Abstract
Tuberculosis (TB) remains a significant global health problem for which rapid diagnosis is critical to both treatment and control. This report describes a multiplex PCR method, the Mycobacterial IDentification and Drug Resistance Screen (MID-DRS) assay, which allows identification of members of the Mycobacterium tuberculosis complex (MTBC) and the simultaneous amplification of targets for sequencing-based drug resistance screening of rifampin-resistant (rifampinr), isoniazidr, and pyrazinamider TB. Additionally, the same multiplex reaction amplifies a specific 16S rRNA gene target for rapid identification of M. avium complex (MAC) and a region of the heat shock protein 65 gene (hsp65) for further DNA sequencing-based confirmation or identification of other mycobacterial species. Comparison of preliminary results generated with MID-DRS versus culture-based methods for a total of 188 bacterial isolates demonstrated MID-DRS sensitivity and specificity as 100% and 96.8% for MTBC identification; 100% and 98.3% for MAC identification; 97.4% and 98.7% for rifampinr TB identification; 60.6% and 100% for isoniazidr TB identification; and 75.0% and 98.1% for pyrazinamider TB identification. The performance of the MID-DRS was also tested on acid-fast-bacterium (AFB)-positive clinical specimens, resulting in sensitivity and specificity of 100% and 78.6% for detection of MTBC and 100% and 97.8% for detection of MAC. In conclusion, use of the MID-DRS reduces the time necessary for initial identification and drug resistance screening of TB specimens to as little as 2 days. Since all targets needed for completing the assay are included in a single PCR amplification step, assay costs, preparation time, and risks due to user errors are also reduced.
INTRODUCTION
The diagnosis and control of tuberculosis (TB) is a very significant problem in global health. A recent report by the World Health Organization (WHO) estimates that Mycobacterium tuberculosis, the causative agent of the disease, has infected one-third of the world's population (71). TB can persist in a latent state within infected individuals for decades (69). Progression to active disease occurs in approximately 5% to 10% of immunocompetent individuals and triggers the frequent cough that enables the organism to be transmitted (11). Moreover, effective treatment of patients with active TB has become more difficult with the increasing prevalence of multidrug-resistant (MDR) strains (7, 70). MDR-TB is resistant to both of the first-line drugs rifampin (RIF) and isoniazid (INH) (8). MDR-TB cases have been estimated to account for one in four new TB cases in some parts of the world (70). In the United States, MDR-TB cases comprise approximately 1.1% of new TB cases, with the majority of these cases imported from regions where the disease is endemic.
All members of the M. tuberculosis complex (MTBC) are capable of causing pulmonary disease, and early diagnosis is the most effective means of TB control. Culture-based identification remains the gold standard method for TB diagnosis (10). However, culture-based identification of MTBC and traditional drug susceptibility testing (DST) can take up to 6 weeks due to the low growth rate of the organism. Molecular assays that use genetic markers such as insertion sequence 6110 (IS6110) allow the sensitive and specific detection of MTBC (16, 18) and can often provide results within a day. DNA sequence-based approaches that screen for mutations associated with drug resistance have become a valuable tool for the early identification of resistant TB isolates (55, 60, 76). For example, over 95% of all rifampin (RIF)-resistant strains of TB contain specific point mutations in the Rifampin Resistance-Determining Region (RRDR) of rpoB (60, 72). With INH resistance, a single mutation in codon 315 of katG is associated with up to 80% of isolates resistant to INH (11). A total of 70% to 80% of TB isolates resistant to pyrazinamide (PZA), another common first-line drug used in TB treatment, contain mutations within pncA (6, 19, 24, 58).
Among nontuberculous mycobacteria (NTM), infections by members of the Mycobacterium avium complex (MAC) are among those most frequently diagnosed in countries with a low TB incidence such as the United States (21). Patients with MAC infections can present with clinical manifestations similar to MTBC infections, such as cough, fever, malaise, and weight loss. MAC infections also pose a serious clinical problem for immunocompromised patients, in whom extensive organ damage and disseminated disease can occur. Furthermore, members of the MAC are naturally resistant to most drugs used to treat MTBC (46); therefore, rapid identification of MAC and the discrimination of MAC species from MTBC species is important for appropriate treatment.
The use of molecular testing is becoming more widespread to meet the need for rapid identification of TB-positive cases, including those harboring drug-resistant strains. Rapid molecular tests for the identification of MAC cases remain less common, with the AccuProbe culture test (Gen Probe, San Diego, CA) currently the most commonly used commercial test in US public health laboratories. For MTBC, the recently released GeneXpert MTB/RIF test from Cepheid (Sunnyvale, CA) shows great utility as a rapid, high-performance molecular assay to identify both TB and many RIF-resistant strains directly from sputum (4). Another new test, the GenoType MTBDR assay (Hain Lifesciences, Germany), permits the screening of multidrug-resistant TB directly from clinical specimens positive for acid-fast bacteria (AFB). However, the availability and use of the GenoType MTBDR assay remains limited primarily to laboratories outside the United States (30, 63). The development and use of laboratory-developed tests (LDT) for TB detection and drug resistance screening have also increased in recent years (6, 16, 41, 64, 75). For public health and other large reference laboratories, molecular-based LDTs often offer additional advantages compared to commercial tests, such as the ability to tailor assays to meet specific testing needs and greater cost-effectiveness, since the existing capital equipment and infrastructure can be utilized.
The MID-DRS assay described in this report utilizes a single multiplexed PCR with two components, representing both rapid mycobacterial identification and, in cases in which MTBC is detected, targets for DNA sequencing-based screening of mutations associated with resistance to the first-line drugs. The MID-DRS assay was developed and evaluated using both bacterial isolates and respiratory specimens, with complete analysis possible using AFB-positive clinical specimens in as little as 2 days.
MATERIALS AND METHODS
Bacterial strains.
Mycobacterial culture was performed using Middlebrook 7H9 liquid medium and incubation at 30°C. All culture isolates were confirmed as MTBC with the AccuProbe MTD assay (Gen Probe, San Diego, CA) and characterized for in vitro drug susceptibility to INH (0.1 μg/ml), RIF (0.2 μg/ml), and PZA (100 μg/ml) with the radiometric Bactec 460 system (57) (Becton Dickinson, Franklin Lakes, NJ). INH (0.2 and 1 μg/ml) and RIF (1 μg/ml) resistance was established by the agar proportion method using Middlebrook 7H10 quadrant plates with impregnated Sensi-Disc antibiotic disks (Becton Dickinson, Franklin Lakes, NJ) (66).
A total of 125 Mycobacterium tuberculosis culture isolates, derived from clinical cases of pulmonary TB, were obtained from the strain collection of the Washington State Public Health Laboratories (WAPHL). MTBC isolates representing at least 28 different spoligotypes were selected based on spoligotyping information provided by the Genotyping Laboratory of the California Department of Health Services (27, 28). Spoligotyping was also used to discriminate M. bovis and M. africanum from other MTBC strains. NTM strains used to assess assay specificity were cultured on Lowenstein-Jensen slants at 37°C and identified by biochemical tests (10, 29). Additionally, other bacterial pathogens associated with respiratory infection were also screened. The limit of detection of the PCR assay was established using 5-fold serial dilutions of logarithmic-phase liquid cultures of M. tuberculosis H37Rv ATCC 27294 and M. avium ATCC 25291 that were equilibrated to a McFarland standard of 1 (approximately 5 × 106 organisms/ml) (40). The numbers of CFU per milliliter were quantified in duplicate for each dilution by the standard plate count method. Sensitivity was determined as the lysate from the lowest dilution of the cell suspension that produced visible bands after PCR amplification.
Clinical specimens.
