Abstract
The recent emergence of the human infection confirmed to be caused by severe fever with thrombocytopenia syndrome virus (SFTSV) in China is of global concern. Safe diagnostic immunoreagents for determination of human and animal seroprevalence in epidemiological investigations are urgently needed. This paper describes the cloning and expression of the nucleocapsid (N) protein of SFTSV. An N-protein-based double-antigen sandwich enzyme-linked immunosorbent assay (ELISA) system was set up to detect the total antibodies in human and animal sera. We reasoned that as the double-antigen sandwich ELISA detected total antibodies with a higher sensitivity than traditional indirect ELISA, it could be used to detect SFTSV-specific antibodies from different animal species. The serum neutralization test was used to validate the performance of this ELISA system. All human and animal sera that tested positive in the neutralization test were also positive in the sandwich ELISA, and there was a high correlation between serum neutralizing titers and ELISA readings. Cross-reactivity was evaluated, and the system was found to be highly specific to SFTSV; all hantavirus- and dengue virus-confirmed patient samples were negative. SFTSV-confirmed human and animal sera from both Anhui and Hubei Provinces in China reacted with N protein in this ELISA, suggesting no major antigenic variation between geographically disparate virus isolates and the suitability of this assay in nationwide application. ELISA results showed that 3.6% of the human serum samples and 47.7% of the animal field serum samples were positive for SFTSV antibodies, indicating that SFTSV has circulated widely in China. This assay, which is simple to operate, poses no biohazard risk, does not require sophisticated equipment, and can be used in disease surveillance programs, particularly in the screening of large numbers of samples from various animal species.
INTRODUCTION
In the latest dozen years, a life-threatening febrile illness has been sporadically reported in China (8, 16). The clinical manifestations of human infection have been characterized by high fever and hemorrhage. Its circulating region mainly covers eastern and central China, including Jiangsu, Anhui, Shandong, Henan, Hubei, and Liaoning Provinces. The causative agent of the disease was recently proven to be a novel bunyavirus (19). The virus, designated severe fever with thrombocytopenia syndrome virus (SFTSV), is a member of Phlebovirus genus in the Bunyaviridae family (19). Like all bunyaviruses, SFTSV has a trisegmented, single-stranded RNA genome with negative (large [L] and medium [M] segments) or ambisense (small [S] segment) polarity. The L segment encodes the RNA-dependent RNA polymerase. The M segment encodes a precursor of glycoproteins (Gn and Gc). The S segment encodes nucleocapsid (N) protein and a nonstructural (NS) protein using an ambisense coding strategy (7). Of all the genome-encoded proteins, N protein is the most immunodominant viral protein, and it is highly conserved in the Bunyaviridae family (9, 15, 17).
As a newly recognized phlebovirus, SFTSV is regarded to be an arbovirus. This means that SFTSV can probably be transmitted by a variety of vectors, such as ticks (19). However, the role of humans and other animals in the epidemiology of the disease during and between epidemic periods and their natural infection statuses is not well understood. Accurate, robust, safe tools for evaluating SFTSV prevalence in humans and other potential host vertebrates are necessary for surveillance purposes.
SFTSV infection is diagnosed in various ways, including virus isolation, nucleic acid amplification, and antibody detection (19). Although SFTSV infection can induce high serum virus titers in individuals, which may facilitate virus isolation and nucleic acid-based diagnosis, viremia is of very short duration, usually 1 to 6 days after onset of symptoms. Some infected patients and animals experience subclinical or mild symptoms (data not shown). Antibody detection techniques are widely used in epidemiological investigations to determine if a given region is disease free (6). Of the various classical serological methods used for the detection of antibodies against many viruses, the serum neutralization test is generally regarded to be the “gold standard.” However, it is laborious and expensive and requires manipulation of live virus, so it can be performed only in specialized reference laboratories housed in high-level biocontainment facilities (11).
