Abstract
Newcastle disease (ND) is one of the most important diseases of poultry, negatively affecting poultry production worldwide. The disease is caused by Newcastle disease virus (NDV) or avian paramyxovirus type 1 (APMV-1), a negative-sense single-stranded RNA virus of the genus Avulavirus, family Paramyxoviridae. Although all NDV isolates characterized to date belong to a single serotype of APMV-1, significant genetic diversity has been described between different NDV isolates. Here we present the complete genome sequence and the clinicopathological characterization of a virulent Newcastle disease virus isolate (NDV-Peru/08) obtained from poultry during an outbreak of ND in Peru in 2008. Phylogenetic reconstruction and analysis of the evolutionary distances between NDV-Peru/08 and other isolates representing established NDV genotypes revealed the existence of large genomic and amino differences that clearly distinguish this isolate from viruses of typical NDV genotypes. Although NDV-Peru/08 is a genetically distinct virus, pathogenesis studies conducted with chickens revealed that NDV-Peru/08 infection results in clinical signs characteristic of velogenic viscerotropic NDV strains. Additionally, vaccination studies have shown that an inactivated NDV-LaSota/46 vaccine conferred full protection from NDV-Peru/08-induced clinical disease and mortality. This represents the first complete characterization of a virulent NDV isolate from South America.
INTRODUCTION
Newcastle disease (ND), is one the most important diseases of poultry, negatively affecting poultry production worldwide (2). ND is caused by Newcastle disease virus (NDV) or avian paramyxovirus type 1 (APMV-1), a negative-sense single-stranded RNA virus of the genus Avulavirus, family Paramyxoviridae (2). The NDV genome is ∼15.2 kb in length and contains six genes encoding at least seven proteins named nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), RNA-dependent RNA polymerase (L), and V protein, which is produced through editing of the phosphoprotein mRNA (2). ND is characterized by a wide range of clinical manifestations, which vary in severity from subclinical respiratory or enteric infections to fatal neurological or enteric hemorrhagic disease (17).
Although all NDV isolates characterized to date belong to a single serotype, the avian paramyxovirus serotype 1 (APMV-1), significant genetic diversity has been recognized among different NDV isolates (16). Historically, NDV isolates have been classified into two major groups (class I and II), based on their genome lengths and the nucleotide sequences of their genomes (1, 4, 14, 15, 32). Class I viruses are distributed worldwide and have been isolated mainly from waterfowl and shorebirds (7, 8). Class II viruses have been divided into 11 genotypes (I to XI) (4, 14), with genotypes V, VI, VII, and VIII being the predominant genotypes circulating worldwide (14, 15). Among these, genotype VII viruses are particularly important given that they have been associated with many of the most recent outbreaks in Asia, Africa, and the Middle East (12, 14, 15, 32, 34). Additionally, a recent outbreak of ND in South America (Venezuela) has been attributed to a genotype VII virus, suggesting that viruses of this genotype are spreading worldwide (20). Notably, recent phylogenetic studies have shown that NDV is continuously evolving, with viruses of different genotypes undergoing simultaneous changes at different geographic locations, which eventually leads to diagnostic failures (14, 21, 23).
Virulent NDV (vNDV) occurs in at least six of the seven continents of the world and is enzootic in several countries, posing a constant threat to the poultry industry (14). In 2010, infection by vNDV was confirmed in 80 countries, including infections in wild birds in Israel, Kenya, Mongolia, Germany, Italy, Canada, and the United States and infections in domestic poultry in countries of North and South America, Europe, Africa, and Asia (18). In South America, vNDV infection is endemic in some of the northern countries, such as Venezuela, Colombia, and Suriname, while in southern countries, including Chile, Argentina, Uruguay, and Brazil, the disease is only occasionally reported (19). Although vNDV circulates and frequently causes outbreaks in South America, very little information is available on the epidemiology and evolutionary trends of the isolates circulating in that continent. Virulent NDV is exotic in poultry in the United States (31), and migration and/or illegal importation of birds from areas where vNDV is endemic represents a constant threat to the U.S. poultry industry (9, 10, 30). The last outbreak of ND in poultry in the U.S., for example, occurred in California during 2002 and 2003, likely as a result of illegal importation of birds and led to depopulation of more than 3 million birds and containment costs of more than U.S. $160 million (31). These observations highlight the importance of constant epidemiological surveillance for NDV and the need for a proactive characterization of the isolates circulating worldwide. Here we present the complete genome and the clinicopathological characterization of a genetically distinct NDV isolate (poultry/Peru/1918-03/2008 [NDV-Peru/08]) obtained during an outbreak of ND reported by the Official Veterinary Services (SENASA) in Peru in 2008.
MATERIALS AND METHODS
Viruses.
The NDV isolate poultry/Peru/1918-03/2008 (NDV-Peru/08) was obtained from the USDA National Veterinary Services Laboratories (NVSL) repository (after authorization by the Official Veterinary Services of Peru [SENASA]). The virus was isolated from swab samples during an outbreak of ND affecting domestic poultry flocks in the region of Pachacutec, Arequipa, Peru, in 2008. Egg passage 2 (EP2) virus stocks were used for RNA extraction and in all pathogenesis experiments in the present study. The NDV strain LaSota/46 was obtained from the SEPRL repository and grown in 9- to 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs (ECE). Allantoic fluid collected from NDV-LaSota/46-inoculated eggs was used for the production of an inactivated vaccine as described below.
Eggs and chickens.
Embryonated chicken eggs and chickens were obtained from the SEPRL SPF white Leghorn flock. Birds were housed in negative pressure isolators under biosafety level 3 (BSL-3) enhanced containment and received food and water ad libitum.
