Abstract
The development of a rapid test to identify Mycobacterium tuberculosis Beijing isolates and specifically strain GC1237, coming from a sub-Saharan country, is needed due to its alarming wide spread on Gran Canaria Island (Spain). A rapid test that detects IS6110 present between dnaA and dnaN in the Beijing strains and in a specific site for GC1237 (Rv2180c) has been developed. This test would be a useful tool in the surveillance of subsequent cases.
TEXT
Beijing is a lineage of Mycobacterium tuberculosis that is dispersed worldwide predominating throughout East Asia and the former Soviet Union. This lineage may have a selective advantage either with higher virulence or transmissibility that led to clonal expansion. The prevalence of this family is low in Spain, and its presence in the population has been associated with the recent increase in the number of tuberculosis (TB) cases among immigrants (5, 9, 11, 16).
The introduction of a Beijing strain, strain GC1237, in Gran Canaria Island, Spain, by an African refugee (from Liberia), and its explosive spread in this community (27.1% of TB cases) over the next few years were reported in 2001 (6). Through review of the literature, an identical restriction fragment length polymorphism (RFLP) pattern of strain GC1237 was found in four isolates from Somalia, Ethiopia, Sierra Leone, and Liberia (6). Three Beijing isolates sharing the RFLP genotype and MIRU-VNTR were identified in Sweden from patients from Ethiopia and Eritrea (12). An M. tuberculosis Beijing strain responsible for an outbreak of streptomycin-resistant TB in Benin that affected at least 17 patients shared an identical mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) pattern, except for two loci that authors failed to amplify (1). All these findings suggest that this clone originated in sub-Saharan Africa and that it is being introduced in countries with a low incidence of TB through immigration.
The purpose of this study was to develop a rapid diagnostic method based on multiplex PCR to identify the Beijing isolates and specifically the GC1237 genotype which could be applied in the laboratories located in the country of origin. The test could help in the subsequent prospective epidemiological studies to better control of the cases.
In order to carry this out, all the isolates from 2007 to 2008 collected in the three main hospitals of the Province of Las Palmas (Gran Canaria Island) were genotyped at the University of Zaragoza. One stored isolate of the original Beijing GC1237 strain was used as a control of the different techniques. First, all 292 isolates were screened by spoligotyping with a commercially available kit (Ocimum Biosolutions Ltd., Hyderabad, India) by the method of Kamerbeek et al. (14). Seventy of the 292 isolates belonged to the Beijing family. Second, IS6110 RFLP was performed on the Beijing isolates detected by spoligotyping, if enough DNA were available (32 of the 70 isolates) according to the standardized protocol (19). Twenty-nine isolates (90.6%) had RFLP patterns similar to the pattern of GC1237, four of which presented an additional band. The other 3 isolates showed a clearly different Beijing pattern. Besides, genotyping based on 15-locus MIRU-VNTR for 3 different isolates of the GC1237 genotype was conducted as previously described (18). The three isolates showed the pattern 442333464465372 (MIRU-VNTR loci ordered according to their position on the M. tuberculosis H37Rv genome).
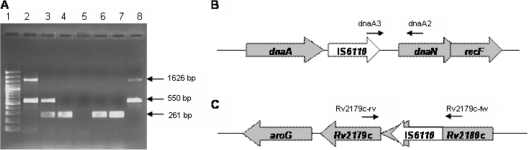
Subsequently, a multiplex PCR detecting two different locations of IS6110 was designed for a rapid identification of isolates related to the GC1237 genotype. The first target recognizes the point of insertion of IS6110 between dnaA and dnaN, which is a characteristic feature of all Beijing strains (15). To accomplish this, primer dnaA3 (5′-GGGCGGTTCAATTGGCTGT), was designed to anneal in the 3′ end of IS6110 and the following sequence of the genome and used together with primer dnaA2 (5′-CCACCCACGACACCGCAT) which had been previously described (10). The second target detected a specific IS6110 of strain GC1237. This target was chosen among the 19 copies of IS6110 recently localized in the genome of GC1237 (3). Eleven of the insertion sites were shared with 210 strains, 2 were located in the variable PPE regions, which could present difficulties in the specificity of the PCR. The other 5 were in hot spots, detected in other non-Beijing clinical isolates or its flanking region was not presented in the reference strain H37Rv. Finally, the copy inserted into the Rv2180c gene was selected as the specific target for rapid diagnosis. To detect this target, primers Rv2179c-fw (fw stands for forward) (5′-CAATACGTGATCGCCGGGAC) and Rv2179c-rv (rv stands for reverse) (5′-CGAATTCCGTGGATACTGCTAG) were designed (3). For this assay, a rapid DNA extraction was made as previously described (2). As a result of the multiplex PCR, two amplicons were expected for isolates with GC1237 genotypes of 1,626 bp and 550 bp, respectively. Two 550-bp fragments and a 261-bp fragment were expected for other Beijing isolates. A non-Beijing isolate was expected to produce only one fragment of 261 bp for the wild-type Rv2180c gene (Fig. 1).