A clinical panel comprised of 100 patient specimens, including sputum (induced and expectorated) and respiratory washes (bronchial and tracheal), was collected from a total of 81 unique cases (no more than 2 specimens per individual were tested). The culture test results were handled in a blinded manner until molecular testing was complete. Specimens were prepared by digestion and decontamination using the N-acetyl l-cysteine NaOH-Na citrate method (10, 29). Each specimen was then centrifuged for 15 min at 2,500 × g to pellet the cells, and the supernatant was discarded. Each pellet was resuspended in 1.5 ml of sterile phosphate-buffered saline (PBS) (pH 6.8). Smears were prepared from the decontaminated specimens by the use of Auramine O, and the load of acid-fast bacteria was evaluated using fluorescence microscopy at ×20 and ×40 magnification. A specimen was considered smear positive when two or more AFB were present per 10 observed microscopic fields. The smear-positive specimens were classified according to conventional guidelines with scores of 1+ to 4+ based on the number of observed AFB-positive cells present (10, 29).
DNA preparation.
Sterile 1.5-ml screw cap microcentrifuge tubes (Perfector Scientific, Atascadero, CA) were filled with equal weights (0.5 g) of <106-μm-diameter and 425- to 600-μm-diameter acid-washed glass beads (Sigma, St. Louis, MO). An aliquot (250 μl) of either processed sputum or culture (equilibrated to a McFarland standard of 1 to 2) was used in each extraction. Cells were heat inactivated by submersion of tubes in a 95°C water bath for 30 min. Following inactivation, samples were frozen at −20°C. Thawed cell lysates were then disrupted for 2 min using a Cell Disruptor Genie (Scientific Industries, Bohemia, NY) and centrifuged at 6,000 × g for 5 min. The clarified lysate was transferred to a new tube and stored at −20°C until use.
Primer design.
PCR primers were designed for each target region (Table 1) by the use of Primer3 (53) and additional analysis to predict secondary structure and primer-dimer interactions when multiplexed in a 14-primer PCR using FastPCR (51), Oligo Analyzer 3.0 (Integrated DNA Technologies), and Mfold (25). Primer designs for the IS6110 and RD9 targets were based on the genome sequence of M. tuberculosis H37Rv (GenBank accession no. NC_000962). To identify MAC species, a consensus region derived from multiple sequence alignments of mycobacterial 16S rRNA gene sequences was created using ClustalW version 1.4 and MAC-specific primers were designed using the resulting consensus region and BioEdit version 7.0.9.0 (15, 61). The 16S DNA sequences from 84 different MAC strains, 9 other NTM species, 18 MTBC strains, and 9 other common bacterial respiratory pathogens were compared during sequence analysis. The putative primer sequences for the MAC assay were further screened for specificity using a large panel of NTM and MAC sequences (Table 2). A mix of two forward primers and one reverse primer was necessary for the specific detection of all the MAC species. Cross-reactivity was observed only with M. chimaera (GenBank accession no. AJ548480), a former member of the MAC (47, 62) for which positive PCR results were deemed correct, since in silico primer specificity studies showed that PCR detection of M. chimaera was expected. The primer sequences used for all other MID-DRS assay targets are outlined in Table 1. The 16S primers used for bacterial species via rRNA gene sequencing were as previously described (67, 73). The sequences of all primers used in this work are listed in Table 1. Primer concentrations between 50 and 250 nmol were evaluated for optimal performance. All primers were purchased from Integrated DNA Technologies (IDT, San Diego, CA).
Table 1.
Primers used in the MDR-TB assaya
| Amplification target | Forward | Reverse | Amplicon (bp) |
|---|---|---|---|
| RD9 | 5′-GTGTAGGTCAGCCCCATCC-3′ | 5′-GTAAGCGCGTGGTGTGGA-3′ | 369 |
| hsp65 | 5′-ACCAACGATGGTGTGTCCAT-3′ | 5′-CTTGTCGAACCGCATACCCT-3′ | 441 |
| katG | 5′-GAGCCCGATGAGGTCTATTG-3′ | 5′-GTCCTTGGCGGTGTATTGC-3′ | 498 |
| pncA PCR | 5′-GACGTATGCGGGCGTTGA-3′ | 5′-CCATCAGGAGCTGCAAACCA-3′ | 569 |
| rpoB PCR | 5′-CGAGGTGCCGGTGGAAAC-3′ | 5′-GTCGTCGTGCTCCAGGAAGG-3′ | 721 |
| rpoB sequencing | 5′-GGTGGAAACCGACGACATC-3′ | 5′-GGCGGTCAGGTACACGATCT-3′ | 508 |
| pncA sequencing | 5′-GTCGACGTGCAGAACGACTT-3′ | 5′-ACACACCCGCTGTCAGGTC-3′ | 472 |
| MAC 16S rRNA gene | 5′-GACCTCAAGACGCATGTCTTC-3′ | 5′-ACCGTCAATCCGAGAAAACC-3′ | 297 |
| 5′-GACCTTTAGRCGCATGTCTT-3′ | |||
| IS6110 | 5′-GGATCCTGCGAGCGTAGGCGTCGG-3′ | 5′-CCTGTCCGGGACCACCCGCGGCAA-3′ | 200 |
| 16S rRNA gene sequencing | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 5′-GGACTACCAGGGTATCTAAT-3′ | 798 |
Table 2.
MID-DRS PCR amplification products for mycobacterial and other bacterial strains used to screen specificitya
| Strain | Source or designation | No. of isolates tested | Target amplicon |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IS6110 | RD9 | rpoB | katG | pncA | MAC 16S | hsp65 | |||
| MTBC | |||||||||
| M. africanum | WAPHL | 1 | + | − | + | + | + | − | + |
| M. bovis | WAPHL | 16 | + | − | + | + | + | − | + |
| M. tuberculosis | ATCC 25177 | 1 | + | + | + | + | + | − | + |
| M. tuberculosis | ATCC 27294 | 1 | + | + | + | + | + | − | + |
| Not identified to the species level | WAPHL | 106 | + | + | + | + | + | − | + |
| NTM (MAC) | |||||||||
| M. avium | ATCC 25291 | 1 | − | − | − | − | − | + | + |
| M. avium | WAPHL | 7 | − | − | − | − | − | + | + |
| M. chimaera | WAPHL | 2 | − | − | − | − | − | + | + |
| M. intracellulare | ATCC 13209 | 1 | − | − | − | − | − | + | + |
| M. intracellulare | ATCC 13950 | 1 | − | − | − | − | − | + | + |
| M. intracellulare | ATCC 35761 | 1 | − | − | − | − | − | + | + |
| M. intracellulare | WAPHL | 3 | − | − | − | − | − | + | + |
| Other NTM | |||||||||
| M. abscessus | WAPHL | 1 | − | − | − | − | − | − | + |
| M. chelonaeb | WAPHL | 5 | − | − | − | − | − | − | + |
| M. colombienseb | WAPHL | 1 | − | − | − | − | − | + | + |
| M. cookiib | WAPHL | 2 | − | − | − | − | − | − | + |
| M. fortuitum | ATCC 6841 | 1 | − | − | − | − | − | − | + |
| M. fortuitum | WAPHL | 1 | − | − | − | − | − | − | + |
| M. gordonaeb | WAPHL | 6 | − | − | − | − | − | − | + |
| M. kansasii | WAPHL | 1 | − | − | − | − | − | − | − |
| M. malmoense | ATCC 29571 | 1 | − | − | − | − | − | + | + |
| M. marinum | ATCC 927 | 1 | − | − | − | − | − | − | + |
| M. mucogenicumb | WAPHL | 1 | − | − | − | − | − | − | + |
| M. nebraskense | WAPHL | 9 | − | − | − | − | − | − | + |
| M. nonchromogenicum | ATCC 19530 | 1 | − | − | − | − | − | − | + |
| M. peregrinum | WAPHL | 1 | − | − | − | − | − | − | + |