ICPI test.
Pathogenicity of NDV-Peru/08 was assessed by using the standard intracerebral pathogenicity index (ICPI) test (3). Day-old chicks were inoculated intracerebrally with 0.1 ml of a 1:10 dilution of infective allantoic fluid. Chicks were monitored during an 8-day observation period and scored as follows: normal, 0; sick or paralyzed, 1; or dead, 2. Total scores were determined, and the mean daily score was calculated to obtain the ICPI (3).
RNA isolation and sequencing.
The NDV-Peru/08 isolate was propagated in SPF embryonated chicken eggs. Total RNA was extracted from allantoic fluids using TRIzol LS (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The F gene sequence was determined by PCR amplification of the complete gene (primers available upon request), followed by sequencing of the amplicon with fluorescent dideoxynucleotide terminators in an ABI 3700 automated sequencer (Applied Biosystems, Inc., Foster City, CA).
The complete genome sequence of NDV-Peru/08 was determined by using a shotgun reverse transcription (RT)-PCR/sequencing approach. Amplification reactions were performed with the one-step RT-PCR kit (Qiagen, Valencia, CA) and the following set of degenerated primers: beg2 (5′-CGCGTCGACTACTACGGGTAGA-3′) and end-r (5′-GTACCCGGGGATCCTTTTTTCTAA-3′) and FR26RV-N (5′-GCCGGAGCTCTGCAGATATC-3′) and FR20RV (5′-GCCGGAGCTCTGCAGATATC-3′). The PCR amplicons were subjected to electrophoresis in 1% agarose gels, and DNA bands with lengths of 500 to 1,000 bp and those longer than 1,001 bp were excised from the gels and purified by using the QuickClean DNA gel extraction kit (Qiagen, Valencia, CA). The purified PCR products were cloned in the TOPO TA vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and subjected to DNA sequencing as described above. Sequence editing and assembly were performed with the LaserGene sequence analysis software package (LaserGene, version 5.07; DNAStar, Inc., Madison, WI).
Phylogenetic analysis.
The complete F and the complete genome sequences were used to construct the phylogenetic trees and to localize NDV-Peru/08 among other class II reference strains. Phylogenetic analysis was performed by using the MEGA5 software (MEGA, version 5) (28). The evolutionary distances were inferred using MEGA 5 (27) and were expressed based on the number of nucleotide substitutions per site. The codon positions included in the analysis were the 1st, 2nd, 3rd, and noncoding. All positions containing gaps and missing data were eliminated from the data set (the “complete deletion” option). Analysis of recombination was performed using RDP3 (13). The numbers used in the phylogenetic trees represent the GenInfo Identifier (GI) sequence identification number in GenBank.
Sequence alignment.
Alignment and comparison of the nucleotide and amino acid sequences between NDV-Peru/08 and selected strains representing established NDV genotypes were performed by using the software ClustalW (29).
Animal inoculations.
The clinicopathological features of NDV-Peru/08 infection were assessed in chickens. Forty 4-week-old SPF white Leghorn chickens were randomly allocated to four experimental groups, consisting of two NDV-Peru/08-infected groups (group 1, n = 10; and group 2, n = 10) and two mock-infected (control) groups (n = 10). Birds were inoculated with a virus suspension (0.1 ml) containing 105 50% embryo infectious doses (EID50)/0.1 ml. Half of the inoculum (0.05 ml) was applied to the conjunctival sac, and half (0.05 ml) to the choanal cleft (26). Mock-infected control birds were inoculated as described above with 0.1 ml of phosphate-buffered saline (PBS). Birds were monitored daily for characteristic clinical signs of Newcastle disease. Two birds of groups 1 (NDV-Peru/08) and 2 (PBS control) were euthanized and necropsied at 2 and 5 days postinoculation (dpi), and all birds in extremis (presenting a severe clinical condition) were euthanized regardless of the sampling schedule. Tissue samples consisting of eyelid, spleen, bursa of Fabricius, thymus, Harderian gland, proventriculus, small intestine, cecal tonsils, large intestine, air sacs, trachea, lung, heart, esophagus, pharynx, crop, brain, liver, pancreas, kidney, comb, head of the femur, and nasal turbinate were collected and fixed in 10% neutral buffered formalin for 52 h. All sampled tissues were processed for histological examination and/or immunohistochemistry (IHC) using standard procedures (26). Oral and cloacal swabs were collected on days 2, 3 and 4 postinoculation (p.i.) and processed for virus isolation as described below. Birds of groups 2 (NDV-Peru/08) and 4 (PBS control) were monitored daily for characteristic clinical signs of Newcastle disease, and the time of death was recorded and used to plot the survival curves and to calculate the mean death time.
Virus isolation and quantitation.
Virus shedding was assessed in oral and cloacal secretions from all inoculated birds. Swab samples were cleared by centrifugation at 1,000 × g for 20 min, and the supernatant was subjected to virus isolation in 9- to 10-day-old embryonated chicken eggs as previously described (26). Positive samples were tested by hemagglutination (HA) and hemagglutination inhibition (HI) to determine the presence of NDV. Additionally, positive samples were subjected to virus quantitation in embryonated chicken eggs. Serial 10-fold dilutions (10−5 to 10−10) were inoculated in 9- to 10-day-old embryonated chicken eggs, and viral titers were determined as EID50/0.1 ml according to the method of Reed and Muench.
IHC.
