Abstract
Pyrazinamide is important in the treatment of tuberculosis. Unfortunately, the diagnosis of pyrazinamide resistance is hampered by technical difficulties. We hypothesized that mutation analysis combined with the mycobacterial growth indicator tube (MGIT) phenotypic method would be a good predictor of pyrazinamide resistance. We prospectively analyzed 1,650 M. tuberculosis isolates referred to our tuberculosis reference laboratory in 2008 and 2009. In our laboratory, the MGIT 960 system was used for pyrazinamide resistance screening. If a pyrazinamide-resistant strain was detected, we performed a pncA gene mutation analysis. A second MGIT 960 susceptibility assay was performed afterwards to evaluate the accuracy of the pncA mutation analysis to detect true- or false-positive MGIT results. We observed pyrazinamide resistance in 69 samples using the first MGIT 960 analysis. In a second MGIT 960 analysis, 47 of the 69 samples proved susceptible (68% false positivity). Sensitivity of nonsynonymous pncA mutations for detecting resistant isolates was 73% (95% confidence interval [CI], 61% to 73%), and specificity was 100% (95% CI, 95% to 100%). A diagnostic algorithm incorporating phenotypic and molecular methods would have a 100% positive predictive value for detecting pyrazinamide-resistant isolates, indicating that such an algorithm, based on both methods, is a good predictor for pyrazinamide resistance in routine diagnostics.
INTRODUCTION
Pyrazinamide (PZA) is one of the key components of primary drug therapy against tuberculosis, especially when multidrug resistance has been diagnosed (28). The first clinical report of its antituberculosis activity dates to 1952 (30). The addition of pyrazinamide and rifampin to existing antituberculosis drug regimens has shortened the therapy duration from 9 to 6 months (31), and not using pyrazinamide is correlated with treatment relapse (12). It is a unique antituberculosis drug because of its activity against slowly growing, semidormant bacilli in acidic environments (31).
The enzyme pyrazinamidase plays a crucial role in the mycobactericidal effect of pyrazinamide. This enzyme is expressed constitutively in the cytoplasm of Mycobacterium tuberculosis (31). Only after conversion of pyrazinamide into pyrazinoic acid by this enzyme is its deleterious effect on the tubercle bacilli expressed, by destabilizing the membrane potential and affecting membrane transport function (32). Consequently, loss of pyrazinamidase activity leads to pyrazinamide-resistant tuberculosis bacilli (11).
Nonsynonymous mutations in the gene encoding pyrazinamidase, the pncA gene, lead to the loss of pyrazinamidase activity and are the major mechanism in the development of pyrazinamidase resistance (19). Mutation analysis could thus be used to indirectly assess susceptibility to pyrazinamide (3). However, assessment of susceptibility based on pncA gene sequence analysis has its shortcomings. Mutations are highly diverse and are widely dispersed throughout the pncA gene, limiting the chances of successful development of simple screening methods, such as line probe assays. Furthermore, not all pyrazinamide-resistant M. tuberculosis isolates have mutations in this gene (9). For instance, mutations in the rpsA gene, encoding ribosomal protein S1, have been described recently as a novel mechanism for pyrazinamide resistance (21).
Phenotypic methods for testing susceptibility of M. tuberculosis to pyrazinamide remain the gold standard but also have their shortcomings. Both false-negative and false-positive resistance results are seen (18). In our experience, false-positive resistance results (major errors) are seen most commonly. This observation has been noted by others also (4, 16). In a study of susceptibility of 743 isolates tested using the Bactec MGIT 960 (mycobacterial growth indicator tube 960) method, Chedore et al. found that 42% of strains that tested as pyrazinamide resistant at first appeared to be in fact susceptible when the test was repeated (4). It is assumed that a large inoculum size impairs pyrazinamidase activity (13) and leads to false-positive cases of pyrazinamide resistance.
Because the gold standard—phenotypic pyrazinamide susceptibility testing—can be hampered by false-positive results and mutation analysis is not yet a validated alternative, diagnosis of pyrazinamide resistance remains difficult. We hypothesized that in routine diagnostics, mutation analysis added to culture-based methods might be a good predictor for pyrazinamide resistance. More specifically, based on the observation that susceptibility testing by the MGIT 960 method is mainly hampered by major errors, we hypothesized that nonsynonymous pncA mutations would be able to differentiate between true resistant and false resistant results.
