Abstract
Quantitative PCR (qPCR) is more sensitive than microscopy for detecting Pneumocystis jirovecii in bronchoalveolar lavage (BAL) fluid. We therefore developed a qPCR assay and compared the results with those of a routine immunofluorescence assay (IFA) and clinical data. The assay included automated DNA extraction, amplification of the mitochondrial large-subunit rRNA gene and an internal control, and quantification of copy numbers with the help of a plasmid clone. We studied 353 consecutive BAL fluids obtained for investigation of unexplained fever and/or pneumonia in 287 immunocompromised patients. No qPCR inhibition was observed. Seventeen (5%) samples were both IFA and qPCR positive, 63 (18%) were IFA negative and qPCR positive, and 273 (77%) were both IFA and qPCR negative. The copy number was significantly higher for IFA-positive/qPCR-positive samples than for IFA-negative/qPCR-positive samples (4.2 ± 1.2 versus 1.1 ± 1.1 log10 copies/μl; P < 10−4). With IFA as the standard, the qPCR assay sensitivity was 100% for ≥2.6 log10 copies/μl and the specificity was 100% for ≥4 log10 copies/μl. Since qPCR results were not available at the time of decision-making, these findings did not trigger cotrimoxazole therapy. Patients with systemic inflammatory diseases and IFA-negative/qPCR-positive BAL fluid had a worse 1-year survival rate than those with IFA-negative/qPCR-negative results (P < 10−3), in contrast with solid-organ transplant recipients (P = 0.88) and patients with hematological malignancy (P = 0.26). Quantifying P. jirovecii DNA in BAL fluids independently of IFA positivity should be incorporated into the investigation of pneumonia in immunocompromised patients. The relevant threshold remains to be determined and may vary according to the underlying disease.
INTRODUCTION
The laboratory diagnosis of Pneumocystis pneumonia (PcP), caused by the opportunistic fungal pathogen Pneumocystis jirovecii, still relies on tinctorial and/or immunofluorescent staining of bronchoalveolar lavage (BAL) fluid samples (27). Nucleic acid amplification tests can overcome the difficulties of microscopic examination (27). Several previously reported PCR studies used nested PCR with a final endpoint reading (16, 20). This format is not intended to give quantitative results, and the test is prone to contamination with previously amplified products, leading to false-positive results. In contrast, real-time quantitative PCR (qPCR) dramatically reduces the risk of false-positive results because of the closed-tube nature of the amplification process (6), and the resulting data are quantitative if guidelines for interpretation of qPCR results are followed (7).
Several qPCR assays have already been described and have shown that qPCR is more sensitive than microscopy (1–3, 5, 10–12, 14, 15, 17, 18, 22, 26). This raises the issue of the clinical significance of P. jirovecii DNA detection for explaining the symptoms observed (24, 25). Quantitative PCR data can also address the issue of the correlation between tissue burden and outcome (16). In particular, we questioned the use of microscopically detectable P. jirovecii cysts as a threshold for therapeutic decision-making, especially if the same criteria are used in decision making for extended to all patients whatever their risk factors for developing PcP. Therefore, we developed a qPCR assay and tested all the bronchoalveolar lavage (BAL) fluids received by our laboratory without selection of the patients from whom samples were obtained. The qPCR results were compared with immunofluorescence assay (IFA) results and assessed in light of medical information, including underlying diseases and outcome.
MATERIALS AND METHODS
BAL fluid sampling and conventional diagnosis of PcP.
This was a retrospective single-center, hospital-based cohort study of all patients who underwent BAL to investigate the cause of pneumonia and/or unexplained fever from 1 January 2006 to 31 December 2007. The study was approved by the Comité de Protection des Personnes, Ile de France, France. Since the qPCR assay was performed blind on stored samples, the results did not alter therapeutic management of the patients.
BAL was performed with three washes (washes 1 to 3) with 50 ml of sterile saline solution. Aliquots of the washes were dispatched to the microbiology and pathology laboratories. Upon arrival in the laboratory, at least 10 slides were prepared, each using 200 μl of wash 2. The routine procedure included May-Grünwald Giemsa (MGG) staining (RAL-555; RAL Reagents, Martillac, France), methenamine silver staining (MSS), and an indirect immunofluorescence assay (IFA) (Monofluo kit, P. jirovecii; Bio-Rad, Marnes la Coquette, France). Information recorded included the total volume of BAL fluid recovered, the lung X-ray or CT (computed tomography) scan conclusion, demographics, underlying conditions at the time of the BAL procedure, outcome at 1 year, and anti-Pneumocystis prophylaxis at the time of BAL procedure, as well as anti-Pneumocystis therapy prescribed after results of the BAL were known.
