spp. (17, 27, 37) reported volatiles not identical to the described M. tuberculosis-specific volatile compounds. Together, these findings indicate an absence of M. tuberculosis-specific compounds from nonmycobacterial microorganisms.
The volatile compounds of a given isolate cultured on artificial medium can differ from those of the same isolate in the host tissue due to differences in growth substrates as reported for the fungus Trichoderma sp. (6, 40). M. tuberculosis bacilli grown in artificial medium also lack characteristic chemical compounds, such as phthioic acid, phthiocol, tuberculostearic acids and polysaccharides found in M. tuberculosis bacilli from host tissue (2). The lipid content of M. tuberculosis bacilli grown in vitro also differs from M. tuberculosis bacilli in tissue (30). These compounds, however, are not among the candidate odor markers of TB, which have recently been reported for M. tuberculosis in both artificial medium and in breath samples of TB patients (24, 33, 34). The absence of the candidate TB volatile compounds in nonmycobacterial species thus suggests that these species do not confound detection of sputum by trained rats. Further studies are needed to precisely determine whether the rats detect candidate volatile compounds of M. tuberculosis produced in artificial medium.
The sensitivity and specificity of TB diagnosis by rats in the present study were high (80.4 and 72.4%, respectively) but lower than the previously reported 86.6 and 93.8%, respectively (38). The lower PPV (41.7%) and higher NPV (93.8%) obtained are largely affected by the prevalence of TB in a given population. Our study indicates that harnessing rats for early TB diagnosis could have a significant impact on TB control. This is supported by the higher NPV (93.8%), which indicates that individuals with rat-negative sputum have a 93.8% likelihood of not having active TB. Consensus results of two rats (two sessions each) are obtained in 32 min for the set of 70 sputa. This is faster than the smear microscopy in which one microscopist is recommended to analyze an average of 20 samples per day (43). The shorter time used by rats to detect TB could enable screening of a larger population and reduce new TB transmissions resulting from undetected TB cases and delayed diagnosis. Further studies are needed to precisely identify the specific volatile compounds detected by rats and their occurrence in diverse Mycobacterium spp. The diagnosis of emerging Nocardia spp. and Rhodococcus spp. pathogens should also be considered, especially when TB is ruled out in patients with pulmonary disease symptoms. Our data further underline the potential value of conditioned rats for rapid, specific, and sensitive diagnosis of TB.
ACKNOWLEDGMENTS
We thank management and technical personnel, as well as patients, in the collaborating TB clinics in Dar es Salaam and Morogoro and SUA-APOPO TB laboratory staff for their cooperation. We thank D. Schad for formatting the figures, J. Weiner for statistical analysis, M. L. Grossman for revising the manuscript, and two anonymous reviewers for their useful comments.
Funding by the UBS Optimus Foundation to S.H.E.K. and B.J.W. is gratefully acknowledged.
Footnotes
Published ahead of print 30 November 2011
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adekambi T, Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA, and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095–2105 [DOI] [PubMed] [Google Scholar]
- 2. Anderson RJ, Reeves RE, Creighton MM, Lothrop WC. 1943. The chemistry of lipids of tubercle bacilli. LXV. An investigation of tuberculous lung tissue. Am. Rev. Tuberc. 48:65–75 [Google Scholar]
- 3. Atmosukarto I, Castillo U, Hess WM, Sears J, Strobel G. 2005. Isolation and characterization of Muscodor albus I-41.3s, a volatile antibiotic producing fungus. Plant Sci. 169:854–861 [Google Scholar]
- 4. Boehme CC, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown JM, Pham KN, McNeil MM, Lasker BA. 2004. Rapid identification of Nocardia farcinica clinical isolates by a PCR assay targeting a 314-base-pair species-specific DNA fragment. J. Clin. Microbiol. 42:3655–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruce A, Wheatley RE, Humphris SN, Hackett CA, Florence MEJ. 2000. Production of volatile organic compounds by Trichoderma in media containing different amino acids and their effect on selected wood decay fungi. Holzforschung 54:481–486 [Google Scholar]
- 7. Buijtels PCAM, et al. 2009. Nontuberculous mycobacteria, Zambia. Emerg. Infect. Dis. 15:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Centers for Disease Control Prevention 2000. Acid-fast direct smear microscopy: a laboratory training program. Centers for Disease Control and Prevention, Atlanta, GA: http://wwwn.cdc.gov/dls/afb/english/english.pdf [Google Scholar]
- 9. Colebunders R, Bastian I. 2000. A review of the diagnosis and treatment of smear-negative pulmonary tuberculosis. Int. J. Tuberc. Lung. Dis. 4:97–107 [PubMed] [Google Scholar]
- 10. Crump JA, van Ingen JAB, Morrissey, et al. 2009. Invasive disease caused by nontuberculous mycobacteria, Tanzania. Emerg. Infect. Dis. 15:53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Djelouadji Z, Arnold C, Gharbia S, Raoult D, Drancourt M. 2008. Multispacer sequence typing for Mycobacterium tuberculosis genotyping. PLoS One 3:e2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flores M, Desmond E. 1993. Opacification of Middlebrook agar as an aid in identification of Nocardia farcinica. J. Clin. Microbiol. 31:3040–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuchs KH, DeMeester TR, Albertucci M. 1987. Specificity and sensitivity of objective diagnosis of gastroesophageal reflux disease. Surgery 102:575–580 [PubMed] [Google Scholar]