Expression and distribution of the viral nucleoprotein (NP) were assessed by immunohistochemistry (IHC). Tissue sections (4 μm) were subjected to deparaffinization followed by antigen retrieval (Vector antigen unmasking solution; Vector Laboratories, Burlingame, CA) and blocking of nonspecific antigens (universal blocking reagent; Biogenex, San Ramon, CA) according to the manufacturer's instructions. Samples were incubated overnight with the anti-NP antibody at 4°C (26), washed, incubated with an alkaline phosphatase-labeled polymer anti-rabbit Fc (LabVision polymer; LabVision, Fremont, CA), and developed by using the chromogen Vector Red substrate (Vector Laboratories, Burlingame, CA) (26). Sections were counterstained with hematoxylin, mounted, and examined under a light microscope.
Vaccine production.
Allantoic fluid collected from ECE inoculated with NDV-LaSota/46 or mock inoculated (sham) was clarified by centrifugation at 1,000 × g for 15 min. The virus titer was determined as EID50/0.1 ml, as described above, and a water-in-oil emulsion inactivated vaccine was prepared as previously described (16).
Vaccination studies.
The efficacy of a traditional inactivated vaccine in conferring protection against NDV-Peru/08 challenge was assessed in chickens. Four-week-old SPF white Leghorn chickens were randomly allocated into two experimental groups consisting of an NDV-LaSota/46-vaccinated group (n = 10) and a control sham-vaccinated group (n = 10). Birds were vaccinated subcutaneously with 0.5 ml of the oil emulsion vaccines. Three weeks after vaccination, all birds were challenged with NDV-Peru/08 as described above and monitored for clinical signs of ND during a 14-day observation period. Serum samples were collected at days 0 and 21 postvaccination and day 14 postchallenge and tested for the presence of NDV antibodies by the hemagglutination inhibition assay.
Nucleotide sequence accession number.
The complete genome sequence of the NDV isolate poultry/Peru/1918-03/2008 is available in GenBank under accession no. JN800306.
RESULTS
NDV-Peru/08 belongs to the velogenic NDV pathotype.
The pathogenicity of NDV-Peru/08 was initially assessed by sequencing of the fusion (F) protein cleavage site and by the standard ICPI test. Sequencing of the F protein cleavage site revealed the presence of three basic amino acid residues at positions 113, 115, and 116 and a phenylalanine at position 117 (112R-R-Q-K-R-F117). Intracerebral inoculation of NDV-Peru/08 in day-old chicks resulted in an ICPI of 1.78, which is typical of velogenic NDV strains (15, 31, 32).
Phylogenetic analysis of the complete F gene and the complete genome sequences demonstrate that NDV-Peru/08 is genetically distinct from other established NDV genotypes.
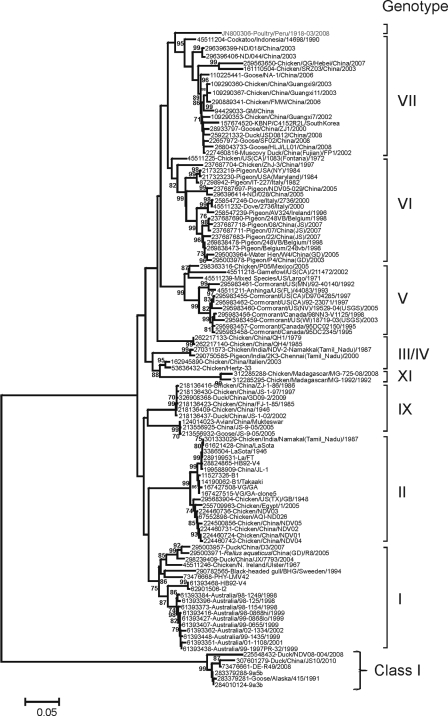
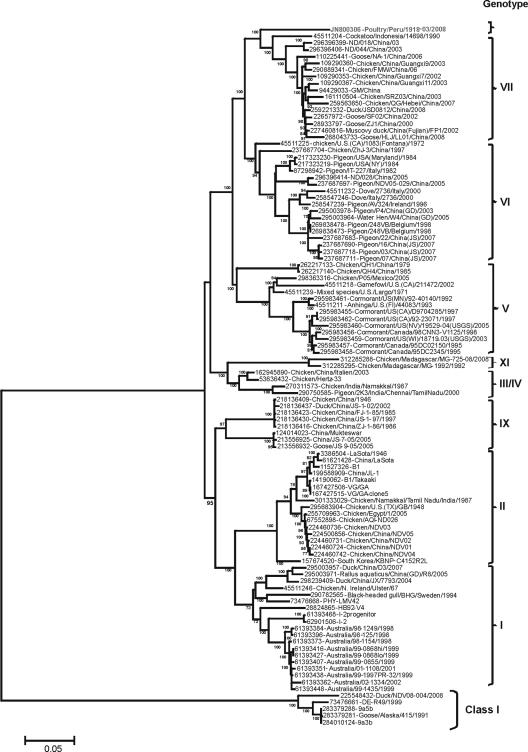
Phylogenetic analysis based on the F gene revealed that NDV-Peru/08 is clearly distinct from viruses representing other known NDV genotypes (Fig. 1). To confirm the results obtained by the analysis of the F gene, we performed a phylogenetic analysis based on the complete genome sequence of NDV-Peru/08 and other reference NDV strains (n = 104). Similar to the results obtained in the analysis of the F gene, the compete genome phylogenetic reconstruction resulted in NDV-Peru/08 forming an isolated branch separate from the viruses of other known class II genotypes (Fig. 2). The viruses that appear most closely related to NDV-Peru/08 are the strains cockatoo/Indonesia/14698/90 (GI 45511204), ND/03/044 (GI 286396406), and ND/03/018 (GI 286396399), which were isolated in Asia and belong to genotype VII and subgenotypes VIIa (cockatoo/Indonesia/14698/90) and VIId (ND/03/044 and ND/03/018). Phylogenetic analysis performed with individual genes of the data set of 104 complete genomes (coding for F, NP, P, M, HN, and L) revealed a similar phylogenetic topology (data not shown). No recombination events were observed in the NDV-Peru/08 genome (data not shown).