Compared to the various diagnostic methods described above, enzyme-linked immunosorbent assay (ELISA) techniques for the detection of virus-specific antibodies are less expensive and less time-consuming. In recent years, various ELISA formats with high diagnostic accuracy and specificity have been developed for the specific detection of IgG, IgM, and total antibodies; in particular, for example, recombinant antigens have been used for accurate, specific detection of antibodies to a number of viruses in the family Bunyaviridae (3, 12, 18). In this study, we took another phlebovirus, specifically, Rift Valley fever virus, as a reference, and we developed a unique double-antigen sandwich ELISA for detection of SFTSV-specific total antibodies in sera from humans and a variety of animals using SFTSV recombinant N (rN) protein.
MATERIALS AND METHODS
Virus.
A confluent monolayer of Vero cells was inoculated with the Jiangsu-014 SFTSV strain, isolated from a patient in Jiangsu Province in 2010. The virus was harvested 7 days after inoculation and stored at −80°C for titration and RNA extraction.
Cloning and expression of SFTSV N protein.
RNA was extracted from cell culture supernatant by TRIzol reagent (Invitrogen). The N-protein-encoding gene was amplified by one-step reverse transcription-PCR (RT-PCR) using specific primers. After sequencing, the N-protein gene was cloned into expression vector pET-28(a)+ (Novagen). The recombinant vector pET-28(a)-NP was transformed into chemically competent Escherichia coli BL21 for expression. The induction of protein expression was performed in LB broth with 50 μg/ml kanamycin and 1 mM isopropyl-β-d-thiogalactopyranoside at 37°C overnight. The culture pellet containing recombinant protein was resuspended in chromatography binding buffer (20 mM sodium phosphate, 500 mM NaCl, 8 M urea, 20 mM imidazole, pH 7.4) and sonicated. The cell lysate was centrifuged, and its supernatant was used to load a nickel ion affinity column (GE Healthcare). After the column was thoroughly washed with binding buffer, recombinant 6His-labeled N protein was eluted from the column by elution buffer (20 mM sodium phosphate, 500 mM NaCl, 8 M urea, 500 mM imidazole, pH 7.4). Purified protein was analyzed by SDS-PAGE and visualized by Coomassie staining. Its identity was confirmed by Western blotting using anti-histidine tag monoclonal antibody (MAb). All operations were performed in accordance with the manufacturer's instructions.
Coupling of rN protein to HRP.
The rN protein was dissolved in phosphate-buffered saline (PBS), pH 7.3, at a concentration of 2 mg/ml, and 100 μl (200 μg) rN protein was coupled to horseradish peroxidase (HRP) using a Lightning-Link HRP conjugation kit (Innova Biosciences, United Kingdom).
Sera. (i) Group A.
Goat sera known to be positive for SFTSV was used as a positive control. They were obtained from 20 goats infected with the Jiangsu-014 strain of SFTSV. Sera collected from 20 goats known to never have been exposed to the virus were tested and found to be negative for SFTSV in a serum neutralization test. These sera were used as a negative control.
(ii) Group B.
A total of 250 human and 304 animal field serum samples were used. Sera were collected in rural parts of Xuyi, Jiangsu Province (human, n = 150; animals, including goats [n = 100], cattle [n = 50], pigs [n = 50], chickens [n = 50], and wild hedgehogs [n = 4]), and Chuzhou, Anhui Province (human, n = 100; goats, n = 50). All sera were taken during the 2010 outbreak and from areas in which several human infections have been reported over the past 5 years (data not shown). All human sera were obtained with the participants' written informed consent.
(iii) Group C.
Thirty-five SFTSV-confirmed sera from convalescent patients in Jiangsu (n = 18), Anhui (n = 15), and Hubei (n = 2) Provinces were used. These patients were clinically diagnosed with severe fever with thrombocytopenia syndrome, and the first blood samples collected from them on the day of the first medical consultation were confirmed to be SFTSV positive by PCR.
(iv) Group D.
