Abstract
HIV-1 group M is classified into 9 subtypes, as well as recombinants favored by coinfection and superinfection events with different variants. Although HIV-1 subtype B is predominant in Europe, intersubtype recombinants are increasing in prevalence and complexity. In this study, phylogenetic analyses of pol sequences were performed to detect the HIV-1 circulating and unique recombinant forms (CRFs and URFs, respectively) in a Spanish cohort of antiretroviral treatment-naïve HIV-infected patients included in the Research Network on HIV/AIDS (CoRIS). Bootscanning and other methods were used to define complex recombinants not assigned to any subtype or CRF. A total of 670 available HIV-1 pol sequences from different patients were collected, of which 588 (87.8%) were assigned to HIV-1 subtype B and 82 (12.2%) to HIV-1 non-B variants. Recombinants caused the majority (71.9%) of HIV-1 non-B infections and were found in 8.8% of CoRIS patients. Eleven URFs (accounting for 13.4% of HIV-1 non-B infections), presenting complex mosaic patterns, were detected. Among them, 10 harbored subtype B fragments. Four of the 11 URFs were found in Spanish natives. A cluster of three B/CRF02_AG recombinants was detected. We conclude that complex variants, including unique recombinant forms, are being introduced into Spain through both immigrants and natives. An increase in the frequency of mosaic viruses, reflecting the increasing heterogeneity of the HIV epidemic in our country, is expected.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) shows great genetic diversity due to its high replication rate, the error-prone reverse transcriptase, and recombination events that may occur during virus replication (57). On the basis of genetic homology, HIV-1 has been classified in four groups: M (main), O (outlier), N (non-M, non-O), and the recently identified group P (36). HIV-1 group M is subdivided into 9 subtypes (A to D, F to H, J, and K), at least 49 circulating recombinant forms (CRFs) (http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html), and multiple unique recombinant forms (URFs). CRFs are defined as intersubtype recombinants for which at least three epidemiologically unlinked variants are monophyletic, sharing an identical genetic structure along their full genomes. URF variants are widely distributed worldwide, with recombination breakpoints different from those found in CRFs. Genetic complexity is not always detected, mainly due to the subtyping of only one genetic region and not of the full genome. Consequently, specimens previously considered “pure” variants may be classified as recombinants when additional viral genes are analyzed. Therefore, the frequency of recombinant variants is underestimated in the pandemic. Recombination, in addition to purifying selection, is involved in the evolution of HIV (42), adaptation to its host, and escape from antiviral treatments (37). Recombination can also increase HIV fitness (50).
HIV-1 subtype B is the prevalent variant in developed areas, such as North America and Western Europe, including Spain (16, 23). However, subtypes other than subtype B and recombinants (HIV-1 non-B variants) are responsible for 90% of the 33 million infections worldwide (20, 46). These variants are increasing in prevalence and heterogeneity in developed countries (3, 15, 23, 45, 49, 54), mainly due to immigration and the movement of populations from areas of endemicity. The coexistence of multiple variants in the same region favors recombination between them after coinfection and/or superinfection events. In Spain, an increase in the frequency of HIV-1 non-B subtypes has been found among native Spaniards and immigrants newly diagnosed with HIV-1 in recent years (23), and the presence of different recombinants has been published (11, 14, 16, 21, 23, 24, 34, 52, 53).
The increasing prevalence of HIV-1 non-B variants could have implications for diagnosis (5), vaccine design (56), and the clinical management of HIV infection (39). HIV-1 non-B variants present clade-specific substitutions in positions related to drug resistance (26, 52). They could accelerate the emergence of drug-resistant viruses, change or induce alternative pathways of resistance (17, 19), influence viral replicative capacity in vitro (25), impair the interpretation of genotypic resistance algorithms (9, 43, 52), reduce the genetic barrier of certain protease inhibitors (47), and affect drug-binding affinity (27). Additionally, patients infected by certain HIV-1 non-B subtypes present accelerated disease progression (2, 48) and higher cognitive impairment (40). Thus, the proper detection and description of HIV-1 variants in representative cohorts is essential for further studies.
CoRIS, the cohort of the Spanish Research Network of Excellence on HIV/AIDS, has recently reported a prevalence of 15.2% for HIV-1 non-B subtypes (16). To further explore the molecular epidemiology of HIV in Spain, the objective of the present study was to characterize the HIV-1 recombinant variants detected in CoRIS by using phylogenetic analysis (phy), the gold-standard method for subtyping and discrimination between subtypes and/or CRFs. We also defined the complex mosaic patterns in variants classified as unique recombinants (not assigned to any known subtype or circulating recombinant form), as well as the phylogenetic clusters including such variants.
METHODS
Study population.
CoRIS is an open, multicenter, prospective cohort of HIV-positive, antiretroviral (ARV)-naïve subjects more than 13 years old seen at 31 HIV units of the 18 Autonomous Regions in Spain from January 2004 on. Ethics approval was obtained from participating sites. The study was designed to protect the rights of all subjects involved under the appropriate local regulations. Written informed consent was obtained at the respective sites from every patient included in the study. A detailed description of the cohort has been published previously (7).