| M. poriferae | ATCC 35087 | 1 | − | − | − | − | − | − | + |
| M. scrofulaceum | ATCC 19981 | 1 | − | − | − | − | − | + | + |
| M. scrofulaceum | WAPHL | 3 | − | − | − | − | − | − | + |
| M. shimoidei | ATCC 27962 | 1 | − | − | − | − | − | − | + |
| M. simiae | ATCC 25273 | 1 | − | − | − | − | − | − | + |
| M. szulgai | ATCC 35799 | 1 | − | − | − | − | − | − | + |
| M. xenopi | WAPHL | 1 | − | − | − | − | − | − | + |
| Nonmycobacteria | |||||||||
| Arcanobacterium haemolyticum | WAPHL | 1 | − | − | − | − | − | − | − |
| Bordetella bronchiseptica | ATCC 19395 | 1 | + | − | + | − | − | − | + |
| Bordetella pertussis | ATCC 9797 | 1 | − | − | + | − | − | − | + |
| Burkholderia cepacia | ATCC 25410 | 1 | − | − | − | − | + | − | + |
| Corynebacterium striatum | WAPHL | 1 | + | − | − | − | − | − | + |
| Escherichia coli | ATCC 25922 | 1 | − | − | − | − | − | − | + |
| Gordonia bronchialis | WAPHL | 1 | − | − | + | − | − | − | − |
| Group A streptococcus | ATCC 12370 | 1 | − | − | − | − | + | − | − |
| Group B streptococcus | ATCC 27591 | 1 | − | − | − | − | − | − | − |
| Haemophilus influenzae | ATCC 49247 | 1 | − | − | − | − | − | − | − |
| Legionella pneumophila | ATCC 33152 | 1 | − | + | − | − | − | − | − |
| Moraxella catarrhalis | ATCC 25238 | 1 | − | − | − | − | − | − | − |
| Neisseria meningitidis | ATCC 13077 | 1 | − | + | − | − | − | − | + |
| Pseudomonas aeruginosa | ATCC 27853 | 1 | − | − | + | − | − | − | − |
| Rhodococcus equi | WAPHL | 1 | − | − | − | + | − | − | + |
| Staphylococcus aureus | ATCC 29213 | 1 | − | − | − | − | − | − | − |
| Staphylococcus epidermidis | ATCC 12228 | 1 | − | − | − | − | + | − | + |
| Streptococcus anginosus | ATCC 33397 | 1 | − | − | − | − | + | − | + |
| Streptococcus pneumoniae | ATCC 49619 | 1 | − | + | − | − | + | − | + |
| Streptococcus salivarius | ATCC 7073 | 1 | − | + | − | − | + | − | + |
| Yersinia pseudotuberculosis | ATCC 23207 | 1 | − | + | − | − | + | + | − |
PCR amplicons observed after agarose gel electrophoresis of MID-DRS reactions. The presence (+) or absence (−) of target genes amplified using the MID-DRS multiplex reaction is noted for each isolate tested.
Results from a small subset of isolates from the blind panel used in validation are included to illustrate PCR amplicons obtained with additional mycobacterial species.
PCR amplification.
Each PCR mixture contained 12.5 μl of 2× PCR mix (ImmoMix Red; Bioline, Taunton, MA), 5 μl of primer mix (containing 2.05 μmol/μl of IS6110 primers and 1 μmol/μl of primers for RD9, hsp65, rpoB, katG, pncA, and the MAC 16S rRNA gene), 2.5 μl of DNA template, and 5 μl of PCR-grade water in a final volume of 25 μl. Genomic DNA from M. tuberculosis ATCC 27294 and from M. avium ATCC 25291 served as positive-control templates. Amplification was performed using a DNA Engine Dyad Peltier thermal cycler (Bio-Rad, Hercules, CA). Thermocycling parameters were 95°C for 15 min; 40 cycles of 95°C for 30 s, 60°C for 50 s, and 72°C for 60 s; and a final step at 72°C for 5 min. The resulting PCR products were analyzed via agarose gel electrophoresis using 1.5% (wt/vol) agarose (NuSieve 3:1; FMC Bioproducts, Rockland, ME). The assay products were run for 60 min at 0.8 mV cm2 in 1× TBE buffer (90 mM Tris, 90 mM borate, 20 mM EDTA [pH 8.0]). DNA was stained with ethidium bromide (1 μg/ml), and the gels were imaged under UV light (AlphaInnotech Corp, San Leandro, CA). PCR amplicon sizes were estimated by comparison to molecular size markers (Hyperladder II; Bioline USA Inc, Taunton, MA). The amplification of 16S rRNA gene for sequence-based identification of bacterial species was performed as previously described (67, 73).
DNA sequencing.
Prior to DNA sequencing, enzymatic cleanup of each PCR was performed using a combination of Exonuclease I (Exo; New England BioLabs, Ipswich, MA) and shrimp alkaline phosphatase (SAP; USB, Affymetrix, Cleveland, OH) as described previously (68). For the sequencing of specific PCR products, 1 μl of the ExoSAP-treated DNA was mixed with 6 μl of 5× sequencing buffer and 2 μl of sequencing reaction mix from a BigDye Terminator cycle sequencing kit, version 1.1 (Applied Biosystems, Foster City, CA), 2 μl of amplicon-specific primer (2 μmol/μl), and 9 μl of PCR-grade water for a final volume of 20 μl. The strand-labeling reactions were performed according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Both strands of each amplicon were sequenced to ensure accuracy of the test data. The labeled DNA extension products were then purified using Sephadex G50 chromatography (GE Healthcare, Piscataway, NJ) and filter plates as described by the manufacturer (Millipore, Billerica, MA). The purified extension products were first dried and then resuspended in 20 μl of Hi-Dye formamide (Applied Biosystems, Foster City, CA). These were then sequenced using an ABI 3130xl genetic analyzer loaded with a 50-cm array according to the manufacturer's instructions (Applied Biosystems, Foster City, CA).
Sequencing analysis.
Sequencing results were evaluated for quality using Sequence Analysis software, version 5.3 (Applied Biosystems, Foster City, CA). “Good quality” readouts were characterized by a continuous read length of greater than 300 bp and with 250 bases exceeding a Phred score of quality value (QV) of >20 (QV) = −10 log10p (where p is the probability of error and the p value for QV > 20 is 1.0%). The average raw signal-to-noise ratio was above 150 for most sequence reads. Forward- and reverse-strand sequences (excluding the primer sequences) for hsp65, rpoB, katG, and pncA were assembled and edited with Sequencher (Gene Codes, Ann Arbor, MI). All sequence reads derived from rpoB, katG, and pncA amplicons were compared to the H37Rv sequences of these regions to identify previously characterized mutations by the use of BioEdit (15). Previously characterized mutations were defined as those associated with drug resistance as listed in the Tuberculosis Drug Resistance Mutation database (54). Unknown mycobacteria sequenced using hsp65 primers (401 bases internal to the 441-bp amplicon) were identified using the BLASTn search hosted by the British Columbia Center for Disease Control (BCCDC; http://hsp65blast.phsa.ca/) as having a minimum identity score of 97.5% (41, 42). All results that were discrepant between sequencing and culture-DST were verified by repeating the sequencing and DST using a fresh culture of the same isolate. 16S rRNA gene sequencing analysis employing the database search engines provided by RIDOM and BLASTN was used to further identify those isolates that hsp65 analysis failed to discriminate using the BCCDC database (64).
RESULTS
MID-DRS assay design.