Sera including 30 hantavirus- and 35 dengue virus-confirmed human serum samples from Jiangsu Province were used for specificity testing.
Serum neutralization test.
Duplicates of serial 2-fold dilutions of sera from 1:10 to 1:5,120 were separately mixed with a suspension containing 100 50% tissue culture infective doses of SFTSV strain Jiangsu-014 in a total volume of 200 μl, and the mixtures were incubated at 37°C for 1 h. The virus-serum mix was then transferred onto Vero cell monolayers in 96-well plates at 37°C for 1 h. The monolayers were washed with minimal essential medium maintenance medium and then incubated at 37°C in a 5% CO2 incubator. Cytopathic effect (CPE) status was observed every 24 h for 6 days. Titers were expressed as the reciprocal of the serum dilution that inhibited ≥75% of viral CPE. A serum sample was considered positive when it had a titer of ≥1:10.
Double-antigen sandwich ELISA procedure.
Costar 96-well EIA/RIA Stripwell immunoplates (Corning) were coated with rN protein diluted to a concentration of 8 μg/ml by carbonate-bicarbonate buffer, pH 9.6 (50 μl/well), and incubated overnight at 4°C. After washing with washing buffer consisting of PBS, pH 7.2, and 0.05% Tween 20, the plates were blocked with 300 μl/well of 10% fat-free milk powder in PBS and incubated overnight at 4°C. They were then washed as described above. Control and test sera were diluted 1:1 in PBS containing 5% bovine serum albumin, and 100 μl of diluted serum was added to the plates. Each test serum sample was assayed in duplicate, and each internal control was tested in quadruplicate. After incubation in a moist chamber for 30 min at 37°C, plates were washed 5 times with washing buffer, and 100 μl per well of the rN protein-HRP conjugate, diluted 1:2,000, was added. Plates were incubated for 30 min at 37°C and washed 5 times, and 100 μl per well of tetramethylbenzidine (TMB) substrate (Thermo) was used for detection. Plates were incubated at room temperature for 10 min, and 100 μl per well of 1 M H2SO4 was added to stop the reaction. Absorbance was read at 450 nm. The results were expressed as a percentage of the positive-control serum (PP) using the following formula: (mean OD of duplicate test serum/mean OD of positive-control serum) × 100, where OD is optical density.
Determination of cutoff values and ELISA diagnostic accuracy.
Cutoff values were determined as the mean value plus 2 or 3 standard deviations (SDs) derived from PP values in the negative-control serum. Estimates of diagnostic sensitivity and specificity, Youden's index, positive predictive value, and negative predictive value were calculated as described previously (12).
RESULTS
Cloning and expression of recombinant SFTSV N protein.
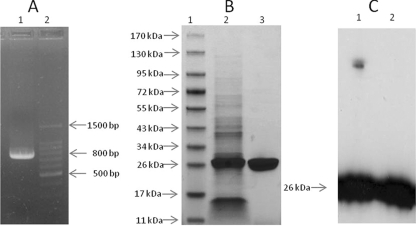
A SFTSV Jiangsu-014 strain N-protein-encoding gene 738 bp in size was successfully amplified (Fig. 1A.). The nucleic acid sequence and its deduced amino acid sequence were aligned with those of the JS-3 strain (GenBank accession no. HQ141603). Five differences between these two DNA sequences were detected. Of these, only one nucleic acid change (N77S) caused a change in the amino acid sequence. The recombinant 6His-labeled N protein with a molecular mass of 26 kDa was detected using SDS-PAGE and evaluated by Coomassie staining of pET-28(a)-NP-transformed bacterial cell extracts. The rN protein was expressed as an inclusion body and purified to homogeneity by affinity chromatography (Fig. 1B). Immunoblotting using anti-histidine tag MAb revealed a polypeptide of about 26 kDa, indicating that the cloned nucleocapsid gene had been translated properly (Fig. 1C). The denatured protein was refolded by dialyzing in PBS with serially decreasing urea concentrations.