Of the 3,351 subjects included from 2004 to 2008, 670 patients provided a FASTA sequence while naïve to antiretroviral treatment (ART) and were included in this study. The alignment including these sequences is available as file S1 in the supplemental material. Overall, 375 patients (56%) were Spanish, 181 (27%) were immigrants (118 Central and South Americans, 17 sub-Saharan Africans, 16 Western Europeans, 11 Eastern Europeans, 10 North Africans, 6 North Americans, and 3 Asians), and 114 were of unknown origin. A pol sequence including the complete protease (codons 1 to 99) and part of the reverse transcriptase (codons 38 to 260 or 1 to 335) was collected from each of the 670 patients included in CoRIS for whom a sequence was deposited in the database. pol sequences were obtained in the course of the clinical routine for drug resistance mutation analysis, before any anti-HIV therapy. Most sequences (69.5%) were obtained within the period of 2007 to 2008.
Phylogenetic analysis.
HIV-1 subtypes and CRFs were identified by phylogenetic analysis (phy) of the 670 pol sequences. The 2008 version of the subtype reference data set provided by the Los Alamos National Laboratory (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) was used. It was updated to include more sequences of CRFs that had been absent or scarcely represented (26_AU, 30_0206, 32_06A1, 34_01B, 38_BF, 41_CD, and 42_BF). Therefore, at least 2 representative sequences of each of the 9 subtypes and 43 CRFs of HIV-1 group M available in GenBank at the moment of the analysis were taken as references. DNA sequences were aligned using the ClustalX program, version 2.0.11. The tree topology was obtained using the neighbor-joining method. The pairwise distance matrix was estimated using the Kimura two-parameter model within the DNAdist program, as implemented in the PHYLIP software package. Bootstrap resampling (1,000 data sets) of the multiple alignments was performed to test the statistical robustness of the tree. We considered the associations of pol sequences showing a bootstrap value higher than 700 in the phylogenetic tree to be “clusters.”
URF characterization.
Recombination analyses of sequences not assigned to any known subtype or CRF by phy were performed using several methods: SimPlot, version 3.5.1, the Recombination Detection Program (RDP; version 3alpha44), and the jumping-profile hidden Markov model (jpHMM) (http://jphmm.gobics.de/jphmm.html). In the SimPlot analysis, which applies the bootscanning method, windows of 300 nucleotides moving in 10-nucleotide increments were used, as recommended by Zhang et al. (55). Sequences obtained from GenBank with the same geographical origin as the sequences analyzed were used as references when possible. After the different recombination analyses, the definitive breakpoints were those confirmed by constructing phylogenetic trees of the subsegments in order to assign them to the parental subtypes involved in the specific recombination event.
RESULTS
HIV-1 subtypes defined by phy in CoRIS: high frequency of recombinants.
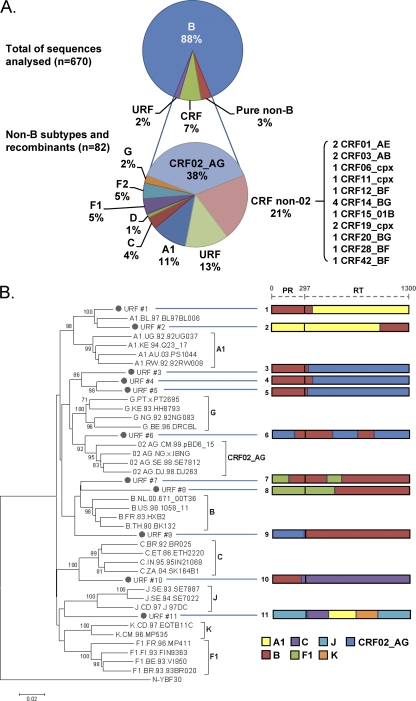
A total of 670 available pol sequences from different patients included in CoRIS were collected. As expected, 588 (87.8%) were assigned to subtype B and 82 (12.2%) to HIV-1 non-B variants. The majority (n = 59 [71.9%]) of HIV-1 non-B sequences were, in fact, viruses recombinant at pol (Fig. 1). Forty-eight viruses (58.5% of HIV-1 non-B variants) were 12 different CRFs (2 CRF01_AE, 31 CRF02_AG, 2 CRF03_AB, 1 CRF06_cpx, 1 CRF11_cpx, 1 CRF12_BF, 4 CRF14_BG, 1 CRF15_01B, 2 CRF19_cpx, 1 CRF20_BG, 1 CRF28_BF, and 1 CRF42_BF). Eleven viruses were unique recombinants (URFs), i.e., not assigned to any subtype or CRF. CRFs and URFs caused 7.2% and 1.6% of total infections, respectively. URF sequences did not cluster with any other known HIV-1 subtype or CRF after phylogenetic analyses of pol sequences, and they presented complex mosaic patterns due to recombination events between different subtypes (Fig. 1). In summary, among the 82 HIV-1 non-B pol sequences, 59 (71.9%) were shown by phy to be recombinants (81.4% CRFs and 18.6% URFs). Recombinant form CRF02_AG and URFs accounted for one-third and one-fifth, respectively, of HIV-1 non-B infections. The data showed that HIV-1 recombinants caused 8.8% of HIV-1 infections (this proportion rose to 14% when only the year 2008 was considered) and represented 71.9% of the HIV-1 non-B variants identified by phy.
Fig 1.
HIV-1 subtypes in the Spanish Cohort (CoRIS). (A) Distribution of HIV-1 variants in CoRIS according to phylogenetic analyses of the 670 pol sequences. (B) Neighbor-joining phylogenetic tree including the 11 HIV-1 unique recombinant forms found in the study population. The URFs are bulleted and are connected by blue horizontal lines to diagrams of the mosaic patterns of their pol sequences (length, 1,300 nucleotides). PR, protease; RT, reverse transcriptase. Bootstrap values of 1,000 repetitions are expressed as percentages. The original tree included all CRFs described at the moment of this work.