MATERIALS AND METHODS
Setting.
The National Institute for Public Health and the Environment (RIVM) is the national tuberculosis (TB) reference center in the Netherlands. It receives all primary Mycobacterium tuberculosis complex isolates from the Netherlands. Annually, around 1,000 TB cases are culture positive, and this is around 70% of all cases (23). Both multidrug resistance (MDR) and pyrazinamide resistance are estimated to be present in around 1% of these culture-positive cases (27, 29).
Drug susceptibility to pyrazinamide.
Susceptibility to pyrazinamide was tested by the MGIT 960 method, according to the manufacturer's instructions (Becton Dickinson, NJ) (1). Briefly, a pyrazinamide susceptibility test was prepared with a positive 7-ml MGIT tube using a direct inoculum obtained 1 to 2 days after the positivity signal. Two 7-ml Bactec MGIT 960 PZA medium tubes were used. One hundred microliters of 8,000-μg/ml pyrazinamide solution was added to one tube to achieve the recommended critical concentration of 100 μg/ml (BD Diagnostics). A 0.5-ml volume of the seed inoculum was aseptically pipetted in this drug-containing tube. A drug-free control tube was inoculated with 0.5 ml of a 1:10 dilution of the seed inoculum. Tubes were placed in the MGIT 960 instrument and automatically read. Readouts were analyzed using the EpiCenter software package (24). Mycobacterium tuberculosis isolates were considered pyrazinamide resistant if the MGIT 960 instrument gave concordant resistant results on two separate occasions.
Amplification and sequencing of the pncA gene.
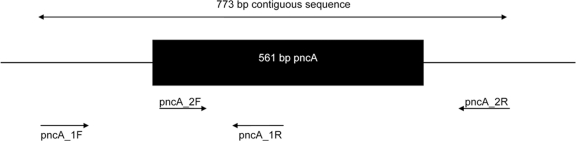
The entire pncA open reading frame, as well as 133 bp upstream and 79 bp downstream, were amplified by PCR. Two overlapping amplicons, covering a 773-bp contiguous sequence (Fig. 1), were generated using the primers described in Table 1. PCR amplifications were carried out in a MBS 0.5S thermal cycler (Thermo Fisher Scientific, Waltham, MA). Each reaction mixture (50 μl) contained 5 μl of 10-ng/μl template DNA, 25 μl of HotStarTaq master mix (Qiagen, Hilden, Germany), 10 μl MilliQ water (Sigma-Aldrich, Irvine, Ayrshire, United Kingdom), and 5 μl of each primer (5 mM). The reaction mixtures were subjected to 15 min at 95°C, followed by 35 cycles of 60 s at 95°C for melting, 120 s at 60°C for annealing, 60 s at 72°C, and an elongation step at 72°C for 10 min. Unincorporated primers and deoxynucleoside triphosphates (dNTPs) were removed from the reaction mixtures using ExoSAP-IT (USB Corporation, Cleveland, OH) according to the manufacturer's instructions. Automated DNA sequencing was performed using BigDye Terminator chemistry according to the protocol supplied by the manufacturer (Applied Biosystems, Foster City, CA). All postrun analyses were performed using the Bionumerics software program, version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium).
Fig 1.
The PCR reading frame for the amplification of the pncA gene.
Table 1.
pncA primer sequences
| Primer name | Sequence (5′ to 3′) | Positiona |
|---|---|---|
| pncA_1F | GGC CGC GAT GAC ACC TCT | −133 |
| pncA_1R | GCC GCA GCC AAT TCA GCA GT | 305 |
| pncA_2F | CGA AGC GGC GGA CTA CCA TCA CG | 180 |
| pncA_2R | CCC CAC CTG CGG CTG CGA ACC | 639 |
Numerical positions of the primers as determined from the start codon of the pncA gene.
Genotyping.
To assess potential associations between M. tuberculosis genotypes and pyrazinamide resistance, we performed IS6110 restriction fragment length polymorphism (RFLP) typing, spoligotyping, and mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) typing of all isolates using previously published methods (10, 25, 26).
RESULTS
Study design.
