Abstract
Hand, foot, and mouth disease (HFMD) is a contagious enteroviral disease occurring primarily in young children and caused by enterovirus 71 (EV71), coxsackievirus A16 (CVA16), and other serotypes of coxsackievirus and echovirus. In this study, a GeXP analyzer-based multiplex reverse transcription (RT)-PCR assay (GeXP assay) consisting of chimeric primer-based PCR amplification with fluorescent labeling and capillary electrophoresis separation was developed to simultaneously identify nine serotypes of enteroviruses associated with HFMD in China, including EV71, CVA16, CVA4, -5, -9, and -10, and CVB1, -3, and -5. The RNAs extracted from cell cultures of viral isolates and synthetic RNAs via in vitro transcription were used to analyze the specificity and sensitivity of the assay. The GeXP assay detected as little as 0.03 tissue culture infective dose (TCID50) of EV71 and CVA16, 10 copies of panenterovirus, EV71, CVA16, CVB1, and CVB5, and 100 copies of 10 (including panenterovirus) premixed RNA templates. A total of 180 stool specimens collected from HFMD patients and persons suspected of having HFMD were used to evaluate the clinical performance of this assay. In comparison with the results of conventional methods, the sensitivities of the GeXP assay for detection of panenterovirus, EV71, and CVA16 were 98.79% (163/165), 91.67% (44/48), and 91.67% (33/36), respectively, and the specificities were 80.00% (12/15), 98.48% (130/132), and 100% (144/144), respectively. The concordance of typing seven other serotypes of enteroviruses with the results of conventional methods was 92.59% (25/27). In conclusion, the GeXP assay is a rapid, cost-effective, and high-throughput method for typing nine serotypes of HFMD-associated enteroviruses.
INTRODUCTION
Hand, foot, and mouth disease (HFMD) is a common acute enteroviral infectious disease which usually affects infants and young children below 10 years old, characterized by a brief febrile illness with a vesicular rash and cutaneous vesicles on the hands, feet, mouth, and buttocks. Some complications such as myocarditis, aseptic meningitis, encephalitis, pulmonary edema, circulatory disturbance, and even death have occurred in a few patients (3, 8, 22). HFMD is caused by enteroviruses, which are members of the picornavirus family (single-stranded RNA, nonenveloped), and is most commonly associated with coxsackievirus A16 (CVA16) and human enterovirus 71 (EV71) (12). Other members of this group, including CVA2, CVA4 to CVA6, CVA9, CVA10, CVB1 to CVB3, CVB5 (1, 3, 5–6, 10, 14, 23), and partial echovirus (ECHO) (2, 9, 22, 32), have also been associated with outbreaks or sporadic cases of HFMD.
The identification and serotyping of enteroviruses have been based on the time-consuming and labor-intensive procedures of viral isolation in cell culture and neutralization with mixed hyperimmune equine serum pools and specific monovalent polyclonal antisera for confirmation (4, 13, 21). Recently, a series of molecular typing methods were developed for rapid identification of enteroviral serotypes, including reverse transcription (RT)-PCR combined amplicon sequencing (17–18), real-time RT-PCR (26–27), nested and seminested PCR (16), PCR-RFLP (restriction fragment length polymorphism) assay for typing of enteroviruses causing aseptic meningitis in Korea (11), microwell oligonucleotide arrays (24), and an RT-PCR-based reverse line blot (RLB) hybridization assay (31). These molecular methods could sensitively identify different serotypes of enteroviruses; however, the real-time RT-PCR and nested and seminested PCR were used for detecting only a limited number of serotypes. The PCR-RFLP, microwell oligonucleotide arrays, and RLB hybridization assay could simultaneously detect several viral serotypes; however, costly, time-consuming, and labor-intensive manual procedures were needed. Therefore, a rapid, cost-effective, and high-throughput method for typing the HFMD associated enteroviruses was needed.
The GeXP analyzer is a multiplex gene expression profiling analysis platform developed by Beckman Coulter Company (Brea, CA) which was previously used in the rapid identification of gene expression prostate cancer biomarker signatures in biological samples, rapid and sensitive detection of 68 unique varicella-zoster virus gene transcripts (15), and detection of pandemic influenza A H1N1 virus (19). The principle of the GeXP multiplex amplification assay is based on the amplification of two sets of primers, the universal primers and the gene-specific chimeric primers (gene-specific primers linked to the universal primer sequences at the 3′ end). During the first few cycles of PCR, amplification is carried out by chimeric forward and reverse primers. In the later stages of PCR, amplification is predominantly carried out by universal forward and reverse primers. All gene targets in the multiplex panel are amplified by universal primers. The forward universal primer is labeled with a fluorescent dye, enabling subsequent fluorescence detection of amplicons by capillary electrophoresis.