HIV-1 non-B variants found in native Spanish individuals.
Of the 375 native Spaniards included in the study, 31 (8.3%) carried HIV-1 non-B variants. Among these, only 9 harbored pure HIV-1 non-B subtypes; 22 were infected with recombinants (18 CRF and 4 URF). It is remarkable that 1 out of 2 CRF01_AE viruses, the CRF12_BF virus, 3 out of 4 CRF14_BG viruses, 1 out of 2 CRF19_cpx viruses, the CRF20_BG virus, and the CRF42_BF virus were found in Spanish individuals. Although the frequency of these variants among native Spaniards was low, native patients accounted for more than one-third (37.8%) of the total number of patients infected with HIV-1 non-B variants.
Origins of CoRIS patients infected by HIV-1 recombinants.
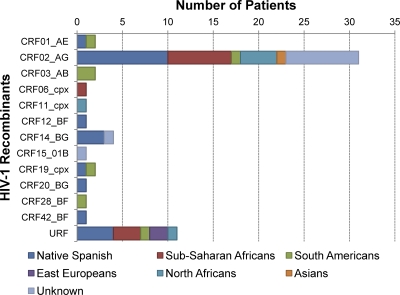
The origins of subjects infected by each HIV-1 recombinant variant are shown in Fig. 2. According to the origins of patients, the prevalences of HIV-1 recombinants were as follows, in descending order: 64.7% (11/17) for sub-Saharan Africans, 60% (6/10) for North Africans, 33.3% (1/3) for Asians, 18.2% (2/11) for Eastern Europeans, 5.9% (7/118) for South and Central Americans, 5.9% (22/375) for Spaniards, and 0% for North Americans (0/6) and Western Europeans other than Spaniards (0/16). Among patients of unknown origin, this prevalence was 8.8% (10/114). Surprisingly, a CRF02_AG variant was isolated from an Asian patient, and CRF03_AB recombinants, first described in Russia, were isolated from South American patients.
Fig 2.
Distribution of patients infected by HIV-1 recombinants in CoRIS according to their geographic origins.
Complex variants in CoRIS and epidemiological data.
The 11 URFs presented mosaic patterns due to recombination events in the pol region between different subtypes and/or CRFs (Fig. 1). Recombinants presented fragments from 7 different pure subtypes (A, B, C, F, G, J, and K) and CRF02_AG. Some carried B/CRF02_AG (4/11), B/F1 (2/11), and B/A1 (2/11) sequences, and one showed a complex pattern including regions of subtypes A1, C, J, and K. Interestingly, 10 of the 11 URFs contained subtype B regions.
Table 1 records the epidemiological features of 11 HIV-1 URF-infected CoRIS patients defined by phy (Fig. 2) and confirmed by bootscanning and other methods (Fig. 1). Seven of these patients came from sub-Saharan Africa (n =3), Eastern Europe or Russia (n =2), South or Central America (n =1), or North Africa (n =1). Of note, the remaining four were native Spaniards (36.4% of cases), demonstrating the increasing heterogeneity of HIV-1 even in native Western Europeans. The modes of transmission for the URF-infected patients were as follows: 6 and 2 acquired HIV infection through heterosexual or homosexual routes, respectively; 2 were injecting drug users (IDU); and for 1 patient, viral transmission took place through a contaminated-blood transfusion. In other words, URF variants in the CoRIS cohort were transmitted mostly through heterosexual contacts.
Table 1.
Epidemiological features of the 23 HIV-1-infected patients included in CoRIS carrying URF sequences at pol
| URF no.a | Patient IDb | Sexc | Origin | Parental strains | Exposure categoryd | City (region) of sampling | Yr of sampling |
|---|---|---|---|---|---|---|---|
| 1 | 21 | F | Eastern Europe | B/A1 | IDU | Granada (East Andalusia) | 2005 |
| 2 | 41 | M | Eastern Europe | A1/B | IDU | Elche (Valencia) | 2008 |
| 3 | 46 | M | Equatorial Guinea | B/CRF02 | Heterosex. | Madrid | 2007 |
| 4 | 52 | M | Equatorial Guinea | B/CRF02 | Heterosex. | Madrid | 2005 |
| 5 | 29 | F | North Africa | B/CRF02 | Heterosex. | Madrid | 2005 |
| 6 | 50 | M | Spain | B/CRF02 | Homosex. | Malaga (South Andalusia) | 2008 |
| 7 | 1 | U | Spain | B/F1 | Heterosex. | Seville (West Andalusia) | 2008 |
| 8 | 76 | M | Spain | B/F1 | Homosex. | Madrid | 2006 |
| 9 | 39 | M | South America | CRF02/B | Other | Madrid | 2006 |
| 10 | 34 | M | Spain | B/C | Heterosex. | Madrid | 2008 |
| 11 | 66 | F | Sub-Saharan Africa | J/C/A1/K | Heterosex. | Terrassa (Catalonia) | 2008 |
Numbered as in Fig. 1.
Patient number according to the sequence alignment provided as File S1 in the supplemental material.
M, male; F, female; U, unknown.
Heterosex., heterosexual risk behavior; Homosex., homo/bisexual risk behavior; IDU, injecting drug user.
Clusters of CoRIS patients carrying complex recombinants.