- 14. Hasegawa T, et al. 2007. Identification of Nocardia farcinica by a PCR primer amplifying a specific DNA band for the bacterium. Jpn. J. Med. Mycol. 48:173–175 [DOI] [PubMed] [Google Scholar]
- 15. Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J. Clin. Microbiol. 35:2854–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones N, Khoosal M, Louw M, Karstaedt A. 2000. Nocardial infection as a complication of HIV in South Africa. J. Infect. 41:232–239 [DOI] [PubMed] [Google Scholar]
- 17. Khoga JM, Toth E, Marialigeti K, Borossay J. 2002. Fly-attracting volatiles produced by Rhodococcus fascians and Mycobacterium aurum isolated from myiatic lesions of sheep. J. Microbiol. Methods 48:281–287 [DOI] [PubMed] [Google Scholar]
- 18. Mahoney AM, et al. 2011. Using giant African pouched rats to detect tuberculosis in human sputum samples: 2010 findings. Pan Afr. Med. J. 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin RS, Sumarah RK, Robart EM. 1975. Comparison of four culture media for the isolation of Mycobacterium tuberculosis: a 2-year study. J. Clin. Microbiol. 2:438–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mfinanga GS, et al. 2007. The quality of sputum smear microscopy diagnosis of pulmonary tuberculosis in Dar es Salaam, Tanzania. Tanzan. Health Res. Bull. 9:164–168 [DOI] [PubMed] [Google Scholar]
- 21. Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. 2010. Resuscitation promoting factors reveal an occult population of tubercle bacilli in sputum. Am. J. Respir. Crit. Care. Med. 181:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niimura Y, Nei M. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One 2:e708 doi:10.1371/journal.pone.0000708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Osoagbaka OU, Njoku-Obi AN. 1985. Nocardiosis in pulmonary diseases in parts of Nigeria. I. Preliminary observations on five cases. J. Trop. Med. Hyg. 88:367–372 [PubMed] [Google Scholar]
- 24. Phillips M, et al. 2010. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb.). 90:145–151 [DOI] [PubMed] [Google Scholar]
- 25. Poling A, et al. 2010. Using giant African pouched rats to detect tuberculosis in human sputum samples: 2009 Findings. Am. J. Trop. Med. Hyg. 83:1308–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richardson ET, Samson D, Banaei N. 2009. Rapid identification of Mycobacterium tuberculosis and nontuberculous mycobacteria by multiplex, real-time PCR. J. Clin. Microbiol. 47:1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schöller CEG, Gürtler H, Pedersen R. 2002. Volatile metabolites from Actinomycetes. J. Agric. Food Chem. 50:2615–2621 [DOI] [PubMed] [Google Scholar]
- 28. Schulz S, Fuhlendorff J, Reichenbach H. 2004. Identification and synthesis of volatiles released by the myxobacterium Chondromyces crocatus. Tetrahedron 60:3863–3872 [Google Scholar]
- 29. Schulz S, Dickschat J. 2007. Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24:814–842 [DOI] [PubMed] [Google Scholar]
- 30. Sheehan HL, Whitwell F. 1949. The staining of tubercle bacilli with Sudan black B. J. Pathol. Bacteriol. 61:269–271 [DOI] [PubMed] [Google Scholar]
- 31. Shrestha NK, et al. 2003. Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J. Clin. Microbiol. 41:5121–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. SUA-APOPO 2011. Video demonstration of tuberculosis detection by rats. SUA-APOPO, Morogoro, Tanzania: http://www.youtube.com/watch?v=KoRvdyuHxdE [Google Scholar]
- 33. Syhre M, Chambers ST. 2008. The scent of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 88:317–323 [DOI] [PubMed] [Google Scholar]
- 34. Syhre M, Manning L, Phuanukoonnon S, Harino P, Chambers ST. 2009. The scent of Mycobacterium tuberculosis. II, Breath. Tuberculosis (Edinb.) 89:263–266 [DOI] [PubMed] [Google Scholar]
- 35. Tell LA, Foley J, Needham ML, Walker RL. 2003. Comparison of four rapid DNA extraction techniques for conventional polymerase chain reaction testing of three Mycobacterium spp. that affect birds. Avian Dis. 47:1486–1490 [DOI] [PubMed] [Google Scholar]
- 36. van den Dool H, Kratz P. 1963. A generalization of the retention index system including linear temperature programmed gas-liquid partition chromatography. J. Chromatogr. 11:463–471 [DOI] [PubMed] [Google Scholar]
- 37. Verhulst NO, et al. 2009. Cultured skin microbiota attracts malaria mosquitoes. Malaria J. 8:1475–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weetjens BJ, et al. 2009. African pouched rats for the detection of pulmonary tuberculosis in sputum samples. Int. J. Tuberc. Lung Dis. 13:1–7 [PubMed] [Google Scholar]
- 39. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wheatley RE, Hackett C, Bruce A, Kundzewicz A. 1997. Effect of substrate composition on the production of volatile organic compounds from Trichoderma spp. inhibitory to wood decay fungi. Intern. Biodet. Biodeg. 39:199–205 [Google Scholar]
- 41. Wilkins K, Schöller C. 2009. Volatile organic metabolites from selected Streptomyces strains. Actinomycetologica 23:27–33 [Google Scholar]
- 42. Wilton S, Cousins D. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. Genome. Res. 1:269–273 [DOI] [PubMed] [Google Scholar]
- 43. World Health Organization 2005. Management of tuberculosis. Training for district TB coordinators. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2006/9789241594417_moduleF.pdf [Google Scholar]
- 44. World Health Organization 1998. Laboratory services in tuberculosis control. III. Culture. Global Tuberculosis Programme, p 18 World Health Organization, Geneva, Switzerland [Google Scholar]