DNA extraction from BAL fluids.
From the portion of BAL fluid wash 2 remaining after microscopy and microbiological cultures, 1.5 ml was centrifuged at 10,000 × g for 10 min. Supernatant was aspirated, and the cell pellet was stored in 200 μl of the washing solution at −40°C until further processing. After thawing, DNA was extracted using a QIAamp DNA minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions, except that total DNA was eluted from the spin columns with 50 μl of elution buffer in order to increase the DNA concentration. Five μl of this DNA extract was used for qPCRs.
Pneumocystis jirovecii qPCR assay.
Two primers (PNC-LSU3 [forward], 5′-TGGTAAGTAGTGAAATACAAATCGG-3′; PNC-LSU4 [reverse], 5′-ACTCCCTCGAGATATTCAGTGC-3′) and a pair of hybridization probes (PNC-LSU5 [5′ LCRed640 labeled, 3′ Ph labeled], 5′-TTCGCAGAAAACCAGCTATATCCTAGT-3′; PNC-LSU6 [3′FITC labeled], 5′-AGAGGAATAAACAATTTGCCAAAACAA-3′) were selected to amplify and to detect a conserved 152-bp region of the mitochondrial large subunit rRNA gene (LSU) of P. jirovecii using real-time PCR (GenBank accession number AJ608260). The amplification was carried out in a LightCycler 1.5 instrument (Roche Diagnostics, Meylan, France). PCR was set up in a final volume of 20 μl with a Faststart DNA master hybridization probes kit (Roche Diagnostics, Meylan, France), 4 mM MgCl2, each primer and probe (Sigma, Paris, France) at a concentration of 0.5 μM and 0.25 μM, respectively, and 0.25 μl of uracil-DNA-glycosylase (UDG) (Biolabs, Courtaboeuf, France). The reaction mixture was initially incubated for 1 min at 50°C, followed by a 8-min step at 95°C. Amplification was performed for 50 cycles of denaturation (95°C for 10 s; ramp rate, 20°C/s), annealing (60°C for 10 s; ramp rate, 20°C/s), and extension (72°C for 15 s; ramp rate, 20°C/s). Results were considered positive when a significant fluorescent signal above the baseline was detected, as determined by the second-derivative algorithm method, and were expressed as quantification cycle (Cq) values. During each run, a P. jirovecii DNA positive control and the elution buffer for DNA extraction as a negative control were used. A residual amplification inhibitory effect in the DNA extract was tested by using a universal internal standard as previously described (8) and as adapted for the LightCycler 1.5 instrument.
A 299-bp fragment of the LSU gene including the targeted PCR fragment was cloned into a pUC57 plasmid (Ecole de Biologie Industrielle, Cergy-Pontoise, France), its concentration was measured with a fluorometer, and the corresponding copy number was calculated. A 10-fold serial dilution series of this plasmid clone ranging from 1 to 109 copies/ml was used to construct the standard curve. Cq values in each dilution were measured in duplicate in three independent runs and were plotted against the logarithm of their initial template copy numbers. Results were expressed as number of copies/μl for each DNA extract from a positive sample determined against the standard curve.
Statistical methods.
Means and standard deviations (SD) are shown when distributions were confirmed as normal or after log10 transformation when needed. The comparisons used Fisher's exact test for categorical variables and the t test or one-way analysis of variance (ANOVA) for continuous variables using Prism 4.0 (GraphPad Software). Overall survival at 12 months was calculated using Kaplan Meier analysis from the date of the first BAL to the 1-year follow-up or death from any cause. When multiple BAL fluid samples with different qPCR results were available for one patient, the first qPCR-positive sample was used for the analysis.
RESULTS
Validation of the qPCR assay.