One cluster was found in the phylogenetic analysis of the 11 sequences assigned to HIV-1 URF variants (Fig. 1). This cluster consisted of three samples harboring sequences from HIV-1 subtype B, mainly at the protease, and CRF02_AG sequences at the reverse transcriptase (Fig. 1, URFs 3 to 5), with very similar breakpoint estimates. The three patients were diagnosed in Madrid, Spain, and were infected by heterosexual transmission. Two of them were from sub-Saharan Africa (Table 1), which could explain the presence of CRF02_AG sequences, given the predominance of this strain in their country of origin (Equatorial Guinea). The third was a North African. The remaining eight URFs did not cluster with any other sample in the study population.
DISCUSSION
One of 10 HIV-1-infections in Spain could represent recombinant variants.
This study was performed within a large and representative Spanish cohort of ART-naïve HIV-infected patients included in the Research Network on HIV/AIDS (CoRIS). CoRIS collects data on patients from different areas of Spain, a country with one of the highest HIV prevalences in the European Union (13, 46). The results presented confirmed that subtype B is still the main HIV-1 variant in Spain, as reported previously for the same sample of subjects from CoRIS (16), and are concordant with data from other Spanish studies (23, 31, 34). What our study adds is the finding that 8.8% of the 670 CoRIS patients (14% of the 164 patients assessed in 2008) were infected with HIV-1 recombinant variants (CRFs and URFs) and the description of the genetic nature of the complex recombinants. The recombinant variants represented almost three-quarters (72%) of the 82 HIV-1 non-B variants found. The highest prevalence of recombinants was found in patients from sub-Saharan Africa, followed by Eastern Europeans. As previously reported in the initial CoRIS description (16) and in other studies (23, 24, 31, 34), recombinant CRF02_AG was the most frequently found HIV-1 non-B variant in our country, accounting for more than one-third of HIV-1 non-B infections. This is due to the high prevalence of this recombinant in Central and West Africa, the most common origin of infected sub-Saharan Africans residing in Spain. Of note, we detected CRF02_AG in an Asian patient, as well as CRF03_AB in two South American patients. To our knowledge, this was the first time that these two variants were reported for patients from those regions in Spain, suggesting that transmission occurred in Spain.
Introduction of complex HIV-1 variants into the native population.
In recent years, increasing prevalences of HIV-1 non-B subtypes and recombinants have been reported across Western Europe (23, 51, 54). The rising prevalence of HIV-1 non-B variants has traditionally been attributed to the growing number of immigrants from developing regions where these variants are prevalent. In fact, one-third of patients newly diagnosed with HIV in Spain in 2008 were immigrants (8, 38). However, the current heterogeneity of the HIV epidemic in Spain caused by recombinants can be only partially explained by immigration. Among the 23 and 59 subjects infected by HIV-1 non-B pure subtypes or recombinants, 9 (39.1%) and 22 (37.3%), respectively, were native patients. Among the 22 native subjects infected by recombinants, 18 and 4 patients carried CRF and URF variants, respectively. The detection in native Spaniards of 37.3% of the recombinant variants found in this study is evidence of the introduction of complex HIV-1 variants into the native population of Spain, which also happens in other developed countries. Thus, the attribution of the increasing HIV-1 heterogeneity exclusively to immigrant populations is a prejudgment that only partially explains this phenomenon.
Origin and transmission of URFs in Spain.
Interestingly, 13% of infections with HIV-1 non-B variants were caused by URFs. These are very frequent in regions where multiple clades cocirculate, such as sub-Saharan Africa (10), and are increasingly present in developed countries (15). However, whether the complex recombinant variants were transmitted directly from the immigrant community to the native population or whether the recombination events took place in native Spaniards cannot be ascertained with the current data.
URF sequences from heterosexually infected sub-Saharan Africans that form a cluster (URFs 3 to 5) resulted from very similar recombination events involving CRF02_AG and subtype B, a phenomenon previously reported in both Spain and France (24, 29). Two of these patients came from Equatorial Guinea, where CRF02_AG is highly prevalent (12), but the third patient was from North Africa, where subtype B is prevalent (1). The different origins of the patients suggest that the infection and subsequent spread occurred in Spain. Of note, URFs 6 and 9 also were B/CRF02_AG recombinants, although these recombinants did not share a common origin with those included in the cluster. In addition, other recombinants, including sequences typically found in Eastern European (URFs 1 and 2, including subsubtype A1) and South American (URFs 7 and 8, including subtypes B and F) countries, were found, reflecting the wide variety of geographical origins of immigrants in Spain.
Despite the pandemic spread of HIV-1 recombinants, their times of origin are not well understood. A recent paper suggests that recombination was common in the early evolutionary history of HIV-1 (44). In this cohort, the oldest URF was sampled in 2005, but complex recombinants have been reported in our country at least since the end of the 1990s (22–24, 30, 33), including the description of the first CRF that originated in Western Europe (CRF14_BG) (11). More epidemiological data about the infection date, risk behavior, and the geographical regions in which these patients have resided or traveled, as well as additional sequencing of longer genetic regions and specific computer programs to study the viruses' genetic evolution, would be necessary to confirm these findings and to define the origin of complex recombinants circulating in Spain.
Possible underestimation of the frequency of HIV-1 URFs in molecular epidemiology studies.