Fig 1.
Phylogenetic analysis based on the complete fusion (F) gene sequence of 104 taxa available in GenBank. The evolutionary history was inferred using the neighbor-joining method (24). The optimal tree with the sum of branch lengths of 2.35304957 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches (6). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (27) and are in units representing the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The differences in the composition bias among sequences were considered in evolutionary comparisons. All positions containing gaps and missing data were eliminated from the data set (“complete deletion” option). There were a total of 1,661 positions in the final data set. Phylogenetic analyses were conducted in MEGA5 (28).
Fig 2.
Phylogenetic analysis based on the complete genome sequence of 104 taxa available in GenBank. The evolutionary history was inferred using the neighbor-joining method (24). The optimal tree with the sum of branch lengths of 2.50688796 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches (6). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (27) and are in units representing the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The differences in the composition bias among sequences were considered in evolutionary comparisons. All positions containing gaps and missing data were eliminated from the data set (“complete deletion” option). There were a total of 15,034 positions in the final data set. Phylogenetic analyses were conducted in MEGA5 (28).
The evolutionary distances between the complete genome of NDV-Peru/08 and those of strains of other class II genotypes (I, II, III/IV, V, VI, and VII) revealed that NDV-Peru/08 is significantly distant from viruses of all other NDV genotypes (Table 1). The distance between NDV-Peru/08 and the viruses of genotype VII, for example, is 0.1255, which is in the range of the distances observed between other genotypes (Table 1) and is markedly larger than the mean distance within genotype VII viruses (0.0406; standard error, 0.0009) (data not shown). Notably, the distances observed between NDV-Peru/08 and viruses of genotypes I and II (vaccine-like viruses) were 0.218 and 0.2432, respectively, and are the largest distances observed among all NDV genotypes (Table 1). Similar results were obtained when the F gene sequences were used to calculate the evolutionary distances (data not shown).
Table 1.
Estimates of evolutionary distances between NDV-Peru/08 and viruses representing other established NDV genotypes
| Genotype | No. of base substitutions/site in genotypea: |
|||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III/IV | V | VI | VII | IX | NDV-Peru/08 | |
| I | (0.0037) | (0.0036) | (0.0043) | (0.0047) | (0.0048) | (0.0030) | (0.0056) | |
| II | 0.1423 | (0.0047) | (0.0051) | (0.0051) | (0.0053) | (0.0041) | (0.0071) | |
| III/IV | 0.1464 | 0.1696 | (0.0041) | (0.0036) | (0.0051) | (0.0030) | (0.0059) | |
| V | 0.2038 | 0.2289 | 0.1641 | (0.0028) | (0.0035) | (0.0039) | (0.0041) | |
| VI | 0.1924 | 0.2226 | 0.1604 | 0.1576 | (0.0033) | (0.0036) | (0.0041) | |
| VII | 0.2042 | 0.2262 | 0.1715 | 0.1655 | 0.1371 | (0.0043) | (0.0036) | |
| IX | 0.1357 | 0.1576 | 0.1183 | 0.1772 | 0.1753 | 0.1864 | (0.0053) | |
| NDV-Peru/08 | 0.218 | 0.2432 | 0.1893 | 0.1804 | 0.1491 | 0.1255 | 0.2011 | |
The number of base substitutions per site is shown for NDV-Peru/08 and genotypes of class II viruses. All results are based on the pairwise analysis of 98 sequences. The numbers of sequences analyzed per group were as follows: I, n = 16; II, n = 18; III/IV, n = 6; V, n = 12; VI, n = 18; VII, n = 16; IX, n = 8; and poultry/Peru/08 (NDV-Peru/08), n = 1. Analyses were conducted using the maximum composite likelihood method in MEGA5. The codon positions included were the 1st + 2nd + 3rd + noncoding. All positions containing gaps, and missing data were eliminated from the data set (“complete deletion” option). There were a total of 15,034 positions in the final data set. Values in parentheses are standard errors, obtained by a bootstrap procedure (500 replicates).
A comparison of the nucleotide and amino acid sequences between NDV-Peru/08 and selected class II reference strains representing genotypes I, II, III, IV, V, VI, VII, and IX is presented in Table 2. Alignment of the complete genome sequences revealed that NDV-Peru/08 shares 89.9% and 89.6% nucleotide identity with NDV strains Fontana/72 (genotype VI) and ZJ1/2000 (genotype VII), respectively. Notably, the lowest nucleotide identity was observed between NDV-Peru/08 and the vaccine strains Ulster/67 (84.4%; genotype I) and LaSota/46 (82.5%; genotype II). Similar to the complete genome nucleotide sequence analysis, comparison of the nucleotide and amino acid sequences of each gene confirmed that NDV-Peru/08 shares the highest nucleotide and amino acid identity with the strains of genotypes VI and VII, while the lowest degree of identity was observed between NDV-Peru/08 and the vaccine-like strains of genotypes I and II (Table 2).
Table 2.