To answer our research question, we first determined the diagnostic accuracy of nonsynonymous pncA mutations for detecting pyrazinamide-resistant isolates in general. We retrospectively extracted from our database all M. tuberculosis isolates with both MGIT 960 data and pncA data from 2007 (Table 2). Mycobacterium bovis isolates were excluded since they are exclusively pyrazinamide resistant and carry only one specific pncA gene mutation. The year 2007 was selected because in that year both MGIT 960 and pncA gene mutation analyses were performed simultaneously in routine diagnostics. Moreover, MGIT 960 analyses were repeated when resistant strains were found, to minimize false-positive results.
Table 2.
Contingency (2 × 2) table of the diagnostic accuracy of nonsynonymous pncA mutations for detecting pyrazinamide-resistant M. tuberculosis isolates
| Mutation observed | No. of isolates with phenotype |
|
|---|---|---|
| Pyrazinamide resistant | Pyrazinamide susceptible | |
| Nonsynonymous pncA mutation | 6 | 4 |
| Wild-type pncA or synonymous mutation | 1 | 155 |
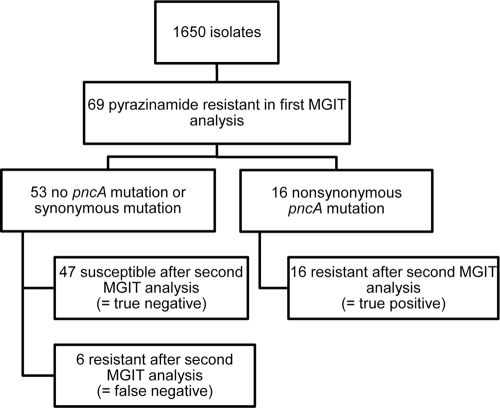
We next asked whether nonsynonymous pncA gene mutations could be used to distinguish between true-resistant and false-resistant MGIT 960 results. To answer this second research question, we prospectively analyzed all M. tuberculosis isolates sent for resistance testing to our laboratory in 2008 and 2009. Again, M. bovis isolates were excluded. This second study was set up as a pragmatic laboratory-based study using a diagnostic algorithm close to routine clinical practice. In our laboratory, the MGIT 960 analysis was used for pyrazinamide resistance screening. If a pyrazinamide-resistant strain was detected, we performed a pncA gene mutation analysis. A second MGIT 960 susceptibility assay was performed afterwards to evaluate the accuracy of the mutation analysis to detect true- or false-positive MGIT results (Fig. 2).
Fig 2.
Diagnostic accuracy of pncA mutation analysis added to bacteriological susceptibility testing in detecting false- and true-positive pyrazinamide-resistant M. tuberculosis isolates.
Diagnostic accuracy of nonsynonymous pncA mutations for detecting pyrazinamide-resistant isolates.
We first determined the diagnostic accuracy of nonsynonymous pncA mutations for detecting pyrazinamide-resistant isolates in general. We collected 166 isolates from 2007, 159 susceptible isolates and 7 pyrazinamide-resistant isolates (Table 2). In 159 susceptible isolates, 4 nonsynonymous pncA mutations were observed (Table 3). We performed both the MGIT 960 analysis and the pncA mutation analysis two times to confirm these results. In 7 resistant isolates, 6 nonsynonymous mutations were seen, 4 of which were identical, but these were different isolates from the same patient (Table 3).
Table 3.
Drug susceptibility results and genotypic characterization of the 10 M. tuberculosis isolates with a nonsynonymous pncA mutation from 2007
| Age of patient (yrs) | Sex of patient (M/F) | Characteristic of isolate |
||||
|---|---|---|---|---|---|---|
| VNTR type | Pyrazinamide susceptibilityc | Resistance to other drugsd |
pncA mutation |
|||
| Nucleotide change | Amino acid change | |||||
| 34 | F | 9001812 | S | H, RFB, E, SPT | G to A at 82 | Ala28Thr |
| 42 | F | 9001951 | S | C to T at 127 | His43Tyr | |
| 29 | M | 9002014 | S | H, RFB, E, SPT | C to G at 147 | Asp49Glu |
| 46 | F | 9000148 | S | G to A at 371 | Gly124Asp | |
| —a | —a | 9001879 | R | Unknowna | C to G at 102 | Frame shift |
| 48b | F | 9002249 | R | H, RFB, E, SPT | A to C at 226 | Thr76Pro |
| 48b | F | 9002249 | R | H, RFB, E, SPT | A to C at 226 | Thr76Pro |
| 31 | F | 9002426 | R | H, RFB, E, SPT | C to T at 169 | His57Tyr |
| 48b | F | 9002249 | R | H, RFB, E, SPT | A to C at 226 | Thr76Pro |
| 48b | F | 9002249 | R | H, RFB, E, SPT | A to C at 226 | Thr76Pro |
—, anonymous research isolate from foreign TB reference center.