This work reveals the circulation and spread of complex HIV-1 variants in a large and representative cohort of HIV-infected persons in Spain. However, the detection of URFs could be even higher if more viral regions were analyzed. For instance, more than one-third of HIV-1 sequences described in the Los Alamos HIV database to date might be found to be recombinant forms if different genes or full-length sequences were analyzed (41), and therefore, complex recombinants could be more frequent than expected in the HIV-1 pandemic (35). Another limitation of our study lies in the representativeness of the data, given the unequal distribution of the patients across the 18 centers and the territory of Spain. Thus, due to the scarcity of data from certain regions and/or hospitals, our findings could not be representative of the HIV-1 epidemic in our country. Nevertheless, this is the largest HIV-1 molecular epidemiology study performed in Spain using phylogeny to determinate the distribution of HIV-1 variants.
Biological consequences of recombination in HIV-1 evolution.
Not only is the frequency of recombinants likely underestimated, but an increasing presence of URFs in developed countries is expected in the coming years due to the movement of populations between countries where different HIV-1 variants are prevalent. In fact, decreasing numbers of pure subtype B viruses and increases in the numbers of unique recombinants including subtype B sequences among HIV-1-seropositive patients have also been reported recently in neighboring countries (15). It has also been suggested that the HIV-1 epidemic could be evolving toward a more complex epidemiological landscape (18). Recombination seems to be very important in the evolution of HIV-1 (42), since it can provide a biological advantage versus parental viruses (28), promoting biological adaptation and enhancing fitness (6). It can also facilitate drug resistance and may allow superinfecting HIV-1 strains to evade preexisting immune responses (32). Thus, the continuous spread of HIV-1 recombinants may have serious implications for efforts to control the AIDS pandemic (including future vaccination trials) and could represent one of the highest barriers to HIV-1 eradication (32). However, despite some cases where URFs are described as highly pathogenic (4), the clinical implications of the presence of URFs for the AIDS pandemic remain to be clarified.
Supplementary Material
ACKNOWLEDGMENTS
This study would not have been possible without the collaboration of all the patients, medical and nursery staff, and data managers who have taken part in the project.
The RIS Cohort (CoRIS) is funded by the Instituto de Salud Carlos III through the Red Temática de Investigación Cooperativa en Sida (RIS C03/173). This work was supported in part by grants from the Fondo de Investigaciones Sanitarias (FIS 09/00284). A.H. is supported by the Agencia Laín Entralgo. G.Y. is supported by the Consejería de Educación de la Comunidad de Madrid and the Fondo Social Europeo (FSE).
Centers and investigators involved in CoRIS. Steering committee: Juan Berenguer, Julia del Amo, Federico García, Félix Gutiérrez, Pablo Labarga, Santiago Moreno, and María Ángeles Muñoz. Data management and statistical analyses: Ana María Caro-Murillo, Paz Sobrino Vegas, Santiago Pérez-Cachafeiro, Victoria Hernando Sebastián, Belén Alejos Ferreras, and Débora Álvarez. BioBank: M Ángeles Muñoz-Fernández, Isabel García-Merino, Coral Gómez Rico, Jorge Gallego de la Fuente, and Almudena García Torre. Centers: Hospital General Universitario de Alicante, Alicante (Joaquín Portilla Sogorb, Esperanza Merino de Lucas, Sergio Reus Bañuls, Vicente Boix Martínez, Livia Giner Oncina, Carmen Gadea Pastor, Irene Portilla Tamarit, and Patricia Arcaina Toledo), Hospital Universitario de Canarias, Santa Cruz de Tenerife (Juan Luis Gómez Sirvent, Patricia Rodríguez Fortúnez, María Remedios Alemán Valls, María del Mar Alonso Socas, Ana María López Lirola, María Inmaculada Hernández Hernández, and Felicitas Díaz-Flores), Hospital Carlos III, Madrid (Vicente Soriano, Pablo Labarga, Pablo Barreiro, Carol Castañares, Pablo Rivas, Andrés Ruiz, Francisco Blanco, Pilar García, and Mercedes de Diego), Hospital Universitario Central de Asturias, Oviedo (Víctor Asensi, Eulalia