In this study, a GeXP analyzer-based multiplex RT-PCR assay (GeXP assay) was developed to simultaneously detect nine common serotypes of enteroviruses associated with HFMD in China, including EV71, CVA16, CVA4, CVA5, CVA9, CVA10, CVB1, CVB3, and CVB5. This assay can be implemented effectively in routine testing environments by allowing users to process more samples in less time than existing assays and platforms.
MATERIALS AND METHODS
Viral isolates and RNA extraction.
Twenty-eight serotypes of cell-cultured enterovirus isolates used for evaluating the sensitivity and specificity of the GeXP assay were obtained from National Laboratory for Poliomyelitis, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention. These isolates were used as control viruses and were identified previously by sequencing or neutralization tests. These control viruses included EV71, CVA16, CV2, CVA4, CVA5, CVA6, CVA9, CVA10, CVA12, CVA14, CVA24v, CVB1 to CVB6, ECHO1 to ECHO7, ECHO11, ECHO13, ECHO19, and ECHO30. Among them, the EV71 isolate (strain FY17.08/AN/CHN/2008; GenBank accession no. EU703812) and CVA16 isolate (strain FY18/AN/CHN/2008; GenBank accession no. EU812514) were used as reference viruses in this study; both reference viruses had an infectivity titer of 106.5 50% tissue culture infective doses (TCID50)/ml on human rhabdomyosarcoma (RD) cells. The viral RNAs were extracted from 140 μl of cell culture of various reference virus stocks using a QIAamp viral RNA minikit (Qiagen) according to the manual and eluted in 50 μl of nuclease-free water. The eluted RNA was aliquoted and stored at −80°C until needed (7).
Primers.
The GeXP multiplex assay consisted of 11 pairs of chimeric primers (including one pair of panenterovirus primers and 10 pairs of human enterovirus serotype-specific primers) and one pair of universal primers (Tag-F/Tag-R) (Table 1.). Each of these chimeric primers consisted of a gene-specific sequence for each virus fused at the 5′ end to the universal sequence. Both the forward and reverse universal sequences were quasi-T7 sequences and selected by default using the GeXP eXpress Profiler software. Tag-F/Tag-R was the same as the forward or reverse universal sequence. This strategy was first developed by Beckman Coulter Company and was reported in our previous study (19). The panenterovirus primers (PE-F and PE-R) were designed in a highly conserved region of the 5′ untranslated region (UTR) of the enterovirus genomes as reported by Yang et al. (28). Ten pairs of human enterovirus serotype-specific primers for the nine serotypes of enteroviruses were designed in relatively conserved VP1 regions of each serotype. Serotype-specific primers sequences were evaluated using the NCBI Primer-Blast, Primer Premier 5.0, and Oligo 6.0 software. Degenerate bases were introduced to cover different strains. The 5′ end of the forward universal primer (Tag-F) was labeled with the fluorescent dye Cy5 and purified with high-pressure liquid chromatography. All chimeric primers and the reverse universal primer (Tag-R) were synthesized and purified by polyacrylamide gel electrophoresis (Invitrogen, Shanghai, China).
Table 1.