Different isolates from the same patient.
S, susceptible; R, resistant.
Drug names: H, isoniazid; RFB, rifampin; E, ethambutol; SPT, streptomycin.
The sensitivity of the nonsynonymous pncA mutation in detecting pyrazinamide-resistant isolates was 86% (95% confidence interval [CI], 53% to 97%), and the specificity was 98% (95% CI, 96% to 98%). The positive predictive value of the nonsynonymous pncA mutations in detecting pyrazinamide resistance was 60% (95% CI, 37% to 68%), and the negative predictive value was 99% (95% CI, 98% to 99%). The positive likelihood ratio was 34.1 (95% CI, 13.5 to 48.6), and the negative likelihood ratio was 0.15 (95% CI, 0.03 to 0.5). The overall accuracy of nonsynonymous pncA mutations for predicting pyrazinamide resistance in M. tuberculosis isolates was high (97% [95% CI, 94% to 98%]).
Predicting a true or false-positive pyrazinamide-resistant 960 MGIT result using mutation analysis.
We next examined whether nonsynonymous pncA mutations could distinguish between true- and false-positive resistant MGIT 960 results. To answer this question, we prospectively analyzed all M. tuberculosis isolates sent for resistance testing to our laboratory in 2008 and 2009.
During 2008 and 2009, 1,650 M. tuberculosis isolates were sent for resistance testing to our laboratory. We observed pyrazinamide resistance in 69 out of 1,650 samples after the first MGIT 960 analysis. However, in the second MGIT 960 test, 47 of the 69 samples proved susceptible and 22 were confirmed to be resistant, indicating a false-positive rate of 68% in the first MGIT 960 test. Sensitivity of the nonsynonymous pncA mutation in detecting pyrazinamide true-resistant isolates was 73% (95% CI, 61% to 73%), and specificity was 100% (95% CI, 95% to 100%). The positive predictive value of the nonsynonymous pncA mutation in detecting pyrazinamide resistance was 100% (95% CI, 85% to 100%), and the negative predictive value was 89% (95% CI, 84% to 89%). The overall accuracy of nonsynonymous pncA mutations for detecting pyrazinamide-resistant isolates was 91% (95% CI, 84% to 92%).
Pyrazinamide-resistant cases in the Netherlands.
Twenty-two isolates from 15 patients from 2008 and 2009 were pyrazinamide resistant; their baseline characteristics, MIRU-VNTR typing results, drug susceptibility profiles, and pncA sequence analyses are described in Table 4. A wide variety of mutations was seen, and one mutation was observed in the putative promoter region (patient NLA000801739). Some of the strains had the same types of mutations but different MIRU-VNTR patterns, indicating that they were truly different strains that had coincidently acquired the same type of mutation. For example, strains from patients NLA000800922 and NLA000800620 had the same mutation of His71 → Gln, yet MIRU-VNTR analysis indicated that they were different M. tuberculosis strains.
Table 4.