Nucleotide and amino acid comparison between the Newcastle disease virus isolate NDV-Peru/08 and viruses representing other genotypes within class IIa
| Protein | % identity for virus by genotypeb: |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I, Ulster/67 |
II, LaSota |
III, JS/7/05/Ch |
IV, Italien |
V, Anhinga/93 |
VI, Fontana/72 |
VII, ZJ1/2000 |
IX, FJ/1/85/Ch |
|||||||||
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| NP | 85 | 93 | 84 | 91 | 87 | 93 | 90 | 95 | 88 | 93 | 88 | 93 | 91 | 96 | 87 | 94 |
| P | 82 | 81 | 81 | 80 | 84 | 82 | 84 | 85 | 84 | 80 | 85 | 80 | 87 | 85 | 85 | 81 |
| M | 85 | 89 | 84 | 88 | 86 | 89 | 88 | 93 | 85 | 92 | 85 | 92 | 91 | 95 | 86 | 91 |
| F | 84 | 90 | 82 | 88 | 84 | 90 | 87 | 92 | 86 | 91 | 86 | 91 | 89 | 93 | 84 | 91 |
| HN | 85 | 89 | 83 | 86 | 84 | 87 | 86 | 89 | 85 | 89 | 86 | 89 | 90 | 93 | 84 | 88 |
| L | 87 | 94 | 85 | 92 | 87 | 94 | 88 | 95 | 88 | 95 | 88 | 95 | 91 | 96 | 87 | 94 |
| Complete genome | 84.8 | 82.5 | 85 | 86.8 | 85.9 | 89.9 | 89.6 | 84.8 | ||||||||
Genomic features of NDV-Peru/08.
The genome of NDV-Peru/08 is 15,192 nucleotides (nt) in length, it contains a 6-nt insertion in the 5′ noncoding region of the NP gene between positions 1738 and 1743 and a G+C content of 46.2%. A summary of the genomic features of NDV-Peru/08, including the gene start, gene end, and intergenic and coding regions is presented in Table 3.
Table 3.
Genomic features of Newcastle disease virus isolate NDV-Peru/08
| Protein | Gene start positions (nt) | 3′ UTR (length nt)a | Coding sequence positions (nt) | 5′ UTR (nt length) | Gene end positions (nt) | Length of: |
||
|---|---|---|---|---|---|---|---|---|
| Intergenic region (nt) | Gene (nt) | Protein (aa) | ||||||
| NP | 56–65 | 66 | 122–1591 | 217 | 1798–1808 | 1 | 1,469 | 489 |
| P | 1810–1819 | 73 | 1883–3080 | 180 | 3250–3260 | 1 | 1,197 | 395 |
| M | 3262–3271 | 34 | 3286–4390 | 112 | 4493–4502 | 1 | 1,073 | 364 |
| F | 4504–4513 | 46 | 4550–6211 | 84 | 6285–6285 | 31 | 1,661 | 553 |
| HN | 6327–6336 | 91 | 6418–8133 | 195 | 8318–8328 | 47 | 1,715 | 571 |
| L | 8376–8387 | 11 | 8387–15001 | 77 | 15069–15078 | 6,614 | 2,204 | |
UTR, untranscribed region.
Infection of chickens with NDV-Peru/08 results in characteristic velogenic viscerotropic Newcastle disease.
The clinicopathological characteristics and the virulence of NDV-Peru/08 were assessed in chickens. All NDV-Peru/08-inoculated birds presented severe prostration, had mucous diarrhea, and died or were euthanized by day 5 postinoculation (mean death time, 4.7 days) (Fig. 3). Gross lesions were initially observed by day 2 p.i. and consisted of conjunctivitis, multifocal necrosis, and hemorrhages in the intestine and cecal tonsils, as well as mottling and multifocal necrosis in the spleen. Additionally, atrophy of the thymus was observed by day 3 p.i. and was characterized by a marked reduction in size and by the presence of gelatinous edema and perithymic hemorrhages. Multifocal hemorrhages were also observed in the pharynx by day 4 p.i. Control group birds did not exhibit any clinical signs or pathological changes.
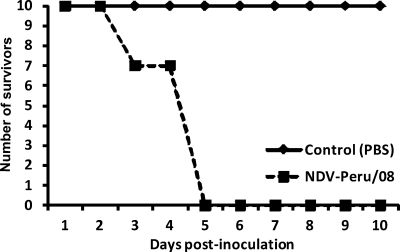
Fig 3.
Survival curve. Three birds of the NDV-Peru/08-inoculated group (group 2) died on day 3 postinoculation, and the remaining birds died on day 5 p.i. (mean death time, 4.7 days).
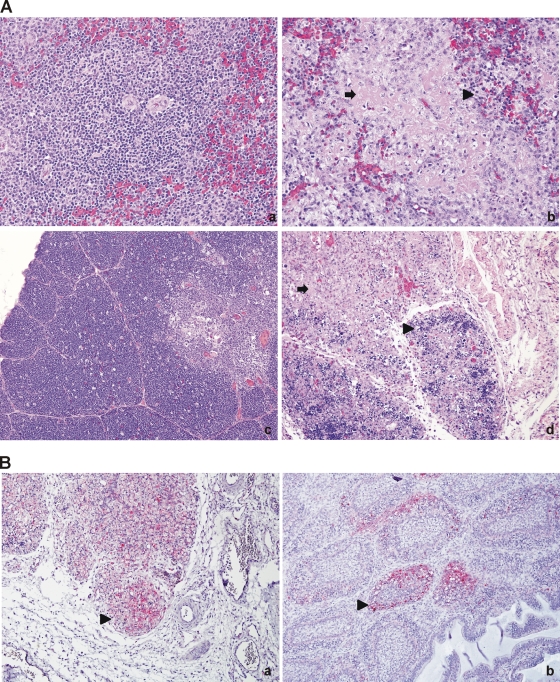
Typical histological changes associated with velogenic viscerotropic NDV infection, including lymphoid depletion and necrosis, accumulation of macrophages, necrotic debris, and scattered heterophils, were observed in multiple organs of NDV-Peru/08-inoculated birds (Fig. 4A, panels b and d; Table 4). Lesions were more severe as the disease progressed and peaked at day 4 p.i. A summary of the severity and distribution of the lesions is presented in Table 4. Control group birds did not present histological changes.