Fig 1.
(A) Results of multiplex PCR visualized with agarose gel electrophoresis. Lane 1, molecular size markers (100-bp DNA ladder); lane 2, Beijing GC1237 genotype (1,626 bp and 550 bp); lane 3, a Beijing genotype different from strain GC1237 (550 bp and 261 bp); lane 4, strain H37Rv (261 bp); lane 5, negative PCR control; lanes 6, 7, and 8, different clinical isolates of M. tuberculosis. (B and C) Schematic representations of the positions and orientations of the two IS6110 insertions used for the diagnostic test, the dnaA-dnaN region in the Beijing strains (B) and the Rv2180c region in strain GC1237 (C). The IS6110 element is represented by the white arrow, with the arrowhead indicating the direction of transcription of the putative transposase. The primers are represented by the small black arrows above the schematic representations.
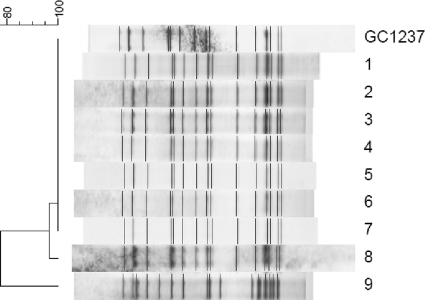
To validate this technique, all 58 isolates of M. tuberculosis from one of the hospitals reported between 2007 and 2008 were tested. Nine isolates were found to belong to the Beijing lineage, and eight of these isolates exhibited the GC1237 pattern. Subsequently, the IS6110 RFLP patterns of the Beijing isolates (9 isolates) were compared. The other 49 isolates presented a non-Beijing genotype. Of the isolates with a non-Beijing genotype, 8 isolates showed a high similarity to strain GC1237, and one had a different pattern, validating the diagnostic test, which showed 100% specificity (Fig. 2).
Fig 2.
Dendrogram showing the IS6110 RFLP fingerprints patterns of M. tuberculosis Beijing isolates used to verify the multiplex PCR technique. Isolates 1 to 8 shows high similarity to strain GC1237, and isolate 9 had a different pattern.
Finally, the multiplex PCR was used to identify the GC1237 genotype among 66 Beijing isolates. All the isolates amplified the specific target of Beijing family. Fifty-six presented the GC1237 genotype, and 10 presented a genotype of a different Beijing strain (Table 1). We confirmed these results by comparison with RFLP in 32 of the 66 isolates supporting the specificity of the test. A group of 50 “non-Beijing” isolates of TB patients diagnosed in the same period previously analyzed by spoligotyping were further analyzed, and all presented a non-Beijing genotype.
Table 1.
Results of the multiplex PCR (validating and applying the method) for detection of the isolates with the GC1237 genotype
| Purpose and method | No. of isolates with the following genotype: |
Total no. of isolates tested by method | ||
|---|---|---|---|---|
| Beijing GC1237 | Beijing but not GC1237 | Not Beijing | ||
| Validation | ||||
| Spoligotyping | 9a | 9a | 49 | 58 |
| Multiplex PCR | 8 | 1 | 49 | 58 |
| RFLP | 8 | 1 | 9 | |
| Test use | ||||
| Spoligotyping | 66a | 66a | 50 | 116 |
| Multiplex PCR | 56 | 10 | 50 | 116 |
Because spoligotyping does not discriminate between the Beijing GC1237 genotype and Beijing but not GC1237 genotype, these data correspond to the same isolates.
Due to the high incidence of this strain in the population studied, a rapid diagnostic method to discriminate non-Beijing subtypes from strain GC1237 would be useful for better control of TB on Gran Canaria island. Considering that the RFLP, spoligotyping, and MIRU methods are difficult to implement in the clinical setting, the described multiplex PCR could bring the possibility of in situ identification of GC1237 genotype isolates.