Oligonucleotide primers for the GeXP assay
| Primer | Sequence (5′–3′)a | GenBank accession no. | Position | Size (bp) |
|---|---|---|---|---|
| PE-F | AGGTGACACTATAGAATATCCGGCCCCTGAATGCGGCTAATCC | FJ713137.1 | 450–474 | 151 |
| PE-R | GTACGACTCACTATAGGGAACACGGACACCCAAAGTAGTCGGTCC | 563–538 | ||
| EV71-F | AGGTGACACTATAGAATAGCAGCCCAAAAGAACTTCAC | FJ713137.1 | 2368–2387 | 263 |
| EV71-R | GTACGACTCACTATAGGGAATTTCAGCAGCTTGGAGTGC | 2594–2575 | ||
| CVA16-F | AGGTGACACTATAGAATAATTGGTGCTCCCACTACAGC | GQ279371.1 | 2519–2539 | 245 |
| CVA16-R | GTACGACTCACTATAGGGATCAGTGTTGGCAGCTGTAGG | 2349–2330 | ||
| CVA4-F2 | AGGTGACACTATAGAATACCTAARCCTGATGCYCGAGA | EU908135.1 | 1–20 | 271 |
| CVA4-F3 | AGGTGACACTATAGAATACCTAAGCCTGATGCCCGAGA | 1–20 | ||
| CVA4-R2 | GTACGACTCACTATAGGGACAACTCTAGCTGRGAATGTYCCT | 210–233 | ||
| CVA5-F4 | AGGTGACACTATAGAATAATGAGCCCAGCYAGYACYTA | AY421763.1 | 3013–3032 | 181 |
| CVA5-R4 | GTACGACTCACTATAGGGAGATACHCCTGASACCATGCG | 3036–3157 | ||
| CVA9-F2 | AGGTGACACTATAGAATATTTGATCAGAAGGGCTCATACGGGT | GQ294574.1 | 604–628 | 251 |
| CVA9-R2 | GTACGACTCACTATAGGGATCTGTGATGGGTGTTGGTGTAAA | 796–818 | ||
| CVA10-F2 | AGGTGACACTATAGAATACGBTGTGTGGTTAAYAGRAATGG | GU947783.1 | 198–221 | 274 |
| CVA10-R2 | GTACGACTCACTATAGGGAGCCTCTCCATTYTTAGTCGTTGT | 412–434 | ||
| CVA10-R3 | GTACGACTCACTATAGGGAGCTTCYCCAYCTTCAGTHGTTGT | 412–434 | ||
| CVB1-F1 | AGGTGACACTATAGAATAGAAAATTTCCTGTGCCGGT | GU949568.1 | 193–221 | 223 |
| CVB1-R1 | GTACGACTCACTATAGGGAGGGTTGTTGTGCACTCGTTA | 359–378 | ||
| CVB1-F2 | AGGTGACACTATAGAATAGAGAATTTCCTGTGCCGGTC | AY373099.1 | 83–102 | 223 |
| CVB1-R2 | GTACGACTCACTATAGGGATAGGTTGTTGGGCGCTTGTTA | 249–269 | ||
| CVB3-F2 | AGGTGACACTATAGAATAATGTCCATACCTTTCCTGAGTATTGG | GQ141875.1 | 2985–3010 | 282 |
| CVB3-R2 | GTACGACTCACTATAGGGATTTGCCTTCTCATACTGGCA | 3210–3229 | ||
| CVB5-F1 | AGGTGACACTATAGAATAGCTCACGCATCAAATCATGT | GQ246507.1 | 414–433 | 193 |
| CVB5-R1 | GTACGACTCACTATAGGGAGCATTGCCTATGCTGATGAA | 550–569 | ||
| Cy5-labeled Tag-F | AGGTGACACTATAGAATA | |||
| Tag-R | GTACGACTCACTATAGGGA |
Universal tag sequences are underlined. Bold type shows degenerate sites. Abbreviations are as follows: M = A or C; R = A or G; W = A or T; S = G or C; Y = C or T; K = G or T; V = A, G, or C; H = A, C, or T; D = A, G, or T; B = G, C, or T.
Evaluation of the specificity of the GeXP assay.
Firstly the mono RT-PCR assay was developed individually with extracted RNA from 28 cell-cultured serotypes of enterovirus strains to evaluate the specificity of each pair of gene-specific primers (including PE-F and PE-R) and ascertain the actual amplicon size of each target region. Both the mono-RT-PCR assay and the multiplex RT-PCR were performed with a Qiagen one-step RT-PCR kit in a 25-μl volume containing 5 μl of 5× Qiagen one-step RT-PCR buffer, 1 μl of RT-PCR enzyme mix, 1 μl of deoxynucleotide triphosphate (10 mM each) mix, 5 units of RNase inhibitor, and 1 μl of extracted viral RNA from each serotype. The mono-RT-PCR assay contained a 50 nM concentration of each pair of gene-specific chimeric primers individually, while the GeXP assay contained 50 nM concentrations of 11 pairs of gene-specific chimeric primers and a 500 nM concentration of the universal tag primers (final concentrations); nuclease-free water was added to a volume of 25 μl. The RT-PCR was performed under the following conditions: 50°C for 30 min (the RT reaction) and 95°C for 15 min, followed by three steps of amplification according to the temperature switch PCR (TSP) strategy (25): step 1, 10 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; step 2, 10 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s; step 3, 20 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s.