Our qPCR assay could systematically detect the dilution containing 1 copy of plasmid per μl, leading to a detection sensitivity of at least 5 copies per PCR. Quantification was linear over an order of magnitude of 10, and the standard curve was generated with a high coefficient of determination (R2 = 0.999). The mean overall coefficient of variation (CV) of the Cq values was 1.5% (range, 0.5 to 3.5) (Table 1). For the P. jirovecii DNA positive control, the mean Cq on 20 runs was 32.41 ± 0.20 (CV = 0.62%).
Table 1.
Overview of data obtained from three runs of standard curves with serial dilutions of a plasmid clone containing the target for P. jirovecii DNA detectiona
| Plasmid concn (copies/μl) | Quantification cycle |
||||
|---|---|---|---|---|---|
| Mean | Minimum | Maximum | SD | CV (%) | |
| 109 | 6.88 | 6.53 | 7.09 | 0.24 | 3.5 |
| 108 | 10.41 | 10.22 | 10.51 | 0.11 | 1.0 |
| 107 | 14.16 | 13.74 | 14.47 | 0.27 | 1.9 |
| 106 | 17.72 | 17.26 | 17.96 | 0.32 | 1.8 |
| 105 | 21.03 | 20.85 | 21.14 | 0.12 | 0.6 |
| 104 | 24.54 | 24.19 | 24.73 | 0.24 | 1.0 |
| 103 | 28.21 | 27.72 | 28.50 | 0.32 | 1.1 |
| 102 | 31.97 | 31.32 | 32.46 | 0.48 | 1.5 |
| 10 | 34.91 | 34.02 | 35.75 | 0.68 | 1.9 |
| 1 | 39.32 | 38.98 | 39.57 | 0.20 | 0.5 |
Each dilution was analyzed in duplicate. The coefficients of variation (CV) show good reproducibility.
Analysis of BAL specimens.
A total of 378 consecutive BAL specimens were collected in our laboratory during the study period, of which 353 had sufficient volume left for qPCR analysis after routine testing. No PCR inhibition was detected, and a correct amplification of the internal control was observed in each sample (mean Cq of the internal control = 37.1 ± 1.1), allowing analysis of the qPCR results. Of these 353 samples, 10 (2.8%), 16 (4.5%), and 17 (4.8%) were microscopically positive for P. jirovecii using MSS, MGG, and IFA, respectively. All the MSS-positive samples were MGG positive, and all the MGG-positive samples were IFA positive. The 17 IFA-positive samples were all qPCR positive.
Overall, three groups of samples were thus available for analysis: IFA-positive/qPCR-positive samples (n = 17; 4.8%), IFA-negative/qPCR-positive samples (n = 63; 17.8%), and IFA-negative/qPCR-negative samples (n = 273; 77.3%). Since the recovered BAL volume could interfere with qPCR quantification, we checked that it did not significantly differ between the three groups (Table 2) (P > 0.05). The copy number was markedly (more than 3 log in magnitude) higher for IFA-positive/qPCR-positive samples than for IFA-negative/qPCR-positive samples (4.2 ± 1.2 versus 1.1 ± 1.1 log10 copies/μl; P < 10−4) (Table 2). With IFA as the standard, the qPCR assay sensitivity was found to be 100% for samples containing ≥2.6 log10 copies/μl, and the specificity was 100% for those with ≥4.0 log10 copies/μl. Among the 13 samples containing ≥2.6 log10 and <4.0 log10 copies/μl, 8 were IFA negative/qPCR positive. These 8 samples were from 8 patients who were not given cotrimoxazole, and 5 of them died at day 6 (solid cancer), day 120 (HIV infection), day 197 (acute myeloid leukemia), day 289 (acute lymphoid leukemia), and day 353 (HIV infection) from the day of the first BAL.
Table 2.