Characteristics of pyrazinamide-resistant tuberculosis cases in the Netherlands, 2008 and 2009
| Age of patient (yrs) | Sex of patient (M/F) | Patient no. | VNTR type(s) | Resistance to other drugsa |
pncAmutation |
|
|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | |||||
| 29 | M | NLA000800326 | 9002568 | H, RFB, E, CIP | A to C at 502 | Thr168Pro |
| 9002582 | ||||||
| 84 | M | NLA000800465 | 9002610 | G deletion at 60 | Frame shift | |
| 24 | M | NLA000800620 | 9002653 | T to A at 213 | His71Gln | |
| 43 | M | NLA000800922 | 9003512 | T to A at 213 | His71Gln | |
| 9003513 | ||||||
| 26 | F | NLA000801739 | 9002811 | H | −12 promoter mutation T → C | Frame shift |
| 41 | M | NLA000801926 | 9002939 | H | GAG deletion at 430 | Glu144 deletion |
| 44 | M | NLA000800594 | 9003531 | H, RFB, E, AMK, CIN | G insertion at 218 | Frame shift |
| 29 | F | NLA000900573 | 9003015 | H, RFB, E, AMK, CIN, KAN, MOX | G insertion at 516 | Frame shift |
| 17 | F | NLA000901644 | 9000061 | H, RFB | G to A at 3 | Met1Ile |
| 25 | M | NLA000902122 | 9000408 | H, RFB, E, Q-D | G to C at 289 | Gly97Arg |
| 16 | M | NLA000800519 | 9003284 | |||
| 26 | M | NLA000901231 | 9002622 | |||
| 24 | F | NLA000801502 | 9002838 | H | ||
| 47 | M | NLA000801595 | 9000055 | H | ||
| 31 | M | NLA000801755 | 9002965 | |||
Drug names: H, isoniazid; RFB, rifampin; E, ethambutol; CIP, clarithromycin; AMK, amikacin; CIN, ciprofloxacin; KAN, kanamycin; MOX, moxifloxacin; Q-D, rifabutin.
Interestingly, 5 patients had pyrazinamide-resistant isolates that did not carry any pncA mutation, either in the gene itself or in the putative pncA promoter region. Isolates from three of these patients were in fact pyrazinamide monoresistant (NLA000800519, NLA000901231, and NLA000801755). We performed both the MGIT 960 analysis and the pncA mutation analysis three times to confirm these results. MIRU-VNTR analysis indicated that these were all different strains (Table 4).
Genotype family of pyrazinamide-resistant cases.
Since we found unusual pyrazinamide-monoresistant cases, we wondered whether the respective M. tuberculosis isolates were clustered in certain genotype families. We therefore determined the genotype family of all resistant isolates (Fig. 3). One spoligotype was not available (patient NLA000801926). Although a relatively high percentage of the Beijing genotype (5 out of 15; 33%) was noted in this sample, no specific genotype family clustering was seen among the pyrazinamide-(mono)resistant cases (Fig. 3).
Fig 3.
Dendrogram illustrating the spoligotyping, IS6110 RFLP pattern, and mutation analysis of the 15 pyrazinamide-resistant cases from the Netherlands, 2008 and 2009.
DISCUSSION
This study shows that mutation analysis added to culture-based methods is a good predictor for true pyrazinamide resistance in routine diagnostics. In our first experiments, we found that DNA sequencing may strongly rule out pyrazinamide resistance (Table 2). In our second experiment, we have shown that in a series of 69 isolates found resistant in the first instance by the MGIT 960 system, 68% were false positive for resistance and nonsynonymous pncA mutations could identify these false-positive results correctly (Fig. 2). Moreover, a combination of the MGIT 960 and mutation analyses could correctly identify pyrazinamide-resistant cases (Fig. 2).
Our study has some important strengths. First, we validated our results by retesting discordant pyrazinamide results, thereby minimizing major errors. Second, it was set up as a pragmatic trial enabling direct applicability of our diagnostic algorithm in everyday clinical diagnostics in a variety of settings.
Our results extend our knowledge of the role of molecular methods in the diagnosis of pyrazinamide resistance. The first studies on the association of pncA mutations and pyrazinamide resistance found mutations in up to 97% of pyrazinamide-resistant cases (8, 20). However, these were mainly selected pyrazinamide-resistant cases with a high MIC. Subsequent studies have shown a lower prevalence of pncA mutations in pyrazinamide-resistant cases (2, 9). We also found a lower prevalence of nonsynonymous pncA mutations (67% of isolates). Given our experimental setup, our results may better reflect pyrazinamide-resistant cases found in the daily routine in a country with a low prevalence of pyrazinamide resistance. Zhang and Mitchison have argued that such a finding may in part reflect incorrect PZA susceptibility testing (31). In our experiments, both the MGIT 960 analysis and the mutation analysis yielded identical results three times on separate occasions. Hence, measurement errors cannot explain these findings. Pyrazinamide resistance in a strain with wild-type pncA sequences can alternatively be explained by the presence of a pncA regulatory gene outside our reading frame or by pyrazinamide resistance mechanisms other than the effect on pyrazinamidase (31), such as the recent finding of mutations in the rpsA gene (21).