Separation by capillary electrophoresis and fragment analysis.
PCR product separation and detection were performed on a GenomeLab GeXP genetic analysis system (Beckman Coulter) by capillary electrophoresis, following the protocols described previously (20). After amplified fragments were separated, the peaks were initially analyzed using the fragment analysis module of the GeXP system software and matched to the appropriate genes. The peak height for each gene was reported in the electropherogram.
Evaluation of the sensitivity of the GeXP assay.
Sensitivity was tested by following the method described previously (16), using titrated reference viruses (EV71 and CVA16) obtained from infected human RD cells. Serial 10-fold dilutions of the EV71 and CVA16 stock were made in Hanks' balanced salt solution, and RNA from 140 μl of each dilution was extracted with the QIAamp viral RNA minikit (Qiagen). The RNA preparations, ranging from 105.5 to 10−1.5 TCID50 (0.03 TCID50) per microliter, were tested with the GeXP assay. The assay at each template concentration was repeated three times.
Absolute sensitivity was measured by using in vitro transcribed synthetic RNAs derived from 10 recombinant plasmids containing the VP1 sequences of the nine serotypes of enteroviruses or the 5′ UTR. The in vitro-synthesized RNA products were purified with an RNeasy MinElute cleanup kit (Qiagen) and quantitated by spectrophotometry (NanoDrop ND-1000). The concentrated product for each serotype was diluted to final concentrations ranging from 105 copies to 1 copy per microliter and then individually subjected to the GeXP assay. The concentrations of specific primers were optimized according to the amplification efficiency of the GeXP assay using a single template. The sensitivity of the optimized GeXP assay was re-evaluated by using 10 premixed RNA templates ranging from 105 copies to 10 copies for each serotype per microliter for three times on three different days.
Application to clinical specimens.
RNAs were extracted from 180 original clinical stool specimens obtained from HFMD patients and persons suspected of having HFMD to illustrate the application of the optimized GeXP assay. All the clinical stool specimens were assayed in parallel by conventional methods, including isolation of the viruses followed by neutralization testing and conventional RT-PCR followed by amplicon sequencing (21, 29–30).
RESULTS
Evaluation of the specificity of the GeXP assay.
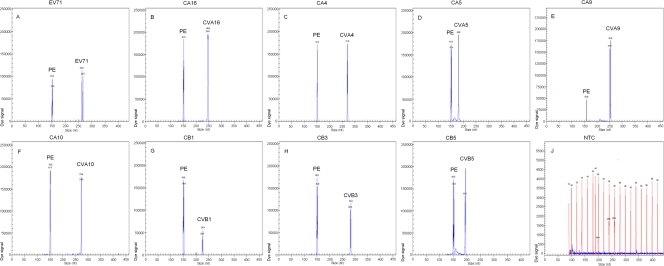
The RNAs of 28 cell-cultured serotypes of enterovirus isolates were individually used as templates to evaluate the specificity of each pair of gene-specific primers. In mono-RT-PCR assays, the panenterovirus universal primers could amplify the target genes of all 28 serotypes of enteroviruses, but each pair of serotype-specific primers generated corresponding VP1 genes only of the targeted serotype, without cross-amplification. The amplicon sizes for the serotypes were as follows: panenterovirus, 150 to 153 bp; EV71, 264 to 267 bp; CVA16, 246 to 248 bp; CVA4, 270 to 272 bp; CVA5, 180 to 181 bp; CVA9, 251 to 254 bp; CVA10, 274 to 276 bp; CVB1, 224 to 225 bp; CVB3, 281 to 284 bp; and CVB5, 193 to 196 bp (electropherograms not shown). The nine serotypes of enteroviruses associated with HFMD were detected via the GeXP assay. Two specific amplification peaks were observed, representing the panenterovirus target amplicon and the serotype-specific target amplicon (Fig. 1).
Fig 1.