Main characteristics of the 353 BAL fluid samples used in this study according to the qPCR results
| Assay results (n [%]) | Mean (SD) |
No. (%) from patients with: |
||
|---|---|---|---|---|
| Recovered vol | Log10 copy no. | Diffuse radiological patterns | Anti-PcP prophylaxis | |
| IFA positive/qPCR positive (17 [4.8]) | 78.5 (18.2) | 4.2 (1.2) | 15 (88.2) | 1 (5.9) |
| IFA negative/qPCR positive (63 [17.8]) | 61.8 (22.2) | 1.1 (1.1) | 27 (42.9) | 2 (3) |
| IFA negative/qPCR negative (273 [77.3]) | 66.2 (25.0) | 94 (34.4) | 51 (18.7) | |
| P | 0.058 | <10−4 | <10−4 | 0.005 |
The presence of diffuse pulmonary lesions with no localized or focal lesions on chest X-ray or CT scan imaging was unequally distributed among the three groups (Table 2). However, although there were more diffuse lesions in the IFA-negative/qPCR-positive group than in the IFA-negative/qPCR-negative one, the difference was not statistically significant (P = 0.24). In contrast, the difference in anti-PcP prophylaxis at the time of the BAL was highly significant, with almost no prophylaxis in patients with qPCR-positive samples (Table 2).
Analysis according to underlying diseases.
The main underlying risk factors and diseases were divided into 6 groups. When several risk factors were present, the samples were classified in the category of the highest-risk factor in the following decreasing order: HIV positivity, acute leukemia, chronic lymphoproliferative disorder, solid-organ transplantation, systemic inflammatory diseases, solid tumor, and other underlying disease.
For the 46 samples from HIV-positive patients, both the CD4+ T cell count and anti-PcP prophylaxis predicted most of the results. Among the 16 samples from HIV-positive patients with a CD4+ T cell count of ≥200 cells/μl, only one (6.2%) was qPCR positive (Table 3). Of the 19 IFA-negative/qPCR-negative samples recovered from patients with CD4+ T cell counts of <200 cells/μl, 15 (78.9%) were from patients undergoing anti-PcP prophylaxis.
Table 3.
Detailed characteristics of the 353 BAL fluids studied, presented according to IFA and qPCR results
| Patient categorya | No. (%) of BAL fluid samples | No. of patients (%) | IFA-positive/qPCR-positive samples |
IFA-negative/qPCR-positive samples |
IFA-negative/qPCR-negative samples |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (%) | Mean log copy no. (SD) | No. from patients with: |
Total (%) | Mean log copy no. (SD) | No. from patients with: |
Total (%) | No. from patients with: |
||||||
| Lesionsb | PcP prophylaxis | Lesionsb | PcP prophylaxis | Lesionsb | PcP prophylaxis | ||||||||
| Total | 353 | 287 | 17 | 15 | 1 | 63 | 27 | 2 | 273 | 94 | 51 | ||
| HIV+ patients | 46 (13) | 39 (14) | 8 (47) | 5.5 (0.9) | 8 (53) | 0 | 4 (6) | 2.1 (1.2) | 3 (11) | 0 | 34 (12) | 15 (16) | 19 (37) |
| ≥200 CD4+ T cells/μl | 16/46 | 13/39 | 0/8 | 0 | 1/4 | 0 | 0 | 15/34 | 8/15 | 4/19 | |||
| <200 CD4+ T cells/μl | 30/46 | 26/39 | 8/8 | 8/8 | 0 | 3/4 | 3/3 | 0 | 19/34 | 7/15 | 15/19 | ||
| Hematology patients | 147 (42) | 111 (39) | 6 (35) | 4.2 (0.8) | 4 (27) | 0 | 24 (38) | 1.3 (0.9) | 8 (30) | 2 (100) | 117 (43) | 36 (38) | 22 (43) |
| Acute leukemia | 73/147 | 49/111 | 3/6 | 3/4 | 0 | 15/24 | 4/8 | 0 | 55/117 | 18/36 | 9/22 | ||
| CLD | 74/147 | 62/111 | 3/6 | 1/4 | 0 | 9/24 | 4/8 | 2/2 | 62/117 | 18/36 | 13/22 | ||
| SOT recipients | 57 (16) | 47 (16) | 2 (12) | 4.6 (0.7) | 2 (13) | 1 | 10 (15.9) | 1.4 (1.0) | 3 (11) | 0 | 45 (16) | 17 (18) | 6 (12) |
| Heart | 11/57 | 11/47 | 0/2 | 0 | 0 | 4/10 | 0 | 0 | 7/45 | 2/17 | 2/6 | ||
| Liver | 17/57 | 13/47 | 2/2 | 2 | 1 | 1/10 | 1/3 | 0 | 14/45 | 5/17 | 2/6 | ||
| Kidney | 29/57 | 23/47 | 0/2 | 0 | 0 | 5/10 | 2/3 | 0 | 24/45 | 10/17 | 2/6 | ||
| Patients with SID | 53 (15) | 47 (16) | 1 (6) | 3.5 | 1 (7) | 0 | 16 (25.4) | 0.7 (1.0) | 6 (22) | 0 | 36 | 10 (11) | 3 (6) |
| Patients with solid tumors | 18 (5) | 16 (6) | 0 | 0 | 0 | 0 | 7 (11.1) | 1.3 (1.4) | 6 (22) | 0 | 11 | 4 (4) | 1 (2) |
| Others | 32 (9) | 27 (9) | 0 | 0 | 0 | 0 | 2 (3) | 0.2 (0.1) | 1 (4) | 0 | 30 | 12 (13) | 0 |
CLD, chronic lymphoproliferative disorders; SOT, solid-organ transplant; SID, systemic inflammatory diseases.