Valle, and José Antonio Cartón), Hospital Clinic, Barcelona (José M. Miró, María López-Diéguez, Christian Manzardo, Laura Zamora, Iñaki Pérez, M. Teresa García, Carmen Ligero, José Luis Blanco, Felipe García-Alcaide, Esteban Martínez, Josep Mallolas, and José M. Gatell), Hospital Doce de Octubre, Madrid (Rafael Rubio, Federico Pulido, Silvana Fiorante, Jara Llenas, Violeta Rodríguez, and Mariano Matarranz), Hospital Donostia, San Sebastián (José Antonio Iribarren, Julio Arrizabalaga, María José Aramburu, Xabier Camino, Francisco Rodríguez-Arrondo, Miguel Ángel von Wichmann, Lidia Pascual Tomé, Miguel Ángel Goenaga, María Jesús Bustinduy, and Harkaitz Azkune Galparsoro), Hospital General Universitario de Elche, Elche (Félix Gutiérrez, Mar Masiá, José Manuel Ramos, Sergio Padilla, Andrés Navarro, Fernando Montolio, Yolanda Peral, and Catalina Robledano García), Hospital Germans Trías i Pujol, Badalona (Bonaventura Clotet, Cristina Tural, Lidia Ruiz, Cristina Miranda, Roberto Muga, Jordi Tor, and Arantza Sanvisens), Hospital General Universitario Gregorio Marañón, Madrid (Juan Berenguer, Juan Carlos López Bernaldo de Quirós, Pilar Miralles, Jaime Cosín Ochaíta, Matilde Sánchez Conde, Isabel Gutiérrez Cuellar, Margarita Ramírez Schacke, and Belén Padilla Ortega), Hospital Universitari de Tarragona Joan XXIII, IISPV, Universitat Rovira i Virgili, Tarragona (Francesc Vidal, Joaquín Peraire, Consuelo Viladés, Sergio Veloso, Marta Sanjuán, Montserrat Vargas, Miguel López-Dupla, Montserrat Olona, Alba Aguilar, Joan Joseph Sirvent, Antoni Soriano, and Rami A. A. Qaneta), Hospital Universitario La Fe, Valencia (José López Aldeguer, Marino Blanes Juliá, José Lacruz Rodrigo, Miguel Salavert, Marta Montero, Eva Calabuig, and Sandra Cuéllar), Hospital Universitario La Paz, Madrid (Juan González García, Ignacio Bernardino de la Serna, José María Peña Sánchez de Rivera, Marta Mora Rillo, José Ramón Arribas López, María Luisa Montes Ramírez, José Francisco Pascual Pareja, Blanca Arribas, Juan Miguel Castro, Francisco Javier Zamora Vargas, and Ignacio Pérez Valero), Hospital de la Princesa, Madrid (Ignacio de los Santos, Jesús Sanz Sanz, Johana Rodríguez, Ana Salas Aparicio, and Cristina Sarriá Cepeda), Hospital San Pedro-CIBIR, Logroño (José Antonio Oteo, José Ramón Blanco, Valvanera Ibarra, Luis Metola, Mercedes Sanz, and Laura Pérez-Martínez), Hospital San Pedro II, Logroño (Javier Pinilla Moraza), Hospital Universitario Mutua de Terrassa, Terrassa (David Dalmau, Angels Jaén Manzanera, Mireia Cairó Llobell, Daniel Irigoyen Puig, Laura Ibáñez, Queralt Jordano Montañez, Mariona Xercavins Valls, Javier Martínez-Lacasa, Pablo Velli, and Roser Font), Hospital de Navarra, Pamplona (Julio Sola Boneta, Javier Uriz, Jesús Castiello, Jesús Reparaz, María Jesús Arraiza, Carmen Irigoyen, and David Mozas), Hospital Parc Taulí, Sabadell (Ferrán Segura, María José Amengual, Eva Penelo, Gemma Navarro, Montserrat Sala, Manuel Cervantes, and Valentín Pineda), Hospital Ramón y Cajal, Madrid (Santiago Moreno, José Luis Casado, Fernando Dronda, Ana Moreno, María Jesús Pérez Elías, Dolores López, Carolina Gutiérrez, Beatriz Hernández, María Pumares, and Paloma Martí), Hospital Reina Sofía, Murcia (Alfredo Cano Sánchez, Enrique Bernal Morell, and Ángeles Muñoz Pérez), Hospital San Cecilio, Granada (Federico García García, José Hernández Quero, Alejandro Peña Monje, Leopoldo Muñoz Medina, and Jorge Parra Ruiz), Centro Sanitario Sandoval, Madrid (Jorge del Romero Guerrero, Carmen Rodríguez Martín, Teresa Puerta López, and Juan Carlos Carrió Montiel), Hospital Universitario Santiago de Compostela, Santiago de Compostela (Antonio Antela, Arturo Prieto, and Elena Losada), Hospital Son Dureta, Palma de Mallorca (Melchor Riera, Javier Murillas, María Peñaranda, María Leyes, María Angels Ribas, Antoni Campins, and Concepción Villalonga), Hospital Universitario de Valme, Seville (Juan Antonio Pineda, Eva Recio Sánchez, Fernando Lozano de León, Juan Macías, José del Valle, Jesús Gómez-Mateos, and Rosario Mata), Hospital Virgen de la Victoria, Málaga (Jesús Santos González, Manuel Márquez Solero, Isabel Viciana Ramos, and Rosario Palacios Muñoz), and Hospital Universitario Virgen del Rocío, Seville (Pompeyo Viciana, Manuel Leal, Luis Fernando López-Cortés, and Mónica Trastoy).