Specificity of the multiplex RT-PCR assay. Cy5-labeled PCR products were separated via GeXP capillary electrophoresis and detected by fluorescence spectrophotometry, given as dye signals in arbitrary units on the y axis. Each peak was identified by comparing the expected to the actual PCR product size on the x axis. EV71, CVA16, CVA4, CVA5, CVA9, CVA10, CVB1, CVB3, and CVB5 were assayed by using enterovirus RNAs extracted from various cell-cultured strains. Nuclease-free water was used as the negative control (NTC) (J).
Sensitivity of the GeXP assay.
The sensitivity of the GeXP assay was measured using titrated reference viruses (EV71 and CVA16 stocks) or in vitro-transcribed RNAs of nine serotypes of enteroviruses and panenterovirus. The GeXP assay detected as little as 0.03 TCID50 of EV71 and CVA16, 10 copies of panenterovirus, EV71, CVA16, CVB1, CVB5, and more than 100 copies of CVA4, CVA5, CVA9, CVA10, and CVB3 with a single RNA template.
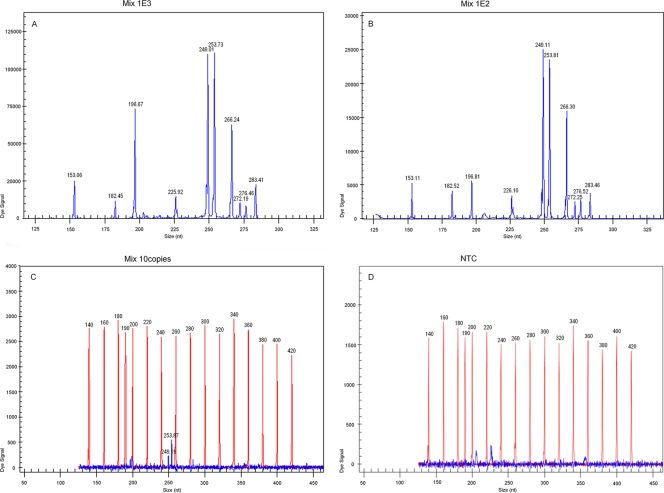
Based on all the sensitivity results of GeXP assay with a single RNA template, the concentration of each chimeric primer in the new GeXP assay was adjusted as follows (F, forward; R, reverse): PE (F/R), 20 nM; EV71 (F/R), 50 nM; CVA16 (F/R), 40 nM; CVA4 (F2, F3/R2), 80 nM; CVA5 (F4/R4), 50 nM; CVA9 (F2/R2), 70 nM; CVA10 (F2/R2, R3), 100 nM; CVB1 (F1/R1; F2/R2), 30 nM; CVB3 (F2/R2), 100 nM; CVB5 (F1/R1), 40 nM. The optimized GeXP assay achieved a sensitivity of 100 copies with 10 premixed RNA templates in three independent experiments on three different days (Fig. 2), and the coefficient of variation was less than 10.15% (not shown).
Fig 2.
Sensitivity of GeXP detection of 10 premixed RNA templates with multiplex primers. All 10 target genes could be detected at 103 copies/μl (A) and 102 copies/μl levels (B); only CVA16 and CVA9 could be detected at 10 copies/μl levels (C). Nuclease-free water was used as the negative control (D).
Application to clinical specimens.
A total of 180 specimens collected from HFMD patients and suspects were assayed simultaneously by both the GeXP assay and the conventional methods, including viral isolation in cell culture, neutralization, and conventional RT-PCR followed by amplicon sequencing (21, 29–30). The numbers of panenterovirus and each serotype of enterovirus detected by different methods are shown in Table 2 and 3. In comparison with the results of conventional methods, the sensitivities of the GeXP assay for detection of panenterovirus, EV71, and CVA16 were 98.79% (163/165), 91.67% (44/48), and 91.67% (33/36), respectively, and the specificities were 80.00% (12/15), 98.48% (130/132), and 100% (144/144), respectively. The concordance between the detection results of GeXP assay and the results of conventional methods for typing seven other serotypes of enteroviruses was 92.59% (25/27).
Table 2.