The MID-DRS utilizes a seven-target multiplex PCR. The presence of specific amplicons produced by the reaction allows the direct identification of either members of the MAC (16S rRNA gene) or members of the MTBC (IS6110 positive), with the ability to also further differentiate M. tuberculosis or M. canetti (IS6110 positive/RD9 positive) from other members of the MTBC, including M. bovis (37, 65). Additional PCR products produced in the same reaction can be used as templates for DNA sequencing (hsp65) for mycobacterial identification or confirmation and, if a MTBC species is identified, to screen for mutations associated with drug resistance (rpoB, katG, pncA) (Fig. 1). Once optimized, each primer set produced a single amplicon corresponding to the predicted product size (Fig. 2). The primer components were then pooled to create a single multiplexed PCR assay, with initial performance assessed using DNA templates derived from M. tuberculosis H37Rv, M. bovis, and M. avium. With M. tuberculosis H37Rv, all PCR amplicons were observed except for the MAC-specific 16S amplicon (Fig. 2). Results from the assay using M. bovis were similar to M. tuberculosis results, with the exception of RD9, which is absent from the M. bovis genome and was therefore not amplified (5, 44). The performance of the MID-DRS assay using M. avium DNA revealed that only the MAC-specific 16S and hsp65 amplicons were produced (Fig. 2). These results indicate that the primers designed for the MID-DRS assay selectively amplify the intended target regions when combined into a multiplex reaction while also allowing straightforward discrimination via amplicon sizes.
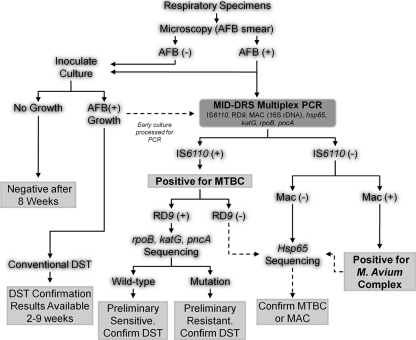
Fig 1.
Proposed MID-DRS testing algorithm. Multiplex PCR is performed using AFB smear-positive processed sputum samples or cultures positive for AFB. Amplification of IS6110 and RD9 prompts preliminary identification of M. tuberculosis and sequencing to screen for mutations associated with RIFr, INHr, and PZAr. Lack of RD9 with IS6110 amplification or lack of IS6110 with RD9 amplification prompts hsp65 sequencing for confirmation of MTBC. All IS6110-negative specimens are evaluated with hsp65 sequencing for confirmation of an (i) MTBC-negative result, (ii) M. avium result, or (iii) NTM identification. rDNA, rRNA gene.
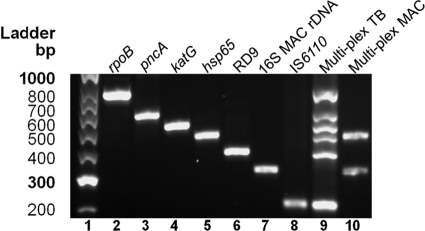
Fig 2.
Amplification products produced with the MID-DRS assay. Results of agarose gel electrophoresis of PCR amplicons produced from the individual and multiplexed primer pairs in the MID-DRS assay using genomic DNA are shown. Lane 1, a Hyperladder II standard; lane 2, rpoB; lane 3, pncA; lane 4, katG; lane 5, hsp65; lane 6, RD9; lane 7, 16S rRNA gene MAC; lane 8, IS6110; lane 9, multiplex PCR (7-plex) of all above-listed targets using TB DNA; lane 10, multiplex PCR (7-plex) of all above-listed targets using MAC DNA.
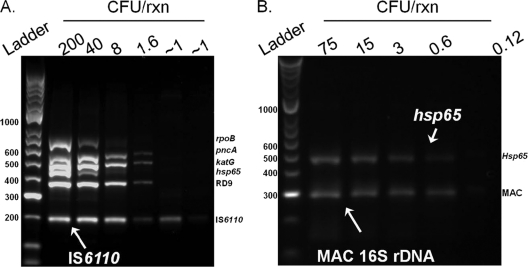
The analytical sensitivity of the MID-DRS reaction to detect MTBC via the IS6110 amplicon was established to be 2 to 4 cells (representing 32 to 64 copies of IS6110, approximately) (Fig. 3A). However, at this concentration the rpoB amplicon could not be clearly detected. Approximately 8 to 40 cells per reaction were required in order to amplify all 6 PCR targets in the MID-DRS assay (Fig. 3A), indicating this may be the minimal mycobacterial cell concentration necessary to consistently amplify adequate quantities of DNA template for use in the downstream sequencing application. The analytical sensitivity of the MID-DRS for detecting MAC was approximately 3 cells (approximately 3 copies of the 16S rRNA gene) (Fig. 3B).
Fig 3.
Limit of detection of MID-DRS multiplex PCR. The analytical sensitivity of the MID-DRS multiplex PCR was determined by comparison of resulting PCR amplification products to numbers of CFU produced with serial dilutions of either M. tuberculosis H37Rv ATCC 27294 (A) or M. avium ATCC 25291 (B). Cell counts were obtained by the direct plate count method.
Performance evaluation of MID-DRS for detection of MTBC and MAC.
MTBC detection by IS6110 was shown to have 100% sensitivity (125 of 125) and 96.8% specificity (61 of 63) using MID-DRS to test the panel of previously characterized culture isolates (Table 2). While no other NTMs were observed to produce an IS6110 amplicon, Corynebacterium striatum and Bordetella bronchiseptica produced weak amplicons of similar size to the 200-bp IS6110 amplicon. In addition, the lysates of these respiratory pathogens generated two unique amplicon profiles after agarose gel electrophoresis upon retesting (Table 2). Specifically, DNA from C. striatum amplified products that matched IS6110, rpoB, and hsp65 whereas B. bronchiseptica DNA amplified IS6110 and hsp65. The absence of the RD9 as well as the katG and pncA amplicons highlighted the results as discordant, because the mycobacterial species screened produce either 6 amplicons (M. tuberculosis and M. canetti) or 5 amplicons (other MTBC). These results were further evaluated using a IS6110 singleplex reaction, which produced only a 200-bp band in the C. striatum reaction. Attempts to obtain sequence data from the hsp65 amplicons produced by these isolates were unsuccessful even upon repeat testing using fresh culture isolates. The identification of these isolates to the species level was finally confirmed via 16S rRNA gene sequence analysis.
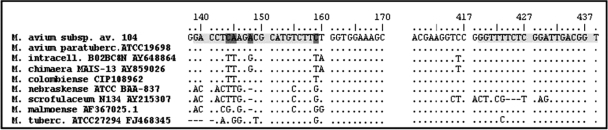
The detection of MAC with the MID-DRS assay is dependent on the targeted PCR amplification of a MAC-specific 16S rRNA gene region (Fig. 2, Fig. 4). The MID-DRS correctly identified 100% (16 of 16) of MAC isolates. The specificity was 98.3%, with the MAC amplicon being absent from 169 of 172 non-MAC isolates (Table 2). Two of the three non-MAC isolates that gave a positive PCR result were confirmed as non-MAC via sequence analysis of the hsp65 amplicon. These were M. malmoense and M. scrofulaceum. Acceptable hsp65 sequence data from the third isolate could not be generated, and the isolate was confirmed to be Yersinia pseudotuberculosis via 16S rRNA gene sequence analysis. Three additional clinical isolates of M. scrofulaceum were available from the WAPHL strain collection and were analyzed to further investigate the potential for cross-reactivity from this group of organisms. No amplification of MAC-specific 16S PCR product was observed in these M. scrofulaceum isolates, indicating that only a subset of M. scrofulaceum strains may cross-react with the MAC-specific primers.
Fig 4.
Specificity of primers for the 16S rRNA gene of MAC. Strains shown represent a subset of all strains used for in silico primer selection. Light gray areas represent the locations of forward and reverse primers with respect to the conventional base pair numbering of the 16S rRNA gene. Dark gray sections represent sites where changes were made to the forward primer for the design of the secondary forward primer used in PCRs. av., avium; paratuberc., paratuberculosis; intracell., intracellulare.
Drug resistance screening using DNA sequencing with MID-DRS products.