Fig 1.
SFTSV nucleocapsid protein gene cloning and expression. (A) N-protein-encoding gene was amplified with a pair of specific primers by one-step RT-PCR. Lanes: 1, amplified N-protein gene with a size of 738 bp; 2, DNA ladder. (B) SDS-PAGE of rN protein. Lanes: 1, protein marker; 2, lysate of pET-28(a)-NP-transformed bacterial cell after overnight induction; the prominent band at 26 kDa constitutes rN protein; 3, His tag-purified rN protein. (C) Western blot of rN protein using anti-histidine tag MAb-HRP conjugate. Lanes: 1, lysate of bacterial cell; 2, purified rN protein.
Internal quality control and assay repeatability.
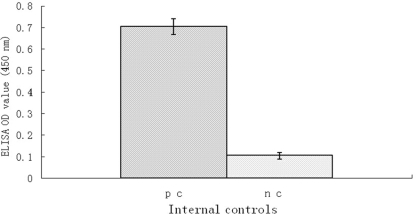
The rN-protein-based double-antigen sandwich ELISA generated minimal background activity and clearly differentiated between all internal controls used. There was no evidence of excessive variation within or between routine runs. The mean OD value and its SD for positive-control sera were 0.705 and 0.037, respectively, and those for negative-control sera were 0.104 and 0.016, respectively (Fig. 2).
Fig 2.
Mean ± SD of OD values of positive-control (pc) and negative-control (nc) sera in rN-protein-based double-antigen sandwich ELISA on 8 plates during routine runs of the assay over a period of 2 months. Each internal control was tested in quadruplicate for every plate.
Cutoff values and diagnostic accuracy of double-antigen sandwich ELISA.
All human and animal serum samples were tested by double-antigen sandwich ELISA and serum neutralization test. The selection of a cutoff value for the double-antigen sandwich ELISA was based on data sets which were dichotomized according to the results of the serum neutralization test (see Table 2). Estimates of sensitivity, specificity, and other combined measures of diagnostic accuracy were determined at cutoffs of 19.29 PP and 21.56 PP (mean + 2× SD and mean + 3× SD, respectively, derived from PP readings in negative-control sera) (Table 1). As shown in Table 1, the use of 21.56 PP as an ELISA threshold value more truly reflects the human and animal infection status decided by the serum neutralization test than a threshold of 19.29 PP.
Table 2.
Double-antigen sandwich ELISA and virus neutralization results
| ELISA result | No. of samples with the following serum neutralization test result: |
||
|---|---|---|---|
| + | − | Total | |
| + | 187 | 2 | 189 |
| − | 0 | 465 | 465 |
| Total | 187 | 467 | 654 |
Table 1.
Diagnostic accuracy of SFTSV double-antigen sandwich ELISA
| Cutoff (PP) | Sensitivity (%) | Specificity (%) | Youden's index | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|
| 19.29a | 100 | 98.72 | 0.987 | 96.89 | 100 |
| 21.56b | 100 | 99.57 | 0.996 | 98.94 | 100 |
Cutoff value based on mean + 2× SD of ELISA PP values in negative-control serum.
Cutoff value based on mean + 3× SD of ELISA PP values in negative-control serum.
Evaluation of double-antigen sandwich ELISA and serum neutralization testing.
At the cutoff 21.56 PP, the mean + 3× SD PP values in negative-control sera, 9 of the 250 human field serum samples (3.6%) and 145 of the 304 animal serum samples (including 123 from goats, 17 from cows, 2 from pigs, 1 from a chicken, and 2 from hedgehogs) (47.7%) were found to be positive by ELISA (Fig. 3A to C). All of the 35 SFTSV-confirmed convalescent-phase patient serum samples were positive, and all of the 65 hantavirus- and dengue virus-confirmed human serum samples were negative (Fig. 3D). All the 187 serum samples that tested positive in the serum neutralization test were also found to be positive by ELISA (Table 2). However, two specimens from Jiangsu, including one from a human (specimen 118) and one from a pig (specimen 33), tested positive by ELISA at a cutoff threshold of 21.56 PP with OD values of 26.52 PP and 26.56 PP, respectively, but were found to be negative by serum neutralization (Fig. 3A and C). Using the Spearman test, a high correlation (R2 = 0.986) between ELISA PP values and serum neutralization titers was demonstrated.