Comparison of GeXP assay and conventional methods for detecting panenterovirus, EV71, and CVA16a
| GeXP assay result and virus | No. of samples with result by conventional methods |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | |||
| Panenterovirus | 163 | 3 | 166 |
| EV71 | 44 | 2 | 46 |
| CVA16 | 33 | 0 | 33 |
| Negative | |||
| Panenterovirus | 2 | 12 | 14 |
| EV71 | 4 | 130 | 134 |
| CVA16 | 3 | 144 | 147 |
| Total | |||
| Panenterovirus | 165 | 15 | 180 |
| EV71 | 48 | 132 | 180 |
| CVA16 | 36 | 144 | 180 |
All 180 stool specimens had been identified previously by cell culture and classical PCR followed by sequencing. A neutralization test was performed for samples positive for EV71 (n = 48) and CVA16 (n = 36). Twelve negative specimens were detected by both the GeXP assay and the conventional methods. Two specimens that were negative by the GeXP assay were enterovirus positive by the conventional methods. The false negatives of GeXP assay might due to the RNA degradation or occurrence of PCR inhibition with the samples. Three specimens that were negative by the conventional methods were enterovirus positive by the GeXP assay, which were confirmed later by independent RT-PCR and sequencing to be true positives. As a measure of agreement, kappa values for panenterovirus (P = 0.000), EV71 (P = 0.000), and CVA16 (P = 0.000) were 0.813, 0.914, and 0.946, respectively (using SPSS13.0).
Table 3.
Comparison of the GeXP assay and conventional methods for detecting seven serotypes of enteroviruses
| Serotype | No. of samples positive by: |
|
|---|---|---|
| GeXP assay | Conventional methods | |
| CVA4 | 7 | 7 |
| CVA5 | 3 | 3 |
| CVA9 | 2 | 3 |
| CVA10 | 3 | 3 |
| CVB1 | 2 | 2 |
| CVB3 | 4 | 4 |
| CVB5 | 4 | 5 |
DISCUSSION
In our study, 10 pairs of serotype-specific primers and one pair of panenterovirus universal primers were designed to develop a GeXP assay for simultaneous identification of nine serotypes of enteroviruses, including EV71, CVA16, CVA4, CVA5, CVA9, CVA10, CVB1, CVB3, and CVB5. The selection of enteroviruses associated with HFMD was based on the research data from the National Notifiable Disease Reporting System (NNDRSe) in China and the previous study (10, 23). The serotype-specific primers were designed to target relatively conserved VP1 regions of each enterovirus serotype, and the panenterovirus universal primers were designed from the 5′ UTR (28). Due to the high degree of diversity among VP1 sequences, some degenerate bases were introduced, and more than two primers for some enterovirus serotypes were designed to cover the majority of the viral sequences, such as the primers for CVA4, CVA10, and CVB1.
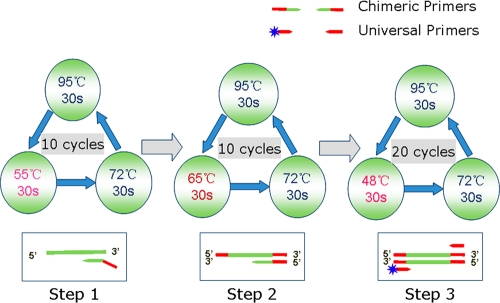
The original standard workflow of GeXP was performed in two separate tubes with an RT reaction and a subsequent PCR amplification, as described in the previous reports (15, 19–20), which is a costly, time-consuming, tedious process and apt to bring carryover contamination. In order to eliminate these problems, a one-step RT-PCR kit (Qiagen) was adopted and replaced the GeXP start kit (Beckman Coulter Company). The amplification procedure of the GeXP assay was improved via temperature switch PCR (TSP) strategy (25), including three steps with different annealing temperatures (Fig. 3): step 1 was carried out with gene-specific sequences of chimeric forward and reverse primers, step 2 was carried out mainly with chimeric forward and reverse primers, and step 3 was carried out predominantly with universal forward and reverse tag primers. The GeXP assay, based on the use of chimeric and fluorescent-dye-labeled universal tag primers and the TSP strategy, offered highly sensitive and specific amplifications of different genes in one multiplex RT-PCR assay, avoided preferred and inferior amplification, and minimized nonspecific reactions. The resolution of GeXP analyzer-based capillary electrophoresis is superior to that of conventional capillary electrophoresis. The forward universal primer was fluorescently labeled in the GeXP assay. The resulting dye-labeled PCR products were separated and detected with a Beckman Coulter GenomeLab GeXP genetic analysis system using capillary electrophoresis. After amplified fragments were separated, the data were initially analyzed using the fragment analysis module of the GeXP system software. Then each amplified fragment would be present as a sole peak with an accurate size on the electropherogram. One can clearly differentiate two peaks with a 3-nucleotide or greater difference in a practical way. In theory, the target PCR productions of CVA4 and CVA10 are 271 bp and 274 bp, respectively. The nucleotide difference between them is 3 bp.