Fig 4.
Histological changes and distribution of Newcastle disease virus nucleoprotein in NDV-Peru/08-infected tissues. (A) Histological changes (panels b and d) consisted of lymphoid depletion and necrosis (arrows), accumulation of macrophages (arrowheads), and scattered heterophils. Panel a, spleen, PBS control, 40×; panel b, spleen, NDV-Peru/08, 40×; panel c, thymus, PBS control, 20×; panel d, thymus, NDV-Peru/08, hematoxylin and eosin staining. (B) Immunohistochemical staining for Newcastle disease virus nucleoprotein. Shown is positive staining in the thymus (panel a, 20×) and bursa of Fabricius (panel b, 20×). Positive cells consisted mainly of lymphocytes and macrophages (arrowheads).
Table 4.
Distribution and severity of lesions and distribution and intensity of viral antigen staining in tissues of NDV-Peru/08-inoculated birds
| Tissuea | Result on postinoculation dayb: |
||
|---|---|---|---|
| 2 | 3 | 4 | |
| Eyelid | |||
| HE | ++ | ++ | ++ |
| IHC | ++ | +++ | +++ |
| Spleen | |||
| HE | ++ | +++ | +++ |
| IHC | ++ | ++ | +++ |
| Thymus | |||
| HE | + | ++ | +++ |
| IHC | − | ++ | ++++ |
| Bursa | |||
| HE | + | + | +++ |
| IHC | + | ++ | ++ |
| Harderian gland | |||
| HE | − | + | − |
| IHC | − | + | +++ |
| Proventriculus | |||
| HE | + | ++ | − |
| IHC | − | ++++ | +++ |
| Pancreas | |||
| HE | + | − | ++ |
| IHC | − | − | ++ |
| Small intestine | |||
| HE | − | − | + |
| IHC | + | − | ++ |
| Mekel's diverticulum | |||
| HE | + | − | ++ |
| IHC | − | − | ++ |
| Cecal tonsils | |||
| HE | + | ++ | +++ |
| IHC | +++ | +++ | ++++ |
| Large intestine | |||
| HE | − | + | + |
| IHC | − | ++ | ++ |
| Air sacs | |||
| HE | − | − | − |
| IHC | − | − | + |
| Trachea | |||
| HE | − | − | − |
| IHC | − | + | − |
| Lung | |||
| HE | − | − | + (BALT) |
| IHC | − | + (BALT) | ++ (BALT) |
| Pharynx | |||
| HE | + | ++ | ++ |
| IHC | − | + | |
| Brain | |||
| HE | − | − | − |
| IHC | − | − | + (cerebellum) |
| Liver | |||
| HE | − | − | + (MALT) |
| IHC | − | − | − |
| Kidney | |||
| HE | − | − | − |
| IHC | − | − | + |
| Comb | |||
| HE | − | − | − |
| IHC | − | − | ++ |
| Femur | |||
| HE | − | − | + |
| IHC | − | − | + |
HE, hematoxylin and eosin staining; IHC, immunohistochemistry staining for NDV nucleoprotein.
BALT, bronchus-associated lymphoid tissue; MALT, mucosa-associated lymphoid tissue. For HE, the results are scored by tissue as follows: (i) spleen, +, moderate hyperplasia, ++, lymphocytic depletion, +++, moderate (<50%) lymphocyte depletion, histiocytic accumulation, and multifocal necrosis, and ++++, severe (>50%) lymphocytic depletion, histiocytosis, and necrosis; (ii) thymus, cecal tonsil, gut-associated lymphoid tissue (GALT), and bursa, +, mild lymphocytic depletion, ++, moderate (<50%) lymphocytic depletion with necrosis and histiocytosis, and +++, severe (>50%) lymphocytic depletion, necrosis, and histiocytosis; (iii) bone marrow, +, mild (<20%) bone marrow necrosis, ++, moderate (20 to 50%) bone marrow necrosis, and +++, severe (>50%) bone marrow necrosis; (iv) pancreas, +, mild (<3 areas) vacuolation and degeneration, and ++, moderate (>3 areas) vacuolation and degeneration; and (v) brain, +, vascular reactivity, ++, vascular reactivity and perivascular cuffing, and +++, vascular reactivity, perivascular cuffing, and gliosis. For IHC staining, the results are scored as follows: −, no IHC signal detected; +, rare cells in the section are positive on IHC; ++, positive cells seen in <50% of all high-magnification fields (HMF); +++, positive signal seen in 50 to 75% of HMF; and ++++, abundant positive signal in more than 75% of HMF.
Immunohistochemical staining for NDV nucleoprotein (NP) revealed a widespread distribution of the virus (Table 4), with antigen load and distribution peaking at 4 dpi. The strongest staining intensity was observed in the eyelids, lymphoid organs, and mucosa-associated-lymphoid tissue (MALT) aggregates of various organs (Table 4; Fig. 4B, panels a and b), with positive cells consisting mainly of lymphocytes and macrophages.
All birds inoculated with NDV-Peru/08 shed the virus in oral and cloacal swabs. The virus was isolated from samples collected on days 2, 3, and 5 p.i., with virus titers ranging from 103.7 to 105.9 EID50/0.1 ml and from 102.9 to 105.4 EID50/0.1 ml in oral and cloacal secretions, respectively.
Inactivated NDV-LaSota/46 vaccine confers protection against NDV-Peru/08-induced disease and mortality.