Diffuse radiological lesions.
Among the 147 samples from the patients with hematological malignancies, only 6 (4.1%) were IFA positive/qPCR positive, whereas 24 (16.3%) were IFA negative/qPCR positive (Table 3). Similarly, only 4% (2/57) and 2% (1/53) of samples were IFA positive/qPCR positive, whereas 18% (10/57) and 30% (16/53) were IFA-positive/qPCR-positive among those from solid-organ transplant (SOT) recipients and patients with systemic inflammatory diseases (7 with vasculitis, 14 with inflammatory rheumatisms, 6 with glomerulonephritis, and 20 with other diseases), respectively (Table 3). A qPCR-positive result was also frequent in samples from solid-cancer patients (7/18, 39%). The two IFA-negative/qPCR-positive samples from the group with none of the above risk factors were from a patient with influenza virus infection and type 2 diabetes and from a patient with chronic obstructive pulmonary disease.
Outcome analysis.
Cotrimoxazole at a curative dose was given only to the patients with IFA-positive results and to 6 other patients based on clinical suspicion [3/63 (5%) in the IFA-negative/qPCR-positive group and 3/273 (1%) in the IFA-negative/qPCR-negative group].
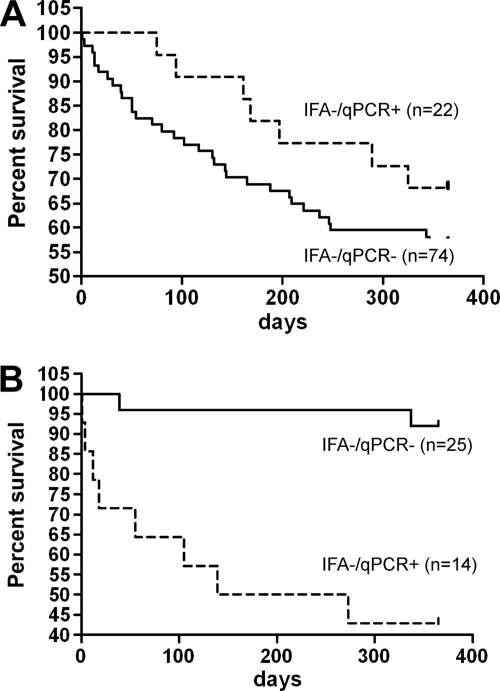
There was no significant difference in survival at 1 year between IFA-negative/qPCR-positive (n = 22) and IFA-negative/qPCR-negative (n = 74) patients with hematological diseases (P = 0.26) (Fig. 1A). Similar results were observed for the SOT recipients, with no significant difference (P = 0.88) between IFA-negative/qPCR-positive (n = 8) and IFA-negative/qPCR-negative (n = 35) patients (data not shown). In contrast, 1-year survival was significantly worse (P < 10−3) (Fig. 1B) for IFA-negative/qPCR-positive (n = 14) than for IFA-negative/qPCR-negative patients (n = 25) with systemic inflammatory diseases. Of note, the mean age (in years, ± SD) was not statistically different between the IFA-negative/qPCR-positive and the IFA-negative/qPCR-negative patients (63 ± 10 versus 58 ± 13; P = 0.22).
Fig 1.