Footnotes
Published ahead of print 7 December 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Annaz HE, et al. 2011. Presence of drug resistance mutations among drug-naïve patients in Morocco. AIDS Res. Hum. Retroviruses 27: 917–920 [DOI] [PubMed] [Google Scholar]
- 2. Baeten JM, et al. 2007. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J. Infect. Dis. 195: 1177–1180 [DOI] [PubMed] [Google Scholar]
- 3. Brennan CA, Yamaguchi J, Devare SG, Foster GA, Stramer SL. 2010. Expanded evaluation of blood donors in the United States for human immunodeficiency virus type 1 non-B subtypes and antiretroviral drug-resistant strains: 2005 through 2007. Transfusion 50: 2707–2712 [DOI] [PubMed] [Google Scholar]
- 4. Bruselles A, et al. 2009. Use of massive parallel pyrosequencing for near full-length characterization of a unique HIV type 1 BF recombinant associated with a fatal primary infection. AIDS Res. Hum. Retroviruses 25: 937–942 [DOI] [PubMed] [Google Scholar]
- 5. Candotti D, et al. 2000. AIDS in an HIV-seronegative Ghanaian woman with intersubtype A/G recombinant HIV-1 infection. J. Med. Virol. 62: 1–8 [DOI] [PubMed] [Google Scholar]
- 6. Carobene MG, Rodrigues CR, De Candia CA, Turk G, Salomon H. 2009. In vitro dynamics of HIV-1 BF intersubtype recombinants genomic regions involved in the regulation of gene expression. Virol. J. 6: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caro-Murillo AM, et al. 2007. Spanish cohort of naive HIV-infected patients (CoRIS): rationale, organization and initial results. Enferm. Infecc. Microbiol. Clin. 25: 23–31 [DOI] [PubMed] [Google Scholar]
- 8. Caro-Murillo AM, et al. 2009. HIV infection in immigrants in Spain: epidemiological characteristics and clinical presentation in the CoRIS Cohort (2004–2006). Enferm. Infecc. Microbiol. Clin. 27: 380–388 [DOI] [PubMed] [Google Scholar]
- 9. Champenois K, et al. 2008. Expected response to protease inhibitors of HIV-1 non-B subtype viruses according to resistance algorithms. AIDS 22: 1087–1089 [DOI] [PubMed] [Google Scholar]
- 10. Delgado E, et al. 2008. High prevalence of unique recombinant forms of HIV-1 in Ghana: molecular epidemiology from an antiretroviral resistance study. J. Acquir. Immune Defic. Syndr. 48: 599–606 [DOI] [PubMed] [Google Scholar]
- 11. Delgado E, et al. 2002. Identification of a newly characterized HIV-1BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29: 536–543 [DOI] [PubMed] [Google Scholar]
- 12. Djoko CF, et al. 2010. HIV type 1 pol gene diversity and genotypic antiretroviral drug resistance mutations in Malabo, Equatorial Guinea. AIDS Res. Hum. Retroviruses 26: 1027–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. European Centre for Disease Prevention and Control/WHO Regional Office for Europe. 2010. HIV/AIDS surveillance in Europe 2009. European Centre for Disease Prevention and Control, Stockholm, Sweden: http://ecdc.europa.eu/en/publications/Publications/101129_SUR_HIV_2009.pdf [Google Scholar]
- 14. Fernández-GarcíA A, et al. 2010. Identification of a new HIV type 1 circulating BF intersubtype recombinant form (CRF47_BF) in Spain. AIDS Res. Hum. Retroviruses 26: 827–832 [DOI] [PubMed] [Google Scholar]
- 15. Frange P, et al. 2008. New and old complex recombinant HIV-1 strains among patients with primary infection in 1996–2006 in France: the French ANRS CO06 PRIMO cohort study. Retrovirology 5: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García F, et al. 2011. Transmission of HIV drug resistance and non-B subtype distribution in the Spanish cohort of antiretroviral treatment naive HIV-infected individuals (CoRIS). Antiviral Res. 91: 150–153 [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez LM, et al. 2008. Impact of HIV-1 protease mutations A71V/T and T74S on M89I/V-mediated protease inhibitor resistance in subtype G isolates. J. Antimicrob. Chemother. 61: 1201–1204 [DOI] [PubMed] [Google Scholar]
- 18. González-Alba JM, et al. 2011. Molecular surveillance of HIV-1 in Madrid, Spain: a phylogeographic analysis. J. Virol. 85: 10755–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grossman Z, et al. 2004. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob. Agents Chemother. 48: 2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holguín Á, Álvarez A, Soriano V. 2005. Differences in the length of gag proteins among different HIV type 1 subtypes. AIDS Res. Hum. Retroviruses 21: 886–893 [DOI] [PubMed] [Google Scholar]
- 22. Holguín Á, Álvarez A, Soriano V. 2002. HIV-1 subtype J recombinant viruses in Spain. AIDS Res. Hum. Retroviruses 18: 523–529 [DOI] [PubMed] [Google Scholar]
- 23. Holguín Á, de Mulder M, Yebra G, López M, Soriano V. 2008. Increase of non-B subtypes and recombinants among newly diagnosed HIV-1 native Spaniards and immigrants in Spain. Curr. HIV Res. 6: 327–334 [DOI] [PubMed] [Google Scholar]
- 24. Holguín Á, et al. 2008. Genetic characterization of complex inter-recombinant HIV-1 strains circulating in Spain and reliability of distinct rapid subtyping tools. J. Med. Virol. 80: 383–391 [DOI] [PubMed] [Google Scholar]
- 25. Holguín Á, Sune C, Hamy F, Soriano V, Klimkait T. 2006. Natural polymorphisms in the protease gene modulate the replicative capacity of non-B HIV-1 variants in the absence of drug pressure. J. Clin. Virol. 36: 264–271 [DOI] [PubMed] [Google Scholar]
- 26. Kantor R, Katzenstein D. 2003. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 5: 25–35 [PubMed] [Google Scholar]
- 27. Kinomoto M, et al. 2005. HIV-1 proteases from drug-naive West African patients are differentially less susceptible to protease inhibitors. Clin. Infect. Dis. 41: 243–251 [DOI] [PubMed] [Google Scholar]
- 28. Konings FAJ, et al. 2006. Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes A and G. J. Med. Virol. 78: 523–534 [DOI] [PubMed] [Google Scholar]