The efficacy of an inactivated NDV-LaSota/46 vaccine in conferring protection against NDV-Peru/08 challenge was assessed in chickens. All NDV-LaSota/46-vaccinated birds seroconverted (HI titers between 64 and 128, geometric mean titer [GMT] of 105.95) and were fully protected from clinical disease and mortality induced by NDV-Peru/08 infection, whereas all birds from the sham-vaccine group presented typical clinical signs of velogenic viscerotropic NDV strains, including severe prostration and diarrhea, and died by day 5 p.i.
DISCUSSION
Virulent Newcastle disease virus (vNDV) is endemic in many countries of North, Central, and South America, and outbreaks of ND are frequently reported to the World Organization for Animal Health (OIE). In June and July of 2008, simultaneous outbreaks of ND were confirmed in domestic poultry in Peru (provinces of Arequipa and Cusco). The high morbidity and mortality rates (75.6%) of the 2008 outbreak and the clinicopathological characteristics of NDV-Peru/08 resemble the clinical presentation described for the highly virulent viruses of genotypes V and VII that circulate in Central America, Africa, Asia, and the Middle East (26). Although the source of the 2008 outbreak in Peru is still unknown, field epidemiological investigations reported to the OIE suggest that the virus was likely introduced through live birds that were added to the affected flocks (19). However, a complete epidemiological investigation, including sampling of domestic and wild bird species, is needed to define the actual source of the outbreak and origin of the virus.
Recent phylogenetic studies have shown that NDV is continuously evolving, with viruses of different genotypes undergoing simultaneous changes in distinct geographic locations (4, 5, 11, 22, 25, 33, 34). The results presented here indicate that NDV-Peru/08 has evolved and significantly diverged from viruses representing all known NDV genotypes. Classification of NDV-Peru/08 into one of the existing NDV genotypes or into a new genotype, however, is still uncertain, and isolation of additional viruses with similar genetic and phylogenetic properties is needed in order to classify this isolate.
Historically, NDV isolates have been classified as lentogenic, mesogenic, or velogenic based on to the clinicopathological outcome of the infection in chickens (3). Clinicopathological characterization of NDV-Peru/08 revealed that this virus behaves as a typical velogenic viscerotropic strain. The mortality rate (100%) and mean death time (4.7 days) observed in NDV-Peru/08-inoculated birds are characteristic of highly virulent strains of genotypes V, VI, and VII (21, 26, 31). Histological examination and immunohistochemical staining of tissue samples collected from multiple organs confirmed the tropism of NDV-Peru/08 for lymphoid tissues associated with the gastrointestinal tract and, to a lesser extent, with the respiratory system. A similar tropism is seen in infections with the highly virulent genotype VII viruses (26).
The data presented here provide evidence that a genetically distinct virus, most closely related to but different from isolates of genotypes VI and VII caused the outbreak of ND in Peru. Notably, the evolutionary distances between NDV-Peru/08 and vaccine-like viruses of genotypes I and II are the largest distances observed between all genotypes. Given the genetic differences observed between NDV-Peru/08 and vaccine-like viruses (Table 1 and 2), vaccination studies were performed with chickens. Results of these experiments demonstrated that, under biosafety level 3 conditions and using SPF birds, a traditional inactivated NDV vaccine strain from genotype II (NDV-LaSota/46) fully protected chickens from NDV-Peru/08-induced clinical disease and mortality. These observations suggest that the genetic diversity of NDV-Peru/08 is not reflected in antigenic changes that significantly affect the immune response elicited by the inactivated NDV-LaSota/46 vaccine. However, whether these findings will be observed under field conditions and with vaccine strains from other NDV genotypes remains to be determined.
In summary, complete genome- and protein-level analysis of the evolutionary distances between NDV-Peru/08 and strains of established NDV genotypes demonstrated that this isolate is distinct from viruses of typical NDV genotypes. Given the lack of epidemiological data from the 2008 outbreak and from other NDV isolates circulating in South America, it is unclear whether similar viruses are still circulating in that continent, or whether that outbreak was a unique episode caused by this isolate. Continuous characterization of novel NDV isolates that occasionally emerge and cause outbreaks or of those that frequently circulate worldwide are important to improve the current understanding of NDV epidemiology and evolution and for the development of improved control and diagnostic strategies.
ACKNOWLEDGMENTS
We thank Dawn Williams-Coplin and Tim Olivier for technical assistance, Roger Brock for help with animal experiments, and the SEPRL sequencing facility personnel for nucleotide sequencing. We also acknowledge the generous support of CEVA Biomune for the vaccination studies and the National Direction of Animal Health of Peru (SENASA) for invaluable collaboration and for sharing information on the outbreak.