One-year survival of patients with hematological malignancies (A) and patients receiving high-dose steroid therapy (B), presented according to qPCR results (patients with microscopy-positive BAL samples who were therefore treated with full-dose cotrimoxazole were excluded). (A) No statistically significant difference was noted between the qPCR-positive and qPCR-negative patients (P = 0.26). (B) A statistically significant difference was observed between the qPCR-positive and qPCR-negative patients (P < 10−3).
Additional BAL samples (n = 66) were obtained from 52 patients (mean [range] = 2 [2 to 6] samples/patient) with a mean interval of 43.5 days (range, 1 to 652 days) between 2 BAL. Thirty-six IFA-negative/qPCR-negative patients remained IFA negative/qPCR negative. Five qPCR-positive patients remained qPCR positive over an interval of <1 month, with a decrease of the fungal load when given cotrimoxazole (3 patients). Three patients who were previously IFA positive/qPCR positive became IFA negative/qPCR negative >2 months after cotrimoxazole therapy. Six patients who were IFA negative/qPCR positive became IFA negative/qPCR negative >2 months later without any known specific therapy. Two IFA-negative/qPCR-negative patients became IFA positive/qPCR positive within 2 and 12 months, respectively.
DISCUSSION
The qPCR assay used here proved to be a reliable tool for quantifying P. jirovecii DNA in BAL fluid samples. We were careful to avoid false-positive samples and unreliable quantification by checking the efficiency of the reaction using an internal control. Under these conditions, all immunofluorescence-positive BAL fluid samples (17/353; 4.8%) were found to be qPCR positive, and 17.8% (63/353) were qPCR positive only.
Our results confirm the higher sensitivity of qPCR over microscopy, as already reported by many authors using qPCR assays. However, there are marked differences among studies, which prevents direct comparison. For instance, the target chosen for amplification was either a multicopy gene [major surface glycoprotein (MSG) gene (10, 11, 17, 18) or ribosomal RNA gene (1, 3, 12, 14, 22)] or a single-copy gene (3, 5, 15, 18, 26). The use of UNG for preventing cross-contamination has been reported only in the present study. An internal control has been effective (1, 5, 11, 14, 17), omitted (2, 3, 12, 15, 18, 22, 26), or inadequately chosen (10). Results have been expressed in copy numbers using a plasmid clone (2, 3, 5, 12, 15, 17, 22) or as Cq values (1, 10, 11, 14, 18, 26). Additionally, cross-study comparison is difficult when the type of clinical specimen sampled is different. Most studies have used BAL fluids only (2, 3, 5, 10, 11, 15, 18, 22, 26), but some have included mixed sputa and BAL fluid (1, 14) or sputa and/or oral washes (12, 17). Even though oral washes or sputum samples can be used for P. jirovecii detection, we used the BAL fluids only to keep the quantification consistent.
Most authors rely on immunofluorescence assay and lung imaging to evaluate the performance of their PCR assays (4, 20). Thus, PCR-positive/immunofluorescence-negative samples that are seen in cases without suggestive lung lesions are considered false-positive results and therefore decrease the overall positive predictive value of PCR assays. In contrast, the negative predictive value is high, since samples with positive immunofluorescence and negative PCR results are extremely rare or even absent, as in the present study. If IFA positivity is the criterion to define PcP, the sensitivity of our qPCR assay was 100% for samples with ≥2.6 log10 copies/μl, and the specificity was 100% for samples with ≥4 log10 copies/μl. Thus, for eight IFA-negative patients, specific therapy was not started, although they had positive qPCR results between our thresholds of sensitivity and specificity. This suggests that IFA positivity cannot be the sole criterion for starting specific therapy. Imaging cannot help in the IFA-negative/qPCR-positive cases, since the lung lesions observed were not different from those seen in the IFA-negative/qPCR-negative group. Also, a strategy based mainly on clinical and radiological signs does not take into account the fungal load, although it is an important issue in other infectious diseases. In hematology, for instance, the treatment of cytomegalovirus (CMV) disease is started above a consensual copy number threshold (19), and for infection with Toxoplasma gondii, the recommendation is to start therapy as soon as a blood PCR result is positive (21).