Genomic regions of TB containing the highest percentage of mutations that are strongly associated with resistance to first-line drugs were selected for the MID-DRS reaction. The targets were the RRDR of rpoB for RIF resistance (RIFr), the region of katG spanning codon 315 for INHr, and a portion of the pncA coding region that is most strongly associated with resistance to PZA (Table 1). Initial evaluation of the sequencing-based drug resistance screening component of MID-DRS was performed using isolates collected at the WAPHL from 1999 to 2010. Screening for RIFr via rpoB sequencing of 114 previously characterized M. tuberculosis isolates resulted in the correct identification of 38 of 39 (97.4%) isolates with RIFr-associated mutations (Table 3). One RIFr isolate had the wild-type sequence of the RRDR of rpoB. With RIFs isolates, 74 of 75 (98.7%) were correctly identified as RIFs (Table 3). A mutation in codon D516Y (GAC/TAC) was observed in a single RIFs isolate, producing a false-positive result (see Table 6). For INH, the screening of codon 315 in katG allowed identification of 60.6% (40 of 66) of the resistant isolates (Table 3). Finally, the ability of MID-DRS to detect PZA resistance was assessed with 76 isolates. Eighteen of 24 (75%) PZAr isolates had mutations in the pncA gene, while 1 of 52 (1.9%) of the PZAs isolates contained a mutation in the same region (Table 3). While the results of recent studies suggest that T61P, the pncA mutation of this isolate, is not a high-confidence mutation for PZAr (54), the classification of this isolate as a false positive was retained for analysis purposes, since no resistance was found by culture-based DST.
Table 3.
Sequencing-based identification of drug-resistant M. tuberculosis complex isolates by the use of MID-DRSa
| MID-DRS PCR result | No. of isolates with indicated culture DST result |
|
|---|---|---|
| Resistant | Sensitive | |
| RIF (rpoB) | ||
| Resistant | 38 | 1 |
| Sensitive | 1 | 74 |
| INH (katG) | ||
| Resistant | 40 | 0 |
| Sensitive | 26 | 48 |
| PZA (pncA) | ||
| Resistant | 18 | 1 |
| Sensitive | 6 | 51 |
For RIF, sensitivity was 97.4%, specificity was 98.7%, the positive predictive value was 97.4%, and the negative predictive value was 98.7%. For INH, sensitivity was 60.6%, specificity was 100.0%, the positive predictive value was 100.0%, and the negative predictive value was 64.9%. For PZA, sensitivity was 75.0%, specificity was 98.1%, the positive predictive value was 94.7%, and the negative predictive value was 89.5%.
Table 6.
Distribution of mutations associated with drug-resistant MTBCa
| Gene | Mutation | No. of isolates screened |
No. of isolates with mutation |
% tested isolates with mutation |
% resistant isolates showing multidrug resistance | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | |||
| rpoB | Q513K (CAA/AAA) | 114 | 39 | 75 | 1 | 0 | 2.6 | 0.0 | 48.7 |
| rpoB | D516V (GAC/GTC) | 114 | 39 | 75 | 2 | 0 | 5.1 | 0.0 | |
| rpoB | D516Y (GAC/TAC) | 114 | 39 | 75 | 0 | 1 | 0.0 | 1.3 | |
| rpoB | H526Y (CAC/TAC) | 114 | 39 | 75 | 4 | 0 | 10.3 | 0.0 | |
| rpoB | H526D (CAC/GAC) | 114 | 39 | 75 | 3 | 0 | 7.7 | 0.0 | |
| rpoB | H526C (CAC/TGC) | 114 | 39 | 75 | 2 | 0 | 5.1 | 0.0 | |
| rpoB | H526L (CAC/CTC) | 114 | 39 | 75 | 3 | 0 | 7.7 | 0.0 | |
| rpoB | S531L (TCG/TTG) | 114 | 39 | 75 | 23 | 0 | 59.0 | 0.0 | |
| katG | S315T (AGC/ACC) | 114 | 66 | 48 | 40 | 0 | 60.6 | 0.0 | 45.7 |
| pncA | H57D (CAC/GAC) | 76 | 24 | 52 | 15 | 0 | 62.5 | 0.0 | 10.5 |
| pncA | H57P (CAC/CCC) | 76 | 24 | 52 | 1 | 0 | 4.2 | 0.0 | |
| pncA | T61P (ACA/CCA) | 76 | 24 | 52 | 0 | 1 | 0.0 | 1.9 | |
| pncA | H82D (CAT/GAC) | 76 | 24 | 52 | 1 | 0 | 4.2 | 0.0 | |
| pncA | C138Y (TGT/TAT) | 76 | 24 | 52 | 1 | 0 | 4.2 | 0.0 | |
Results indicate the prevalence of various alleles associated with RIFr, INHr, and PZAr that were detected when clinical isolates were screened during the evaluation of the MID-DRS assay.
Validation of MID-DRS for MTBC and MAC identification with culture isolates.
The performance of the MID-DRS was validated using a set of 93 clinical isolates of M. tuberculosis and NTM and a blinded approach. MTBC detection using IS6110 correctly identified 59 out 59 (100%) of MTBC isolates (Table 4). Twelve of 13 (92.3%) of the NTM species were correctly identified as MAC by the use of MID-DRS (Table 5). Further evaluation of the false-negative NTM by hsp65 sequencing resulted in the identification of M. chimaera. Additionally, one NTM that was not identified as a MAC species by culture amplified an amplicon equal in size to the MAC 16S rRNA gene target amplicon and was thus considered to represent a false positive. This isolate had an hsp65 sequence without a match in the BCCDC database. Upon 16S rRNA gene sequence analysis, the isolate showed 100% identity to M. peregrinum/M. septicum.
Table 4.
Validation of MID-DRS PCR for identification of M. tuberculosis complexa
| MID-DRS PCR (IS6110) resulta | No. of isolates or samples with indicated MGIT culture result |
|
|---|---|---|
| Positive | Negative | |
| Isolates | ||
| Positive | 59 | 0 |
| Negative | 0 | 34 |
| AFB-positive sputum samples | ||
| Positive | 35 | 3 |
| Negative | 0 | 11 |
For the isolates (blinded test), sensitivity was 100.0%, specificity was 100.0%, the positive predictive value was 100.0%, and the negative predictive value was 100.0%. For the AFB-positive sputum samples, sensitivity was 100.0%, specificity was 78.6%, the positive predictive value was 92.1%, and the negative predictive value was 100.0%.
Table 5.
Validation of MID-DRS PCR for identification of M. avium complexa
| MID-DRS PCR (MAC 16S rRNA gene) result | No. of isolates or samples with indicated MGIT culture result |
|
|---|---|---|
| Positive | Negative | |
| Isolates | ||
| Positive | 12 | 1 |
| Negative | 1 | 79 |
| AFB-positive sputum samples | ||
| Positive | 4 | 1 |
| Negative | 0 | 44 |
For the isolates (blinded test), sensitivity was 92.3%, specificity was 98.8%, the positive predictive value was 92.3%, and the negative predictive value was 98.8%. For the AFB-positive sputum samples, sensitivity was 100.0%, specificity was 97.8%, the positive predictive value was 80.0%, and the negative predictive value was 100.0%.
Validation of MID-DRS for use with clinical specimens.
A total of 49 AFB smear-positive specimens submitted to the WAPHL between 2009 and 2010 were used to evaluate the performance of MID-DRS with clinical specimens. The detection of MTBC from AFB smear-positive specimens had a sensitivity of 100% and a specificity of 78.6% (Table 4). Among specimens collected from TB patients undergoing treatment during or prior to our study, two were classified as false positive, since no TB growth was observed (23). Another specimen was also considered a false positive, as it showed simultaneous products corresponding to both TB and MAC species, and the performance of the MID-DRS assay has not been evaluated for use with dual infections. This specimen was collected from a patient previously treated for TB in 2007; however, only MAC was isolated from the patient's sputum specimen tested in this study. Sequence-based screening for drug resistance was performed on a subset of 26 of the 49 AFB-positive sputum specimens with AFB concentrations ranging from 1+ to 4+ as determined by smear microscopy. Fourteen of the specimens with more than 10 AFB per field (3+ or more) gave reliable sequencing results with consistently high-quality reads as defined in Materials and Methods. The remaining 12 specimens with lower AFB smear concentrations did not consistently provide reads of good quality using the MID-DRS amplification products. The results from this limited sample set suggest that clinical specimens with more than 10 AFB per field may be necessary to produce MID-DRS products of reliable quantity for use as sequencing templates in drug resistance screening or that a better specimen concentration and DNA extraction protocol is required.