This work was supported by USDA funding CRIS 6612-32000-049.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Aldous EW, Mynn JK, Banks J, Alexander DJ. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32: 239–256 [DOI] [PubMed] [Google Scholar]
- 2. Alexander DJ, Senne DA. 2008. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p 75–98 In Saif M, et al. (ed), Diseases of poultry, 12th ed. Blackwell Publishing, Ames, IA [Google Scholar]
- 3. Alexander DJ, Swayne DE. 1998. Newcastle disease virus and other avian paramyxoviruses, p 156–163 In Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM. (ed), A laboratory manual for the isolation and identification of avian pathogens, vol 4 American Association of Avian Pathologists, Kennett Square, PA [Google Scholar]
- 4. Czegledi A, et al. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120: 36–48 [DOI] [PubMed] [Google Scholar]
- 5. Ding Z, et al. 2010. Genetic analysis of avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from swine populations in China related to commonly utilized commercial vaccine strains. Virus Genes 41: 369–376 [DOI] [PubMed] [Google Scholar]
- 6. Felsenstein J. 1992. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet. Res. 60: 209–220 [DOI] [PubMed] [Google Scholar]
- 7. Kim LM, King DJ, Suarez DL, Wong CW, Afonso CL. 2007. Characterization of class I Newcastle disease virus isolates from Hong Kong live bird markets and detection using real-time reverse transcription-PCR. J. Clin. Microbiol. 45: 1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim LM, Suarez DL, Afonso CL. 2008. Detection of a broad range of class I and II Newcastle disease viruses using multiplex real-time reverse transcription polymerase chain reaction assay. J. Vet. Diagn. Invest. 20: 414–425 [DOI] [PubMed] [Google Scholar]
- 9. Kinde H, et al. 2005. The isolation of exotic Newcastle disease (END) virus from nonpoultry avian species associated with the epidemic of END in chickens in southern California: 2002–2003. Avian Dis. 49: 195–198 [DOI] [PubMed] [Google Scholar]
- 10. Lancaster JE, Alexander DJ. 1975. Newcastle Disease virus and spread, p 1–79 Canada Department of Agriculture, monograph no. 11 Canada Department of Agriculture, Ottawa, Canada [Google Scholar]
- 11. Linde AM, et al. 2010. Complete genome characterisation of a Newcastle disease virus isolated during an outbreak in Sweden in 1997. Virus Genes 41: 165–173 [DOI] [PubMed] [Google Scholar]
- 12. Liu XF, Wan HQ, Ni XX, Wu YT, Liu WB. 2003. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985–2001. Arch. Virol. 148: 1387–1403 [DOI] [PubMed] [Google Scholar]
- 13. Martin DP. 2009. Recombination detection and analysis using RDP3. Methods Mol. Biol. 537: 185–205 [DOI] [PubMed] [Google Scholar]
- 14. Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10: 26–35 [DOI] [PubMed] [Google Scholar]
- 15. Miller PJ, Kim LM, Ip HS, Afonso CL. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391: 64–72 [DOI] [PubMed] [Google Scholar]
- 16. Miller PJ, King DJ, Afonso CL, Suarez DL. 2007. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine 25: 7238–7246. [DOI] [PubMed] [Google Scholar]
- 17.OIE 2009. Manual of diagnostic tests and vaccines for terrestrial animals: mammals, birds and bees, 5th ed, vol 1, part 2, chapter 2.3.14, p 576–589 Biological Standards Commission, World Organization for Animal Health, Paris, France [Google Scholar]
- 18.OIE 2011. World Animal Health Information Database (WAHID) Interface. http://web.oie.int/wahis/public.php?page=home&WAHIDPHPSESSID=4d95e8777ff8b8a615b31617ac4c7835
- 19.OIE 2008. World Animal Health Information Database (WAHID) Interface. Event summary: Newcastle disease, Peru. World Organization for Animal Health, Paris, France: http://web.oie.int/wahis/public.php?page=event_summary&reportid=7326 [Google Scholar]
- 20. Perozo F, Afonso RMCL, Fernandez R, Rojo F. 2011. Genotype VII velogenic viscerotropic Venezuelan Newcastle disease virus isolate: live (AvinewR) and killed (Gallimune NDR) vaccination, abstr 169P, p 49 Abstr. 2011 Int. Poultry Scientific Forum, Atlanta, GA, 24 to 25 January 2011 [Google Scholar]
- 21. Perozo F, Merino R, Afonso CL, Villegas P, Calderon N. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52: 472–479 [DOI] [PubMed] [Google Scholar]
- 22. Qiu X, et al. 2011. Entire genome sequence analysis of genotype IX Newcastle disease viruses reveals their early-genotype phylogenetic position and recent-genotype genome size. Virol. J. 8: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rue CA, et al. 2010. Evolutionary changes affecting rapid identification of 2008 Newcastle disease viruses isolated from double-crested cormorants. J. Clin. Microbiol. 48: 2440–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- 25. Seal BS. 2004. Nucleotide and predicted amino acid sequence analysis of the fusion protein and hemagglutinin-neuraminidase protein genes among Newcastle disease virus isolates. Phylogenetic relationships among the Paramyxovirinae based on attachment glycoprotein sequences. Funct. Integr. Genomics 4: 246–257 [DOI] [PubMed] [Google Scholar]
- 26. Susta L, Miller PJ, Afonso CL, Brown CC. 2010. Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet. Pathol. 48: 349–360 [DOI] [PubMed] [Google Scholar]
- 27. Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101: 11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Utterback WW, Schwartz JH. 1973. Epizootiology of velogenic viscerotropic Newcastle disease in southern California, 1971–1973. J. Am. Vet. Med. Assoc. 163: 1080–1088 [PubMed] [Google Scholar]
- 31. Wakamatsu N, King DJ, Kapczynski DR, Seal BS, Brown CC. 2006. Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002–2003. Vet. Pathol. 43: 925–933 [DOI] [PubMed] [Google Scholar]
- 32. Wang Z, et al. 2006. Genotyping of Newcastle disease viruses isolated from 2002 to 2004 in China. Ann. N. Y. Acad. Sci. 1081: 228–239 [DOI] [PubMed] [Google Scholar]
- 33. Wu S, et al. 9 November 2010, posting date Genetic diversity of Newcastle disease viruses isolated from domestic poultry species in Eastern China during 2005–2008. Arch. Virol. [Epub ahead of print.] doi:10.1007/s00705-010-0851-5 [DOI] [PubMed] [Google Scholar]
- 34. Yu L, Wang Z, Jiang Y, Chang L, Kwang J. 2001. Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39: 3512–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]