Therapeutic decisions are often made on the basis of the ability to distinguish infection from colonization. The concept of carriage (25) or colonization (24) refers to the presence of the fungus or its DNA in the absence of clinical pneumonia. Since most of the BAL procedures here were performed for investigating fever and/or pneumonia, the detection of P. jirovecii DNA cannot be regarded as simple carriage. For some authors, cases corresponding to positive qPCR and negative microscopy warrant specific therapy when immunosuppression is ongoing (27). Our results suggest that this approach may need to be assessed according to the underlying disease, keeping in mind the limits of our present study. Because of the retrospective design of the study, the qPCR results were not available to guide clinical decision-making, which was done based on IFA results alone. Furthermore, prescription and observance of anti-PcP prophylaxis, doses of steroids, and concomitant immunosuppressive drugs introduced after the BAL results could not be analyzed in the absence of a common follow-up. Likewise, the cause of death in this immunocompromised population could not be determined retrospectively because of the lack of prestudy-defined criteria. For HIV-positive patients, PcP risk largely depends on the CD4+ T cell count, and both treatment and prophylaxis are well established (27). For patients with hematological diseases and SOT recipients, the presence of P. jirovecii DNA did not seem to impact the overall survival at 1 year. However, cotrimoxazole at a prophylactic dose can be resumed whatever the qPCR result in patients with hematological malignancy and SOT recipients (9, 13). In contrast, among patients receiving high doses of steroids for inflammatory diseases, the IFA-negative/qPCR-positive patients had a worse overall 1-year survival than the qPCR-negative ones. There is currently no recommendation for anti-PcP prophylaxis in patients with autoimmune diseases, with the exception of Wegener's granulomatosis (13). Despite this, some authors have encouraged anti-PcP prophylaxis in patients receiving immunosuppressive therapy for rheumatoid arthritis (23). The opposing argument is that some patients would probably recover without specific therapy, as observed in our study.
Given its high reproducibility and with the emergence of commercially available tests (14), there is no doubt that qPCR has the potential to replace microscopy with the help of the necessary quality control panels. More importantly, qPCR has the potential to give reliable quantification and can address the issue of correlation between tissue burden and outcome (16). Then, qPCR-based therapeutic strategies could be evaluated in homogenous groups of patients, keeping in mind the potential toxicity of anti-PcP treatments. Therefore, operational thresholds should be established from prospective studies.
ACKNOWLEDGMENTS
We thank all the clinicians from the hospital for their cooperation in collecting BAL samples and F. Dromer and K. Bruce for their critical reading of the manuscript.
We have no potential conflicts of interest to report.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Alanio A, et al. 2011. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin. Microbiol. Infect. 17: 1531–1537 [DOI] [PubMed] [Google Scholar]
- 2. Arcenas RC, et al. 2006. A real-time polymerase chain reaction assay for detection of Pneumocystis from bronchoalveolar lavage fluid. Diagn. Microbiol. Infect. Dis. 54: 169–175 [DOI] [PubMed] [Google Scholar]
- 3. Bandt D, Monecke S. 2007. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jiroveci. Transpl. Infect. Dis. 9: 196–202 [DOI] [PubMed] [Google Scholar]
- 4. Bollee G, et al. 2007. Clinical picture of Pneumocystis jiroveci pneumonia in cancer patients. Chest 132: 1305–1310 [DOI] [PubMed] [Google Scholar]
- 5. Brancart F, Rodriguez-Villalobos H, Fonteyne PA, Peres-Bota D, Liesnard C. 2005. Quantitative TaqMan PCR for detection of Pneumocystis jiroveci. J. Microbiol. Methods 61: 381–387 [DOI] [PubMed] [Google Scholar]
- 6. Bretagne S. 2011. Advances and prospects for molecular diagnostics of fungal infections. Curr. Infect. Dis. Rep. 12: 430–436 [DOI] [PubMed] [Google Scholar]
- 7. Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622 [DOI] [PubMed] [Google Scholar]