Four specimens were identified as MAC by both culture and MID-DRS (Table 5). An additional specimen was considered a false positive, as it produced an amplicon equal in size to the 16S rRNA gene target amplicon. Due to its low AFB load (1+), hsp65 sequencing results were not available for species identification directly from this specimen. However, the isolate was identified as M. chelonae by culture methods. The sequencing of hsp65 from isolates identified as MAC by the MAC 16S rRNA gene PCR confirmed the presence of MAC species, including M. avium, M. intracellulare, and M. chimaera.
Identification of NTM via hsp65 sequence analysis.
The 441-bp hsp65 amplicon was sequenced for all mycobacterial isolates that did not amplify IS6110 in either the original verification panel or the blinded validation study (Table 2). Using the BCCDC hsp65 sequence-based identification database, 26 of 29 (89.7%) MAC 16S rRNA gene-positive isolates were confirmed as MAC specimens and 43 other NTM were also identified to the species level (41, 42). 16S rRNA gene sequencing-based identification was conducted on four isolates for which conclusive identification using hsp65 could not be obtained. These isolates were identified as M. xenopi, M. peregrinum/septicum, and M. cookii (2 isolates). The 16S sequencing results matched the biochemical identification of these isolates, with the exception of the M. cookii isolates, which were identified as M. scrofulaceum. In total, culture-based identification was in agreement with hsp65 sequencing results for 72 of 76 (94.7%) NTM isolates.
Identification using hsp65 sequencing was also performed on 27 AFB-positive processed sputum samples, of which 21 (77.8%) produced hsp65 amplification with subsequent good-quality sequencing reads as defined in Materials and Methods. Eighteen of these specimens contained two or more AFB per field (≥2+ AFB), while 3 of the 9 specimens with 1+ AFB scores produced hsp65 sequences of sufficient quality. The results of biochemical identification matched the results of hsp65 sequence identification for all 21 (100%) specimens.
DISCUSSION
The rapid detection of MTBC is essential for outbreak control, and the early identification of drug-resistant strains improves patient management decisions during TB therapy and aids public health responses. The MID-DRS assay offers a rapid, sensitive, and cost-effective molecular approach to accurately identify TB and other mycobacteria in addition to screening for the most common mutations associated with resistance to RIF, INH, and PZA. The assay utilizes equipment for conventional PCR and capillary-based DNA sequencing methods that are available to the majority of large clinical laboratories. Furthermore, the assay facilitates the simultaneous amplification of all targets needed for mycobacterial identification and, if an MTBC species is detected, first-line drug resistance screening reduces labor and reagent costs as well as additional PCR amplification steps that could introduce user errors (Fig. 1).
The IS6110-based TB identification component of the MID-DRS assay was highly specific for MTBC, as it did not cross-react with over 40 NTM isolates and 19 other common respiratory pathogens tested in the study (56) (Table 2). However, two nonmycobacteria, C. striatum and B. bronchiseptica, produced PCR products that were indistinguishable from the IS6110 amplicon by the use of agarose gel electrophoresis. Corynebacterium spp. are chemotaxonomically related to the mycobacteria and have been associated with opportunistic infections in patients recovering from invasive surgery (39). B. bronchiseptica is a common respiratory pathogen of swine and thus could be associated with zoonotic transmission (3). Infection with C. striatum or B. bronchiseptica and their etiologies preclude them from being found in the sputum of patients with pulmonary TB disease. Conversely, by following the MID-DRS test algorithm (Fig. 1), in the event that a nonspecific amplicon was detected by PCR, the lack of other MDR-TB PCR targets would prompt subsequent hsp65 sequencing. This in turn would reduce the potential misidentification of an organism as MTBC. Use of the complete testing algorithm also ensured the proper identification of the small proportion of MAC-positive specimens (less than 2% in the current study) that were incorrectly identified as MAC by the use of the 16rRNA gene PCR component of the MID-DRS assay. Therefore, the proposed experimental flow, which incorporates hsp65 sequencing, can confirm and therefore enhance the specificity of the initial PCR results.
The potential to rapidly identify MAC and other NTM species in countries with a low TB incidence such as the United States is required within many health care systems to rule out MTBC infections and inform critical decisions, including the imposition of quarantine (21). The testing of a limited number of MAC-positive clinical specimens with MID-DRS demonstrated a specificity of 97.8%, indicating that the rapid-detection component for MAC could be a useful tool for high-throughput screening. Moreover, the MID-DRS assay allows further species identification of MAC complex members as well as the identification of other NTM species through downstream sequencing of the hsp65 amplicon included in the multiplex reaction. Therefore, the prompt identification of other NTM and exclusion of MTBC in cases with syndromic TB diagnosis is possible with this assay (41, 42).
Applying the MID-DRS assay for detection of TB from patient specimens with AFB-positive smears further improved turnaround time to a single day while retaining acceptable sensitivity and specificity, especially for patients with new TB cases who were not currently under treatment. As with all DNA-based diagnostic methods for TB, the results generated by MID-DRS PCR using samples from patients undergoing TB treatment require careful consideration in cases of syndromic diagnosis. In the present study, we detected TB via DNA in two specimens that were TB culture negative. Such individuals may continue to shed DNA while harboring nonviable organisms after starting treatment (1, 23, 38). This time period appears to correlate with drug effectiveness not only for reaching the infected area but also for inactivating bacteria (1, 23, 38). Some organisms are able to withstand and shield themselves from treatment better than others, and slow-growing mycobacteria are among the most difficult to target. For example, studies have shown that leprosy patients (with M. leprae disease) continue to test positive by PCR 3 months posttreatment compared to patients diagnosed with infections by chlamydia, whose DNA is no longer detected after the second week of treatment (23, 38).
A limited set of smear-negative, culture-positive specimens evaluated with the MIDR-DRS method indicated specificity of almost 100% but reduced sensitivity (data not shown). Such a small study set made a statistically valid evaluation of the method with this specimen type impossible. However, the low sensitivity of the method with smear-negative specimens was not unexpected, given the reduced performance of most nucleic acid amplification tests for TB with this specimen type (16, 18), including commercially available tests such as the Amplified Mycobacterium tuberculosis Direct Test (AMTD) (32). The sensitivity of testing with smear-negative specimens may be improved by enriching the specimens via short-term culture as previously described (41).
The results of preliminary drug resistance screening using MID-DRS were possible within 2 days upon receipt of a smear-positive specimen. The analysis of the sequence quality for templates amplified directly from clinical specimens indicated that specimens with moderate AFB content (3+ or more) are capable of yielding high-quality sequence data without additional amplification steps. Others have also been able to obtain sequencing data directly from clinical specimens (9, 26, 45). A reduction in the quality of DNA sequences is commonly encountered with specimens containing low numbers of AFB, because these specimens typically display suboptimal amplification of the templates required for sequencing analysis. This is a result of the typically low yield of purified DNA in addition to the carryover of inhibitory compounds from sputum specimens (2, 33).
In this study, the distribution of mutations within rpoB observed in TB isolates was similar to the distribution found globally, with the S531L mutation in RpoB the most prevalent (59.0%), followed by mutations in codon 526 (30.8%) and codon 516 (5.1%) (Table 6) (6, 17, 48, 72). In one instance, we detected a mutation in codon D516Y in an isolate that was sensitive to RIF (72). Previous reports suggest D516V is the most common mutation for this codon in RIFr strains (22, 43, 72). Further investigation is necessary to assess the strength of association with RIFr of different amino acid substitutions at this codon.