Fig 3.
Distribution of ELISA PP values (■) and serum neutralization titers (♢) in all 654 serum samples. (A) Results for 250 human field serum samples, including 150 from Jiangsu and 100 from Anhui; (B) results for 150 goat field serum samples, including 100 from Jiangsu and 50 from Anhui; (C) results for 154 animal field serum samples from 50 cattle, 50 pigs, 50 chickens, and 4 hedgehogs, all from Jiangsu; (D) results for 35 SFTSV-confirmed patient convalescent-phase serum samples, including 18 from Jiangsu, 15 from Anhui, and 2 from Hubei, and 30 hantavirus- and 35 dengue virus-confirmed human serum samples from Jiangsu. Two specimens from Jiangsu from a human (specimen 118) and a pig (specimen 33) (indicated with arrows in panels A and C) showed inconsistency in ELISA and serum neutralization tests. The horizontal lines represent the cutoff for serum neutralization test (—; l g10 [i.e., log1010 with a value of 1] equivalent to a serum dilution of ≥1:10), the cutoff for ELISA based on mean + 2× SD of PP values in negative-control sera (– – –; 1.29, equivalent to 19.29 PP of positive-control sera), and the cutoff for ELISA based on mean + 3× SD of PP values in negative-control sera (- - -; 1.33, equivalent to 21.56 PP).
Specificity of double-antigen sandwich ELISA.
Hantavirus- and dengue virus-confirmed human sera were used to determine the specificity of the sandwich ELISA system. Both groups of sera consistently tested negative for SFTSV, indicating no cross-reaction pattern between SFTSV and hantavirus or dengue virus (Fig. 3D; Table 3).
Table 3.
Specificity of sandwich ELISA with hantavirus- and dengue virus-confirmed human sera
| ELISA result | No. of human serum samples confirmed to have antibody against: |
|
|---|---|---|
| Hantavirus | Dengue virus | |
| + | 0 | 0 |
| − | 30 | 35 |
Effect of derivation of rN protein on ELISA results.
The rN protein, derived from a Jiangsu virus isolate, readily reacted with SFTSV-positive sera (validated by serum neutralization test) collected from Anhui and Hubei Provinces (Fig. 3D; Table 4), indicating a lack of major antigenic variation in N protein between geographically disparate virus isolates in China.
Table 4.
Utility of N-protein-based double-antigen sandwich ELISA on diverse geographically specific sera
| ELISA result | No. of serum samples from the following locations with indicated ELISA (SNTa) result: |
|
|---|---|---|
| Anhui | Hubei | |
| + | 59 (59) | 2 (2) |
| − | 106 (106) | 0 (0) |
| Total | 165b | 2c |
SNT, serum neutralization test.
The serum samples included 100 from humans, 50 from goats in the field, and 15 from clinically convalescent humans.
Two serum samples from clinically convalescent humans.
DISCUSSION
A unique sandwich ELISA format was set up to determine SFTSV seroprevalence in humans and animals in China. Compared to the traditional indirect ELISA, which detects class-specific antibodies (IgG and IgM), this double-antigen sandwich ELISA system detects total antibodies, which improves sensitivity (2). More importantly, this new ELISA format is species independent and can be used to detect pathogen-specific antibodies in different animal species (5). This is invaluable for identifying potential virus reservoir hosts, especially in wildlife, for which very few kits are currently commercially available. However, our ELISA control samples came from only one animal species (goats), and this should be taken into account in later studies (see Materials and Methods).