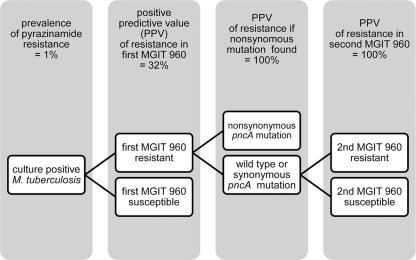
In a recent meta-analysis, Chang and coworkers have summarized the value of pncA mutation in the diagnosis of pyrazinamide resistance (3). Similar to the results of our first experiments, they also found that DNA sequencing may strongly rule out pyrazinamide resistance in a low-prevalence (non-MDR) setting. Second, they concluded that in high-prevalence settings (31 to 89%), pncA mutations may reliably detect true pyrazinamide resistance. The prevalence of true pyrazinamide resistance among the 69 isolates with a first “resistant” MGIT result was 32% (16/69 isolates) (Fig. 2), confirming the conclusion of Chang and coworkers that nonsynonymous pncA mutations can detect pyrazinamide resistance properly (Fig. 2).
Based on our results, we propose an algorithm, depicted in Fig. 4, to assess pyrazinamide resistance in routine clinical diagnostics. After a first round of MGIT 960 testing, all isolates labeled resistant should undergo pncA gene sequence analysis. A nonsynonymous mutation has a positive predictive value of 100% for a true pyrazinamide-resistant isolate. If a synonymous mutation or a wild-type pncA gene is found, the MGIT 960 analysis should be repeated. Given the shorter turnaround time of mutation analysis, incorporating molecular methods has the potential of shortening the diagnosis of pyrazinamide resistance. It is to be seen if such shortening will optimize treatment for tuberculosis patients, especially for multidrug-resistant patients for which pyrazinamide susceptibility testing is essential (28).
Fig 4.
Flow diagram to assess pyrazinamide resistance in M. tuberculosis isolates in routine diagnostics, combining phenotypic and molecular methods.
Using the diagnostic algorithm proposed in this study (Fig. 4), we found 15 cases of pyrazinamide resistance in the Netherlands in a 2-year period (Table 4). The total number of cases diagnosed with culture-confirmed tuberculosis in these 2 years was 1,504 (23). The prevalence of pyrazinamide resistance among M. tuberculosis cases in the Netherlands was therefore 1.0%. Five out of 15 (33%) cases had MDR TB, which is significantly higher than the general prevalence of MDR TB in the Netherlands. Such higher prevalence of pyrazinamide resistance in multidrug-resistant tuberculosis is commonly seen (15, 17). A relatively high number of Beijing genotype strains was seen because the Beijing genotype is associated with MDR TB in Europe (5). Three of 15 pyrazinamide cases found in this study were pyrazinamide monoresistant. Pyrazinamide monoresistance has been described previously (6, 7) but is rare. For instance, in a study from the United States, only 3 out of 1,916 isolates proved pyrazinamide monoresistant (7), which is a percentage comparable to our findings (3 out of 1,650 isolates). However, though repeated analysis yielded the same result, we cannot rule out that we made a systematic measurement error giving repeated false-resistant MGIT results.
A limitation of this study is the use of the MGIT 960 system as the gold standard. Though the proper gold standard for pyrazinamide resistance is not established yet, it is accepted that the Bactec radiometric method is probably most reliable (31). We choose the MGIT 960 system as the gold standard since current meta-analyses suggest that the MGIT has comparable test performances and could therefore be used as reference drug susceptibility assay (3, 18). Moreover, there is a legitimate concern about the disposal of radioactive waste when the Bactec radiometric method is used. Third, Bactec 460 machinery is phased out, and supplies will no longer be available, limiting the applicability of a diagnostic algorithm incorporating this method. Last, other candidates for the gold standard are scarce. The 7H10 agar-based testing methods are considered less reliable (14). Susceptibility testing on solid Löwenstein-Jensen medium is acceptable according to some experts (31) but is not used for drug susceptibility testing in most laboratories in the Western world anymore. The Wayne method might be another alternative (3). However, the Wayne method requires a sufficient number of bacilli for detecting pyrazinamidase activity, making this test prone to false-positive resistance results, which was exactly what we tried to minimize in our diagnostic setup.