The sensitivity of detection of INH resistance by screening for mutations in katG codon 315 was 60.6%. This is within the previously reported range of 20% to 80% noted for this mutation (49, 52, 59). The detection of INHr associated with mutations in katG codon 315 was incorporated into the assay to provide the highest level of association with drug resistance for a single target. However, other mutations are known to confer INHr, notably, mutations in the promoter region of inhA and nod (34, 49). We are investigating the expansion of the MID-DRS method to include additional amplicons that would permit these targets to be analyzed as well. The overall sensitivity of INHr screening was increased from 60.1% to 79.7% by the further interrogation of the inhA promoter region of 12 INHr isolates that had the wild-type katG allele within the sample set (data not shown).
Approximately 70% of the resistance in PZAr isolates occurs as a result of mutation in the pncA promoter or coding region (19, 55). We observed comparable levels of sensitivity with 75.0% of PZAr isolates harboring mutations in pncA. Resistance to PZA is also a common marker for M. bovis (5), a member of the MTBC which is naturally resistant to PZA and is occasionally implicated in human-to-human airborne transmission of bovine TB (13, 14, 36). In the United States, rates of PZAr are low at 3.1%; therefore, screening for PZAr may be useful when targeting patients with a history of close contact with cattle and/or consumption of unpasteurized milk and milk products from regions with high M. bovis rates (12, 20, 35). In addition, M. bovis BCG is given intravesicularly to bladder cancer patients as a treatment strategy. Monitoring for potential complications resulting from this procedure is sometimes necessary, and public health laboratories play an essential role in this effort (31, 50, 74). During our study, IS6110-positive isolates, lacking RD9 and positive for a pncA H57D mutation (5), were correlated to 100% (16 of 16) of isolates identified as M. bovis or M. bovis BCG by spoligotyping, making our detection strategy a simple preliminary test for this organism.
In conclusion, MID-DRS assists in the rapid confirmation of TB, MAC, and other NTM species in smear-positive specimens and reduces the time and number of steps necessary to assess resistance to first-line TB drugs. The utilization of conventional PCR and capillary sequencing for drug resistance screening makes the assay amenable to future expansion to interrogate other targets. Globally, approximately 5.4% of MDR-TB cases had extensively drug-resistant TB (XDR-TB) in 2008 and an estimated 25,000 people acquire XDR-TB every year (7, 70). Expansion of the assay to include other targets associated with drug resistance, especially ones correlated with resistance to second-line drugs indicative of XDR-TB strains, could quickly provide public health laboratories with necessary tools to respond rapidly to these rare but increasing threats.
ACKNOWLEDGMENTS
We thank Karen Hiltbruner, Cathy Lisle, and Calley Vandegrift at the Washington State Public Health Laboratories for assistance with laboratory testing and data mining. We thank Jonathan Frye for his critical review during preparation of the manuscript.
We also acknowledge the Emerging Infectious Diseases (EID) Fellowship Program administered by the Association of Public Health Laboratories (APHL) and funded by the Centers for Disease Control and Prevention (CDC) for support of A.C.P.-O. and the Expansion of Nucleic Acid Amplification Testing for TB in Public Health Laboratories grant administered by the APHL and the CDC for funding to support a portion of this work.
The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the Washington State Department of Health.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Aberer E, Bergmann AR, Derler AM, Schmidt B. 2007. Course of Borrelia burgdorferi DNA shedding in urine after treatment. Acta Derm. Venereol. 87: 39–42 [DOI] [PubMed] [Google Scholar]
- 2. Amicosante M, et al. 1995. Inactivation of polymerase inhibitors for Mycobacterium tuberculosis DNA amplification in sputum by using capture resin. J. Clin. Microbiol. 33: 629–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baron S. 1996. Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX: [PubMed] [Google Scholar]
- 4. Boehme CC, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brosch R, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99: 3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell PJ, et al. 2011. Molecular detection of mutations associated with first and second-line drug resistance compared with conventional drug susceptibility testing in M. tuberculosis. Antimicrob. Agents Chemother. 55: 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 2010. Decrease in reported tuberculosis cases—United States, 2009. MMWR Morb. Mortal. Wkly. Rep. 59: 289–294 [PubMed] [Google Scholar]
- 8. Centers for Disease Control and Prevention 1990. Nosocomial transmission of multidrug-resistant tuberculosis to health-care workers and HIV-infected patients in an urban hospital—Florida. MMWR Morb. Mortal. Wkly. Rep. 39: 718–722 [PubMed] [Google Scholar]
- 9. Choi JH, et al. 2010. Clinical efficacy of direct DNA sequencing analysis on sputum specimens for early detection of drug-resistant Mycobacterium tuberculosis in a clinical setting. Chest 137: 393–400 [DOI] [PubMed] [Google Scholar]
- 10. CLSI 2008. Laboratory detection and identification of mycobacteria: approved guideline. CLSI document M48-A Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Cole ST, Davis Eisenach K, McMurray DN, Jacobs WR., Jr 2005. Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC [Google Scholar]
- 12. de Kantor IN, LoBue PA, Thoen CO. 2010. Human tuberculosis caused by Mycobacterium bovis in the United States, Latin America and the Caribbean. Int. J. Tuberc. Lung Dis. 14: 1369–1373 [PubMed] [Google Scholar]
- 13. Evans JT, et al. 2007. Cluster of human tuberculosis caused by Mycobacterium bovis: evidence for person-to-person transmission in the UK. Lancet 369: 1270–1276 [DOI] [PubMed] [Google Scholar]
- 14. Guerrero A, et al. 1997. Nosocomial transmission of Mycobacterium bovis resistant to 11 drugs in people with advanced HIV-1 infection. Lancet 350: 1738–1742 [DOI] [PubMed] [Google Scholar]
- 15. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- 16. Halse TA, et al. 2010. Combined real-time PCR and rpoB gene pyrosequencing for rapid identification of Mycobacterium tuberculosis and determination of rifampin resistance directly in clinical specimens. J. Clin. Microbiol. 48: 1182–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heep M, et al. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39: 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helb D, et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48: 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. 1997. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuberc. Lung Dis. 78: 117–122 [DOI] [PubMed] [Google Scholar]
- 20. Hlavsa MC, et al. 2008. Human tuberculosis due to Mycobacterium bovis in the United States, 1995–2005. Clin. Infect. Dis. 47: 168–175 [DOI] [PubMed] [Google Scholar]
- 21. Horsburgh CR, Jr, et al. 1991. Survival of patients with acquired immune deficiency syndrome and disseminated Mycobacterium avium complex infection with and without antimycobacterial chemotherapy. Am. Rev. Respir. Dis. 144: 557–559 [DOI] [PubMed] [Google Scholar]
- 22. Isfahani BN, Tavakoli A, Salehi M, Tazhibi M. 2006. Detection of rifampin resistance patterns in Mycobacterium tuberculosis strains isolated in Iran by polymerase chain reaction-single-strand conformation polymorphism and direct sequencing methods. Mem. Inst. Oswaldo Cruz 101: 597–602 [DOI] [PubMed] [Google Scholar]
- 23. Job CK, Jayakumar J, Kearney M, Gillis TP. 2008. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am. J. Trop. Med. Hyg. 78: 518–521 [PubMed] [Google Scholar]
- 24. Jonmalung J, Prammananan T, Leechawengwongs M, Chaiprasert A. 2010. Surveillance of pyrazinamide susceptibility among multidrug-resistant Mycobacterium tuberculosis isolates from Siriraj Hospital, Thailand. BMC Microbiol. 10: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalendar R, Lee D, Shulman AH. 2011. Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analyses. Genomics 98: 137–144 [DOI] [PubMed] [Google Scholar]