As the most abundant, highly conserved, and immunogenic viral component in the virions of the members of the Bunyaviridae family, N protein has been heavily exploited in diagnostic applications (15, 18). Rift Valley fever virus N protein, for instance, has been used in a variety of ELISA formats to detect antibodies in both humans and other animals, and it has shown excellent diagnostic accuracy (11, 12). The application of N-protein-based ELISA formats also avoids the risk of laboratory infection and of residual virus in the test reagents, making them safe for routine use in areas of nonendemicity (13). In the present work, the recombinant SFTSV N protein expressed in E. coli was used as capture antigen for the first time. It served as a labeled detector in this double-antigen sandwich ELISA to detect the total antibodies. The diagnostic performance of this assay with regard to accuracy, specificity, and other factors has been well characterized.
We used the serum neutralization test as a standard to validate the performance of this ELISA. First, the fact that a clinically compatible outbreak in humans has been retrospectively identified by serum neutralization test to have occurred in as early as 1996 (1, 8) and the fact that N protein derived from a Jiangsu virus isolate could readily react with SFTSV-confirmed sera from Anhui and Hubei (Table 4) demonstrated that infection with SFTSV could induce long-term neutralizing immunity and that there would be no serological subgroups between virus isolates from disparate geographic origins. This confirmed that the serum neutralization test could serve as a diagnostic discriminator in this study. At ELISA cutoff values of 21.56 PP, all 187 serum samples that had tested positive in the serum neutralization test were also positive in the ELISA, showing a high correlation between the ELISA and the serum neutralization test (Table 2). However, two serum samples that had tested positive in the ELISA were found to be negative by serum neutralization. In this context, there are at least two factors that could account for this possible false-positive result. Because these two serum samples had lower titers in ELISA than the other positive samples (Fig. 3A and C), it may be that sera at low dilutions are toxic to cell cultures when they are subjected to testing in the serum neutralization test. This would hamper standardization at the interface between negative and positive sera (11). Second, the antibodies measured by ELISA and those measured by the serum neutralization test have different levels of antigenic specificity (14). In this study, an ELISA system that can detect antibodies against all epitopes of the N protein would be more sensitive than the serum neutralization test, which detects only antibodies to virus-neutralizing epitopes. This may explain the higher sensitivity of the ELISA relative to the virus neutralization test in detection of SFTSV-specific antibody. Additionally, dengue fever and hantavirus-borne hemorrhagic fever are endemic in the eastern part of China (4, 10). Patients with SFTSV present signs and clinical symptoms similar to those of patients with these two conditions. The preliminary data provided in this study indicating that the 30 hantavirus- and 35 dengue virus-confirmed serum samples tested negative by this ELISA show that there would be no cross-reactivity between SFTSV and hantaviruses or dengue viruses (Table 3). Although serological cross-reactions with antibodies against unknown phleboviruses cannot be definitely ruled out, to date, this double-antigen ELISA for SFTSV antibodies has shown excellent specificity.
ELISA results showed overall SFTSV-specific antibody prevalence rates of 3.6% in humans and 47.7% in other animals, with values of 82% in goats, 50% in hedgehogs, 34% in cattle, 6% in pigs, and 2% in chickens. This strongly suggests for the first time that SFTSV has circulated widely in China, although the epidemiological cycle and the modalities of this circulation remain unknown. The higher prevalence of total antibodies in goats, cattle, and hedgehogs relative to that in humans, pigs, and chickens may be due to higher exposure to SFTSV vectors, which are not completely understood. Further investigations must uncover the putative ecological cycle of SFTSV, including vector species and the roles of animal reservoirs in the viral transmission network.
In conclusion, this N-protein-based double-antigen sandwich ELISA system not only is compatible with but also has the potential to replace currently established methods. It is a safe, robust, and highly accurate diagnostic tool suitable for disease surveillance programs, particularly with regard to screening large numbers of samples from different species.