- 8. Costa JM, Ernault P, Gautier E, Bretagne S. 2001. Prenatal diagnosis of congenital toxoplasmosis by duplex real-time PCR using fluorescence resonance energy transfer hybridization probes. Prenat. Diagn. 21: 85–88 [DOI] [PubMed] [Google Scholar]
- 9. de Boer MG, Kroon FP, le Cessie S, de Fijter JW, van Dissel JT. 2011. Risk factors for Pneumocystis jirovecii pneumonia in kidney transplant recipients and appraisal of strategies for selective use of chemoprophylaxis. Transpl. Infect. Dis. doi:10.1111/j.1399–3062.2011.00645.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10. Fillaux J, et al. 2008. Accuracy of a routine real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. J. Microbiol. Methods 75: 258–261 [DOI] [PubMed] [Google Scholar]
- 11. Flori P, et al. 2004. Comparison between real-time PCR, conventional PCR and different staining techniques for diagnosing Pneumocystis jiroveci pneumonia from bronchoalveolar lavage specimens. J. Med. Microbiol. 53: 603–607 [DOI] [PubMed] [Google Scholar]
- 12. Fujisawa T, et al. 2009. Real-time PCR is more specific than conventional PCR for induced sputum diagnosis of Pneumocystis pneumonia in immunocompromised patients without HIV infection. Respirology 14: 203–209 [DOI] [PubMed] [Google Scholar]
- 13. Green H, Paul M, Vidal L, Leibovici L. 2007. Prophylaxis of Pneumocystis pneumonia in immunocompromised non-HIV-infected patients: systematic review and meta-analysis of randomized controlled trials. Mayo Clin. Proc. 82: 1052–1059 [DOI] [PubMed] [Google Scholar]
- 14. Hauser PM, et al. 2011. Multicenter, prospective clinical evaluation of respiratory samples from subjects at risk for Pneumocystis jirovecii infection by use of a commercial real-time PCR assay. J. Clin. Microbiol. 49: 1872–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huggett JF, et al. 2008. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 63: 154–159 [DOI] [PubMed] [Google Scholar]
- 16. Khot PD, Fredricks DN. 2009. PCR-based diagnosis of human fungal infections. Expert Rev. Anti-infect. Ther. 7: 1201–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larsen HH, et al. 2002. Development of a rapid real-time PCR assay for quantitation of Pneumocystis carinii f. sp. carinii. J. Clin. Microbiol. 40: 2989–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Linssen CF, et al. 2006. Inter-laboratory comparison of three different real-time PCR assays for the detection of Pneumocystis jiroveci in bronchoalveolar lavage fluid samples. J. Med. Microbiol. 55: 1229–1235 [DOI] [PubMed] [Google Scholar]
- 19. Ljungman P, et al. 2008. Management of CMV, HHV-6, HHV-7 and Kaposi-sarcoma herpesvirus (HHV-8) infections in patients with hematological malignancies and after SCT. Bone Marrow Transplant. 42: 227–240 [DOI] [PubMed] [Google Scholar]
- 20. Lu Y, et al. 2011. PCR diagnosis of Pneumocystis pneumonia: a bivariate meta-analysis. J. Clin. Microbiol. 49: 4361–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martino R, et al. 2005. Early detection of Toxoplasma infection by molecular monitoring of Toxoplasma gondii in peripheral blood samples after allogeneic stem cell transplantation. Clin. Infect. Dis. 40: 67–78 [DOI] [PubMed] [Google Scholar]
- 22. Meliani L, et al. 2003. Real time quantitative PCR assay for Pneumocystis jirovecii detection. J. Eukaryot. Microbiol. 50(Suppl.): 651. [DOI] [PubMed] [Google Scholar]
- 23. Mori S, Cho I, Sugimoto M. 2009. A follow-up study of asymptomatic carriers of Pneumocystis jiroveci during immunosuppressive therapy for rheumatoid arthritis. J. Rheumatol. 36: 1600–1605 [DOI] [PubMed] [Google Scholar]
- 24. Morris A, Wei K, Afshar K, Huang L. 2008. Epidemiology and clinical significance of pneumocystis colonization. J. Infect. Dis. 197: 10–17 [DOI] [PubMed] [Google Scholar]
- 25. Peterson JC, Cushion MT. 2005. Pneumocystis: not just pneumonia. Curr. Opin. Microbiol. 8: 393–398 [DOI] [PubMed] [Google Scholar]
- 26. Rohner P, Jacomo V, Studer R, Schrenzel J, Graf JD. 2009. Detection of Pneumocystis jirovecii by two staining methods and two quantitative PCR assays. Infection 37: 261–265 [DOI] [PubMed] [Google Scholar]
- 27. Thomas CF, Jr, Limper AH. 2004. Pneumocystis pneumonia. N. Engl. J. Med. 350: 2487–2498 [DOI] [PubMed] [Google Scholar]