Though the MGIT can be regarded as a surrogate gold standard, this study confirms an earlier observation by others that the MGIT reports isolates as false positive for resistance (4, 16). Comparable high numbers of false-positive resistant culture results have been reported by other researchers using the MGIT 960 technique. For instance, during pyrazinamide susceptibility testing of 743 M. tuberculosis isolates, Chedore et al. found that 24 out of 57 isolates (42%), initially found resistant in the MGIT 960 test, were in fact susceptible on the basis of wild-type pncA gene sequences, Wayne's method, and the Bactec 460 system, used as reference methods (4). False phenotypic resistance is caused mainly by a well-known difficulty in pyrazinamide susceptibility testing: the use of large inoculums. A large inoculum size increases the pH and thereby inactivates pyrazinamidase (13). An alternative explanation for the high false resistance rate could be the use of a relatively low breakpoint in the MGIT (100 mg/liter) (3), thereby wrongly labeling susceptible or intermediately susceptible isolates resistant. Others have suggested that 200 mg/liter or 300 mg/liter would be a more appropriate resistance breakpoint. Because we wanted to stay close to routine practice, we choose 100 mg/liter as prescribed by the manufacturer (1). Given the high rates of false-positive results, a first notification of pyrazinamide resistance in liquid medium, such as the MGIT 960 system, should be interpreted with caution. We would recommend repeating the MGIT 960 test, giving special attention to the inoculum size and constitution. The present study highlights another possibility, namely, the use of mutation analysis as an adjunct to the MGIT 960 system as depicted in Fig. 4.
This study focused on resolving concerns regarding the high false resistance rates of the MGIT. We did not look into possible false susceptibility results of the MGIT system owing to the fact that this is considered less of a problem when using the MGIT (18). In our first experiments, however, we found four susceptible isolates carrying a nonsynonymous pncA mutation. Though several mutations have been identified in susceptible isolates, we cannot exclude that we made a systematic measurement error and that these isolates were in fact pyrazinamide resistant. We repeated analyses to minimize this error. We did not use the Wayne method to assess pyrazinamide activity to determine whether the mutations led to an impairment of pyrazinamidase activity, because at the time we undertook our study this assay was considered less sensitive for diagnosing pyrazinamide resistance (22). Moreover, a large quantity of bacilli is mandatory for this assay, leading to falsely resistant results.
The issue underlying these aforementioned difficulties in interpreting pyrazinamide resistance testing is the paucity of adequate in vivo data. Although pyrazinamide has been available for 50 years, there are currently no studies published that linked in vitro pyrazinamide resistance to important in vivo clinical outcomes. There is some evidence that the treatment outcome is worse in the presence of initial resistance in general, but this is not specified for pyrazinamide (12). The most important obstacles for sound clinical data are the technical difficulties and the limited standardization in drug susceptibility testing as described earlier. Our proposed flow diagram may help in standardizing pyrazinamide resistance testing and may be used in future studies of clinical outcome.
In summary, we have shown that a combination of MGIT 960 phenotypic pyrazinamide susceptibility testing and pncA mutation analysis provides a good predictor of pyrazinamide resistance in routine diagnostics. Nonsynonymous pncA mutations are able to differentiate between true-resistant and falsely resistant MGIT 960 results. Given the high number of false-positive results from phenotypic methods, our findings suggest that using mutation analysis improves and speeds pyrazinamide susceptibility testing. Based hereupon, we propose a diagnostic algorithm combining both phenotypic and molecular tests for the assessment of pyrazinamide resistance.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Becton Dickinson and Company. 1999. Bactec MGIT 960 system user's manual. Becton Dickinson and Company, Sparks, MD [Google Scholar]
- 2. Bishop KS, et al. 2001. Characterisation of the pncA gene in Mycobacterium tuberculosis isolates from Gauteng, South Africa. Int. J. Tuberc. Lung Dis. 5:952–957 [PubMed] [Google Scholar]