ACKNOWLEDGMENT
This work was supported by Jiangsu Province's Outstanding Medical Academic Leader Program (RC2011082).
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Bao C, Qi X, Jiao Y, Wang H. A nosocomial outbreak of novel bunyavirus infection in eastern China-serological evidence. Emerg. Infect. Dis., in press [Google Scholar]
- 2. Constantine NT, Zink H. 2005. HIV testing technologies after two decades of evolution. Indian J. Med. Res. 121:519–538 [PubMed] [Google Scholar]
- 3. Fafetine JM, et al. 2007. Cloning and expression of Rift Valley fever virus nucleocapsid (N) protein and evaluation of a N-protein based indirect ELISA for the detection of specific IgG and IgM antibodies in domestic ruminants. Vet. Microbiol. 121:29–38 [DOI] [PubMed] [Google Scholar]
- 4. Gao X, Nasci R, Liang G. 2010. The neglected arboviral infections in mainland China. PLoS Negl. Trop. Dis. 4:e624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu WP, et al. 2008. Double-antigen enzyme-linked immunosorbent assay for detection of hepatitis E virus-specific antibodies in human or swine sera. Clin. Vaccine Immunol. 15:1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jansen van Vuren P, Potgieter AC, Paweska JT, van Dijk AA. 2007. Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J. Virol. Methods 140:106–114 [DOI] [PubMed] [Google Scholar]
- 7. Li DX. 2011. An outline of severe fever with thrombocytopenia syndrome virus. Chin. J. Exp. Clin. Virol. 25:81–84 [Google Scholar]
- 8. Lin Y, Yao Z, Miao X, Tu X. 1998. An investigation of epidemic outbreak with unknown etiology. Dis. Surveill. 13:29–30 (In Chinese.) [Google Scholar]
- 9. Magurano F, Nicoletti L. 1999. Humoral response in Toscana virus acute neurologic disease investigated by viral-protein-specific immunoassays. Clin. Diagn. Lab. Immunol. 6:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niklasson BS. 1992. Haemorrhagic fever with renal syndrome, virological and epidemiological aspects. Pediatr. Nephrol. 6:201–204 [DOI] [PubMed] [Google Scholar]
- 11. Paweska JT, et al. 2008. Recombinant nucleocapsid-based ELISA for detection of IgG antibody to Rift Valley fever virus in African buffalo. Vet. Microbiol. 127:21–28 [DOI] [PubMed] [Google Scholar]
- 12. Paweska JT, Mortimer E, Leman PA, Swanepoel R. 2005. An inhibition enzyme-linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in humans, domestic and wild ruminants. J. Virol. Methods 127:10–18 [DOI] [PubMed] [Google Scholar]
- 13. Pepin M, Bouloy M, Bird B, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saunders GC, Clinard EH, Bartlett ML, Sanders WM. 1977. Application of the indirect enzyme-labeled antibody microtest to the detection and surveillance of animal diseases. J. Infect. Dis. Suppl. 136:S258–S266 [DOI] [PubMed] [Google Scholar]
- 15. Schwarz TF, Gilch S, Pauli C, Jäger G. 1996. Immunoblot detection of antibodies to Toscana virus. J. Med. Virol. 49:83–86 [DOI] [PubMed] [Google Scholar]
- 16. Stone R. 2010. Rival teams identify a virus behind deaths in central China. Science 330:20–21 [DOI] [PubMed] [Google Scholar]
- 17. Swanepoel R, et al. 1986. Comparative pathogenicity and antigenic cross-reactivity of Rift Valley fever and other African phleboviruses in sheep. J. Hyg. (Lond.) 97:331–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vapalahti O, et al. 1995. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J. Med. Virol. 46:293–303 [DOI] [PubMed] [Google Scholar]
- 19. Yu XJ, et al. 2011. Fever with thrombocytopenia associated with a novel Bunyavirus in China. N. Engl. J. Med. 364:1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]