Until now, the incidence of this strain was retrospectively screened by spoligotyping in reference laboratories. This technique is a good method to detect those isolates of the Beijing lineage; however, information about clonality in this group was not provided. In addition, the spoligotyping method is better used for the study of 40 isolates at a time (13). RFLP analysis based on IS6110 gives us this information, but a larger amount of DNA is required, it is time-consuming and technically demanding, and the results among laboratories are difficult to compare (19). The rapid proposed technique 24-locus MIRU-VNTR could be also a tool for detecting this genotype, but it requires 24 single PCRs or 8 multiplex PCRs. Several PCR-based methods for the identification of Beijing strains have also been developed targeting different MIRU loci, specific deletions, or single-nucleotide polymorphisms (SNPs). However, most of these methods for identification are not specific, expensive, or difficult to perform (4, 7, 8, 17).
The results obtained in this work show that our multiplex PCR has demonstrated the specificity to identify the GC1237 genotype isolates. In contrast to spoligotyping and RFLP, this multiplex PCR can be used to identify one or more positive isolates. It is a quick, straightforward, and inexpensive technique. It can be completed in 1 day, and it does not require hybridization to a panel of species-specific probes, as RFLP does. Moreover, sample preparation is simple (simple heating of the bacteria), and trained personnel can interpret the gels with minimal difficulty. In contrast to 24-locus MIRU-VNTR, a single reaction is needed. It can be used by laboratories lacking a real-time PCR machine and easily implemented for a large-scale GC1237 strain screening. This designed PCR would allow its use in the clinical setting and facilitate the detection of new cases aiding a more detailed supervision of patients and discrimination from cases infected with other Beijing genotypes.
ACKNOWLEDGMENTS
This study was partially funded by the Fondo de Investigaciones Sanitarias (FIS09/0051), Instituto de Salud Carlos III (CM09/000123), and Gobierno de Aragón (B034/09) in Spain.
We thank Dessislava Marinova for her kind help revising the English and for valuable comments on the manuscript.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Affolabi D, et al. 2009. Possible outbreak of streptomycin-resistant Mycobacterium tuberculosis Beijing in Benin. Emerg. Infect. Dis. 15: 1123–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldous WK, Pounder JI, Cloud JL, Woods GL. 2005. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J. Clin. Microbiol. 43: 2471–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alonso H, et al. 2011. Deciphering the role of IS6110 in a highly transmissible Mycobacterium tuberculosis Beijing strain, GC1237. Tuberculosis 91: 117–126 [DOI] [PubMed] [Google Scholar]
- 4. Alonso M, et al. 2011. A novel method for the rapid and prospective identification of Beijing Mycobacterium tuberculosis strains by high-resolution melting analysis. Clin. Microbiol. Infect. 17: 349–357 [DOI] [PubMed] [Google Scholar]
- 5. Alonso M, et al. 2010. Characterization of Mycobacterium tuberculosis Beijing isolates from the Mediterranean area. BMC Microbiol. 10: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caminero JA, et al. 2001. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164: 1165–1170 [DOI] [PubMed] [Google Scholar]
- 7. Chen J, et al. 2007. Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis 87: 446–449 [DOI] [PubMed] [Google Scholar]
- 8. Chin PJ, Chiu CC, Jou R. 2007. Identification of Beijing lineage Mycobacterium tuberculosis with combined mycobacterial interspersed repetitive unit loci 26, 31, and ETR-A. J. Clin. Microbiol. 45: 1022–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis 2006. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg. Infect. Dis. 12: 736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fennelly KP, et al. 2004. Cough-generated aerosols of Mycobacterium tuberculosis: a new method to study infectiousness. Am. J. Respir. Crit. Care Med. 169: 604–609 [DOI] [PubMed] [Google Scholar]
- 11. Garcia de Viedma D, Chaves F, Iñigo J. 2006. New route of importation of Mycobacterium tuberculosis Beijing genotype. Emerg. Infect. Dis. 12: 169–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghebremichael S, et al. 2010. Drug resistant Mycobacterium tuberculosis of the Beijing genotype does not spread in Sweden. PLoS One 5: e10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hillemann D, Warren R, Kubica T, Rüsch-Gerdes S, Niemann S. 2006. Rapid detection of Mycobacterium tuberculosis Beijing genotype strains by real-time PCR. J. Clin. Microbiol. 44: 302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35: 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kremer K, et al. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42: 4040–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samper S, et al. 2005. Systematic molecular characterization of multidrug-resistant Mycobacterium tuberculosis complex isolates from Spain. J. Clin. Microbiol. 43: 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun JR, Lee SY, Dou HY, Lu JJ. 2009. Using a multiplex polymerase chain reaction for the identification of Beijing strains of Mycobacterium tuberculosis. Eur. J. Clin. Microbiol. Infect. Dis. 28: 105–107 [DOI] [PubMed] [Google Scholar]
- 18. Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44: 4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Embden JD, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31: 406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]