- 3. Chang KC, Yew WW, Zhang Y. 2011. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis: a systematic review with meta-analyses. Antimicrob. Agents Chemother. 55:4499–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chedore P, Bertucci L, Wolfe J, Sharma M, Jamieson F. 2010. Potential erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J. Clin. Microbiol. 48:300–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Devaux I, Kremer K, Heersma H, van Soolingen D. 2009. Clusters of multidrug-resistant Mycobacterium tuberculosis cases. Europe. Emerg. Infect. Dis. 15:1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doustdar F, Khosravi AD, Farnia P. 2009. Mycobacterium tuberculosis genotype diversity in pyrazinamide-resistant isolates of Iran. Microb. Drug Resist. 15:251–256 [DOI] [PubMed] [Google Scholar]
- 7. Hannan MM, Desmond EP, Morlock GP, Mazurek GH, Crawford JT. 2001. Pyrazinamide-monoresistant Mycobacterium tuberculosis in the United States. J. Clin. Microbiol. 39:647–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. 1997. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber. Lung Dis. 78:117–122 [DOI] [PubMed] [Google Scholar]
- 9. Huang TS, et al. 2003. Correlation between pyrazinamide activity and pncA mutations in Mycobacterium tuberculosis isolates in Taiwan. Antimicrob. Agents Chemother. 47:3672–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Konno K, Feldman FM, McDermott W. 1967. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 95:461–469 [DOI] [PubMed] [Google Scholar]
- 12. Lew W, Pai M, Oxlade O, Martin D, Menzies D. 2008. Initial drug resistance and tuberculosis treatment outcomes: systematic review and meta-analysis. Ann. Intern. Med. 149:123–134 [DOI] [PubMed] [Google Scholar]
- 13. McDermott W, Thompson R. 1954. Activation of pyrazinamide and nicotinamide in acidic environment in vitro. Am. Rev. Tuberc. 70:748–754 [DOI] [PubMed] [Google Scholar]
- 14. Morlock GP, et al. 2000. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2291–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mphahlele M, et al. 2008. Pyrazinamide resistance among South African multidrug-resistant Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 46:3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pandey S, Newton S, Upton A, Roberts S, Drinkovic D. 2009. Characterisation of pncA mutations in clinical Mycobacterium tuberculosis isolates in New Zealand. Pathology 41:582–584 [DOI] [PubMed] [Google Scholar]
- 17. Parwati I, Van Crevel R, Van Soolingen D. 2010. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10:103–111 [DOI] [PubMed] [Google Scholar]
- 18. Piersimoni C, Olivieri A, Benacchio L, Scarparo C. 2006. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. J. Clin. Microbiol. 44:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scorpio A, Zhang Y. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662–667 [DOI] [PubMed] [Google Scholar]
- 20. Scorpio A, et al. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi W, et al. 2011. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science 16:1630–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh P, et al. 2007. Comparative evaluation of Löwenstein-Jensen proportion method, BacT/ALERT 3D system, and enzymatic pyrazinamidase assay for pyrazinamide susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 45:76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slump E, Erkens CGM, Kalisvaart NA, Van Rest J, Šebek M, Van Soolingen D. 2010. Tuberculosis in the Netherlands 2009. KNCV Tuberculosis Foundation, the Hague, the Netherlands: (In Dutch.) [Google Scholar]
- 24. Springer B, Lucke K, Calligaris-Maibach C, Ritter C, Böttger EC. 2009. Quantitative drug susceptibility testing of Mycobacterium tuberculosis by the use of MGIT 960 and EpiCenter Instrumentation. J. Clin. Microbiol. 47:1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Supply P, et al. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Embden JD, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Klingeren B, Dessens-Kroon M, Van der Laan TT, Kremer K, Van Soolingen D. 2007. Drug susceptibility testing of Mycobacterium tuberculosis complex by use of a high-throughput, reproducible, absolute concentration method. J. Clin. Microbiol. 45:2662–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. 2010. Treatment of tuberculosis: guidelines, 4th ed WHO, Geneva, Switzerland: [PubMed] [Google Scholar]
- 29. Wright A. 2009. Epidemiology of antituberculosis drug resistance 2002-07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance. Surveillance 373:1861–1873 [DOI] [PubMed] [Google Scholar]
- 30. Yeager RL, Munroe WGC, Dessau FI. 1952. Pyrazinamide (aldinamide) in the treatment of pulmonary tuberculosis. Am. Rev. Tuberc. 65:523–546 [PubMed] [Google Scholar]
- 31. Zhang Y, Mitchison D. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6–21 [PubMed] [Google Scholar]
- 32. Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetic by pyrazinoic acid. J. Antimicrob. Chemother. 52:790–795 [DOI] [PubMed] [Google Scholar]