- 26. Kapur V, et al. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119: 131–138 [PubMed] [Google Scholar]
- 27. Kato-Maeda M, et al. 2011. Strain classification of Mycobacterium tuberculosis: congruence between large sequence polymorphisms and spoligotypes. Int. J. Tuberc. Lung Dis. 15: 131–133 [PMC free article] [PubMed] [Google Scholar]
- 28. Kato-Maeda M, Metcalfe JZ, Flores L. 2011. Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol. 6: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kent PT, Kubica GP. 1985. Public health mycobacteriology. U.S. Dept of Health and Human Services, Public Health Service, Centers for Disease Control, Atlanta, GA [Google Scholar]
- 30. Lalvani A. 2007. Diagnosing tuberculosis infection in the 21st century: new tools to tackle an old enemy. Chest 131: 1898–1906 [DOI] [PubMed] [Google Scholar]
- 31. Lamm DL. 1992. Complications of bacillus Calmette-Guerin immunotherapy. Urol. Clin. North Am. 19: 565–572 [PubMed] [Google Scholar]
- 32. Lebrun L, Mathieu D, Saulnier C, Nordmann P. 1997. Limits of commercial molecular tests for diagnosis of pulmonary tuberculosis. Eur. Respir. J. 10: 1874–1876 [DOI] [PubMed] [Google Scholar]
- 33. Leckie GW, et al. 1998. Method for reduction of inhibition in a Mycobacterium tuberculosis-specific ligase chain reaction DNA amplification assay. J. Clin. Microbiol. 36: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee AS, Teo AS, Wong SY. 2001. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45: 2157–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. LoBue PA, Betacourt W, Peter C, Moser KS. 2003. Epidemiology of Mycobacterium bovis disease in San Diego County, 1994–2000. Int. J. Tuberc. Lung Dis. 7: 180–185 [PubMed] [Google Scholar]
- 36. LoBue PA, et al. 2004. Identification of a familial cluster of pulmonary Mycobacterium bovis disease. Int. J. Tuberc. Lung Dis. 8: 1142–1146 [PubMed] [Google Scholar]
- 37. Lok KH, et al. 2002. Molecular differentiation of Mycobacterium tuberculosis strains without IS6110 insertions. Emerg. Infect. Dis. 8: 1310–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Markey B, Wan C, Hanger J, Phillips C, Timms P. 2007. Use of quantitative real-time PCR to monitor the shedding and treatment of chlamydiae in the koala (Phascolarctos cinereus). Vet. Microbiol. 120: 334–342 [DOI] [PubMed] [Google Scholar]
- 39. Martínez-Martínez L, Suarez AI, Winstanley J, Ortega MC, Bernard K. 1995. Phenotypic characteristics of 31 strains of Corynebacterium striatum isolated from clinical samples. J. Clin. Microbiol. 33: 2458–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McFarland J. 1907. Nephelometer. JAMA 49: 1176–1178 [Google Scholar]
- 41. McNabb A, Adie K, Rodrigues M, Black WA, Isaac-Renton J. 2006. Direct identification of mycobacteria in primary liquid detection media by partial sequencing of the 65-kilodalton heat shock protein gene. J. Clin. Microbiol. 44: 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McNabb A, et al. 2004. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 42: 3000–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Negi SS, et al. 2009. Characterization of RPO B gene for detection of rifampicin drug resistance by SSCP and sequence analysis. Indian J. Med. Microbiol. 27: 226–230 [DOI] [PubMed] [Google Scholar]
- 44. Parsons LM, et al. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40: 2339–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patnaik M, Liegmann K, Peter JB. 2001. Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampin resistance with DNA extracted directly from slides. J. Clin. Microbiol. 39: 51–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peloquin CA. 1993. Mycobacterial infections. Pharmacotherapy 13: 634–639 [PubMed] [Google Scholar]
- 47. Piersimoni C, Scarparo C. 2008. Pulmonary infections associated with non-tuberculous mycobacteria in immunocompetent patients. Lancet Infect. Dis. 8: 323–334 [DOI] [PubMed] [Google Scholar]
- 48. Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79: 3–29 [DOI] [PubMed] [Google Scholar]
- 49. Ramaswamy SV, et al. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rawls WH, et al. 1990. Fatal sepsis following intravesical bacillus Calmette-Guerin administration for bladder cancer. J. Urol. 144: 1328–1330 [DOI] [PubMed] [Google Scholar]
- 51. Rouillard JM, Zuker M, Gulari E. 2003. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 31: 3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rouse DA, Morris SL. 1995. Molecular mechanisms of isoniazid resistance in Mycobacterium tuberculosis and Mycobacterium bovis. Infect. Immun. 63: 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- 54. Sandgren A, et al. 2009. Tuberculosis drug resistance mutation database. PLoS Med. 6: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2: 662–667 [DOI] [PubMed] [Google Scholar]
- 56. Shamputa IC, Rigouts And L, Portaels F. 2004. Molecular genetic methods for diagnosis and antibiotic resistance detection of mycobacteria from clinical specimens. APMIS 112: 728–752 [DOI] [PubMed] [Google Scholar]
- 57. Siddiqi SH. 1989. Bactec TB system. Product and procedure manual. Revision B. Becton Dickinson Diagnostic Instruments System, Towson, MD [Google Scholar]
- 58. Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. 1997. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob. Agents Chemother. 41: 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Telenti A, et al. 1997. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 35: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Telenti A, et al. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341: 647–650 [DOI] [PubMed] [Google Scholar]
- 61. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tortoli E, et al. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54: 1277–1285 [DOI] [PubMed] [Google Scholar]
- 63. Traore H, van Deun A, Shamputa IC, Rigouts L, Portaels F. 2006. Direct detection of Mycobacterium tuberculosis complex DNA and rifampin resistance in clinical specimens from tuberculosis patients by line probe assay. J. Clin. Microbiol. 44: 4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Turenne CY, Tschetter L, Wolfe J, Kabani A. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39: 3637–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Warren RM, et al. 2006. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 10: 818–822 [PubMed] [Google Scholar]
- 66. Wayne LG, Krasnow I. 1966. Preparation of tuberculosis susceptibility testing mediums by means of impregnated disks. Tech Bull. Regist. Med. Technol. 36: 115–117 [PubMed] [Google Scholar]
- 67. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Werle E, Schneider C, Renner M, Volker M, Fiehn W. 1994. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 22: 4354–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. WHO 2009. 2009 update, tuberculosis facts—Stop TB Partnership. World Health Organization, Geneva, Switzerland: http://www.stoptb.org/assets/documents/resources/factsheets/tbfactsheet_2009update_one_page.pdf [Google Scholar]
- 70. WHO 2010. Fact sheet: multidrug and extensively drug-resistant TB (M/XTR-TB). 2010 Global report on surveillance and response. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2010/9789241599191_eng.pdf [Google Scholar]
- 71. WHO 2008. The top 10 causes of death. Fact sheet no. 310 World Health Organization, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs310_2008.pdf [Google Scholar]
- 72. Williams DL, et al. 1994. Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38: 2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wilson KH, Blitchington RB, Greene RC. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28: 1942–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Witjes JA, vd Meijden AP, Debruyne FM. 1990. Use of intravesical bacillus Calmette-Guerin in the treatment of superficial transitional cell carcinoma of the bladder: an overview. Urol. Int. 45: 129–136 [DOI] [PubMed] [Google Scholar]
- 75. Yang Z, et al. 2005. Simultaneous detection of isoniazid, rifampin, and ethambutol resistance of Mycobacterium tuberculosis by a single multiplex allele-specific polymerase chain reaction (PCR) assay. Diagn. Microbiol. Infect. Dis. 53: 201–208 [DOI] [PubMed] [Google Scholar]
- 76. Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358: 591–593 [DOI] [PubMed] [Google Scholar]