- 29. Leoz M, et al. 2011. Circulation of multiple patterns of unique recombinant forms B/CRF02_AG in France: precursor signs of the emergence of an upcoming CRF B/02. AIDS 25: 1371–1377 [DOI] [PubMed] [Google Scholar]
- 30. Lospitao E, Álvarez A, Soriano V, Holguín A. 2005. HIV-1 subtypes in Spain: a retrospective analysis from 1995 to 2003. HIV Med. 6: 313–320 [DOI] [PubMed] [Google Scholar]
- 31. McConnell MJ, et al. 2008. Molecular epidemiology of HIV type 1 in newly diagnosed patients in southern Spain. AIDS Res. Hum. Retroviruses 24: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Onafuwa-Nuga A, Telesnitsky A. 2009. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73: 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pérez-Álvarez L, et al. 2003. High incidence of non-B and recombinant HIV-1 strains in newly diagnosed patients in Galicia, Spain: study of genotypic resistance. Antivir. Ther. 8: 355–360 [PubMed] [Google Scholar]
- 34. Pérez-Álvarez L, et al. 2006. Prevalence of transmitted drug resistance and HIV-1 genetic forms in newly diagnosed individuals from Galicia and the Basque Country, Spain, abstr. 23. 4th European HIV Drug Resistance Workshop, Monte Carlo, Monaco, 29 to 31 March 2006 [Google Scholar]
- 35. Piantadosi A, Ngayo MO, Chohan B, Overbaugh J. 2008. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res. Hum. Retroviruses 24: 1221–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plantier JC, et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15: 871–872 [DOI] [PubMed] [Google Scholar]
- 37. Ramirez BC, Simon-Loriere E, Galetto R, Negroni M. 2008. Implications of recombination for HIV diversity. Virus Res. 134: 64–73 [DOI] [PubMed] [Google Scholar]
- 38. Registro Nacional de Casos de Sida. 2010. Vigilancia epidemiológica del VIH en España: nuevos diagnósticos de VIH en España, período 2003–2009. Actualización 30 de junio de 2010. Centro Nacional de Epidemiología, Madrid, Spain: http://www.msps.es/ciudadanos/enfLesiones/enfTransmisibles/sida/vigilancia/SINIVIH_junio2010.pdf [Google Scholar]
- 39. Rouet F, et al. 2007. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J. Acquir. Immune Defic. Syndr. 45: 380–388 [DOI] [PubMed] [Google Scholar]
- 40. Sacktor N, et al. 2009. HIV subtype D is associated with dementia, compared with subtype A, in immunosuppressed individuals at risk of cognitive impairment in Kampala, Uganda. Clin. Infect. Dis. 49: 780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sagoe KW, Dwidar M, Adiku TK, Arens MQ. 2009. HIV-1 CRF 02 AG polymerase genes in Southern Ghana are mosaics of different 02 AG strains and the protease gene cannot infer subtypes. Virol. J. 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simon-Loriere E, et al. 2009. Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog. 5: e1000418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Snoeck J, et al. 2006. Discordances between interpretation algorithms for genotypic resistance to protease and reverse transcriptase inhibitors of human immunodeficiency virus are subtype dependent. Antimicrob. Agents Chemother. 50: 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tee KK, et al. 2009. Estimating the date of origin of an HIV-1 circulating recombinant form. Virology 387: 229–234 [DOI] [PubMed] [Google Scholar]
- 45. Thomson MM, Nájera R. 2007. Increasing HIV-1 genetic diversity in Europe. J. Infect. Dis. 196: 1120–1124 [DOI] [PubMed] [Google Scholar]
- 46. UNAIDS. 2010. Global report: UNAIDS report on the global AIDS epidemic 2010. UNAIDS, Geneva, Switzerland: http://www.unaids.org/documents/20101123_GlobalReport_em.pdf [Google Scholar]
- 47. van de Vijver DAMC, et al. 2005. Differences in the frequency of minor substitutions between HIV-1 subtypes and their potential impact on the genetic barrier for resistance to protease inhibitors. Antivir. Ther. 10 (Suppl. 1): S145 [Google Scholar]
- 48. Vasan A, et al. 2006. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin. Infect. Dis. 42: 843–852 [DOI] [PubMed] [Google Scholar]
- 49. Vercauteren J, et al. 2009. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J. Infect. Dis. 200: 1503–1508 [DOI] [PubMed] [Google Scholar]
- 50. Vijay NN, Vasantika, Ajmani R, Perelson AS, Dixit NM. 2008. Recombination increases human immunodeficiency virus fitness, but not necessarily diversity. J. Gen. Virol. 89: 1467–1477. [DOI] [PubMed] [Google Scholar]
- 51. Wensing AM, et al. 2005. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J. Infect. Dis. 192: 958–966 [DOI] [PubMed] [Google Scholar]
- 52. Yebra G, de Mulder M, del Romero J, Rodríguez C, Holguín Á. 2010. HIV-1 non-B subtypes: high transmitted NNRTI-resistance in Spain and impaired genotypic resistance interpretation due to variability. Antiviral Res. 85: 409–417 [DOI] [PubMed] [Google Scholar]
- 53. Yebra G, et al. 2009. Clinical differences and viral diversity between newly HIV type 1-diagnosed African and non-African patients in Spain (2005–2007). AIDS Res. Hum. Retroviruses 25: 37–44 [DOI] [PubMed] [Google Scholar]
- 54. Yerly S, et al. 2007. Transmission of HIV-1 drug resistance in Switzerland: a 10-year molecular epidemiology survey. AIDS 21: 2223–2229 [DOI] [PubMed] [Google Scholar]
- 55. Zhang C, Ding N, Wei JF. 2008. Different sliding window sizes and inappropriate subtype references result in discordant mosaic maps and breakpoint locations of HIV-1 CRFs. Infect. Genet. Evol. 8: 693–697 [DOI] [PubMed] [Google Scholar]
- 56. Zhang M, et al. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhuang J, et al. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76: 11273–11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.