Fig 3.
Diagram of the GeXP amplification workflow using a temperature switch PCR strategy.
The improved GeXP assay was further optimized by adjusting the concentration of each chimeric primer in the reaction based on the individual sensitivity results of the GeXP assay with a single RNA template to overcome the potential interference due to preferential amplification in mixed infections. In this study, the relative and absolute sensitivities were analyzed to evaluate the detection limit of the GeXP assay. The optimal GeXP assay detected as little as 0.03 TCID50 of EV71 and EVA16, which is comparable to the detection sensitivity of the real-time RT-PCR assay published recently (26) and slightly lower than the sensitivity reported for seminested RT-PCR (16). The absolute sensitivity of the optimal GeXP assay was 100 copies for simultaneously detecting 10 target genes without cross or nonspecific amplification. In a test of 180 samples, the sensitivities of the GeXP assay for detection of panenterovirus, EV71, and CVA16 were 98.79% (163/165), 91.67% (44/48), and 91.67% (33/36), respectively, and the specificities were 80.00% (12/15), 98.48% (130/132), and 100% (144/144), respectively, compared with conventional methods, revealing a high sensitivity and specificity in the detection of these viruses. Twelve negative specimens were identified by both the GeXP assay and the conventional methods. Two specimens that were negative by the GeXP assay were enterovirus positive by the conventional methods. The false-negative results in the GeXP assay might be due to RNA degradation or to the occurrence of PCR inhibition with the samples. Three specimens that were negative by the conventional methods were enterovirus positive by the GeXP assay, and these were confirmed later by independent RT-PCR and sequencing to be true positives. Because of the limited number of positive samples for the other seven serotypes of enteroviruses (CVA4, CVA5, CVA9, CVA10, CVB1, CVB3, and CVB5), the GeXP assay needs to be validated with a larger number of clinical samples for these viruses.
Two distinct advantages of the GeXP assay are the time savings and cost effectiveness. The cost of the GeXP assay for simultaneous detection of 9 serotypes of enteroviruses is approximately $8 per test, versus $8 per test for each virus using a commercial real-time RT-PCR kit. The whole reaction was completed in one tube in a one-step multiplex RT-PCR within 2.5 h, followed by capillary electrophoresis separation. In addition, two 96-well plates can be placed in parallel in a GeXP machine at the same time to further increase the throughput of the samples.
Conclusion.
This study has demonstrated that the GeXP assay is a rapid, cost-effective, high-throughput method with high sensitivity and specificity for typing most HFMD-associated enteroviruses, which may be adopted for general use in the Department of Viral Disease Control and Prevention for molecular epidemiologic survey of enteroviruses.
ACKNOWLEDGMENTS
This work was supported by the China Mega-Project for Infectious Disease (2011ZX10004-001, 2009ZX10004-101, 202, 2008ZX10004-002, 001, 004).
A Chinese patent (application number 201110089404.9) has been filed for the combination of all the listed primers specific to 10 HFMD associated enteroviruses and experimental parameters. Xuejun Ma and Wenbo Xu are inventors on the patent application. The technology is available for research-only purposes.
Footnotes
Published ahead of print 23 November 2011
REFERENCES
- 1. Abubakar S, Chee HY, Shafee N, Chua KB, Lam SK. 1999. Molecular detection of enteroviruses from an outbreak of hand, foot and mouth disease in Malaysia in 1997. Scand. J. Infect. Dis. 31:331–335 [DOI] [PubMed] [Google Scholar]
- 2. Apisarnthanarak A, Kitphati R, Pongsuwann Y, Tacharoenmueng R, Mundy LM. 2005. Echovirus type 11: outbreak of hand-foot-and-mouth disease in a Thai hospital nursery. Clin. Infect. Dis. 41:1361–1362 [DOI] [PubMed] [Google Scholar]
- 3. Barriere H, Berger M, Billaudel S. 1976. Hand, foot and mouth disease. Sem. Hop. 52:2215–2220 [In French.] [PubMed] [Google Scholar]
- 4. Bell EJ, McCartney RA, Basquill D, Chaudhuri AK. 1986. Mu-antibody capture ELISA for the rapid diagnosis of enterovirus infections in patients with aseptic meningitis. J. Med. Virol. 19:213–217 [DOI] [PubMed] [Google Scholar]
- 5. Blomqvist S, Klemola P, Kaijalainen S, Paananen A, Simonen ML, Vuorinen T, Roivainen M. 2010. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J. Clin. Virol. 48:49–54 [DOI] [PubMed] [Google Scholar]
- 6. Chen SP, et al. 2010. Comparison of clinical features between coxsackievirus A2 and enterovirus 71 during the enterovirus outbreak in Taiwan, 2008: a children's hospital experience. J. Microbiol. Immunol. Infect. 43:99–104 [DOI] [PubMed] [Google Scholar]
- 7. Chen TC, et al. 2006. Combining multiplex reverse transcription-PCR and a diagnostic microarray to detect and differentiate enterovirus 71 and coxsackievirus A16. J. Clin. Microbiol. 44:2212–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frydenberg A, Starr M. 2003. Hand, foot and mouth disease. Aust. Fam. Physician 32:594–595 [PubMed] [Google Scholar]
- 9. Han JF, et al. 2011. Echovirus 30 in EV71-associated hand, foot and mouth disease outbreak, Guangxi, China. J. Clin. Virol. 50:348–349 [DOI] [PubMed] [Google Scholar]
- 10. Hu YF, et al. 2011. Complete genome analysis of coxsackievirus A2, A4, A5, and A10 strains isolated from hand, foot, and mouth disease patients in China revealing frequent recombination of human enterovirus A. J. Clin. Microbiol. 49:2426–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YS, et al. 2002. PCR-RFLP based molecular typing of enteroviruses isolated from patients with aseptic meningitis in Korea. Arch. Virol. 147:1711–1720 [DOI] [PubMed] [Google Scholar]
- 12. Li L, et al. 2005. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 43:3835–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin TL, et al. 2008. Rapid and highly sensitive coxsackievirus A indirect immunofluorescence assay typing kit for enterovirus serotyping. J. Clin. Microbiol. 46:785–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindenbaum JE, Van Dyck PC, Allen RG. 1975. Hand, foot and mouth disease associated with coxsackievirus group B. Scand. J. Infect. Dis. 7:161–163 [DOI] [PubMed] [Google Scholar]
- 15. Nagel MA, Gilden D, Shade T, Gao B, Cohrs RJ. 2009. Rapid and sensitive detection of 68 unique varicella zoster virus gene transcripts in five multiplex reverse transcription-polymerase chain reactions. J. Virol. Methods 157:62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oberste MS, Nix WA, Maher K, Pallansch MA. 2003. Improved molecular identification of enteroviruses by RT-PCR and amplicon sequencing. J. Clin. Virol. 26:375–377 [DOI] [PubMed] [Google Scholar]
- 19. Qin M, et al. 2010. Detection of pandemic influenza A H1N1 virus by multiplex reverse transcription-PCR with a GeXP analyzer. J. Virol. Methods 168:255–258 [DOI] [PubMed] [Google Scholar]
- 20. Rai AJ, Kamath RM, Gerald W, Fleisher M. 2009. Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal. Bioanal. Chem. 393:1505–1511 [DOI] [PubMed] [Google Scholar]
- 21. Rigonan AS, Mann L, Chonmaitree T. 1998. Use of monoclonal antibodies to identify serotypes of enterovirus isolates. J. Clin. Microbiol. 36:1877–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russo DH, Luchs A, Machado BC, Carmona RC, Timenetsky MC. 2006. Echovirus 4 associated to hand, foot and mouth disease. Rev. Inst. Med. Trop. Sao Paulo 48:197–199 [DOI] [PubMed] [Google Scholar]
- 23. Shi HJ, et al. 2010. Biological characters of a coxsackievirus B3 variant strain isolated from a hand-foot-mouth disease patient with severe clinical symptoms. Zhonghua Yi Xue Za Zhi 90:1141–1144 [PubMed] [Google Scholar]
- 24. Susi P, et al. 2009. Typing of enteroviruses by use of microwell oligonucleotide arrays. J. Clin. Microbiol. 47:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tabone T, Mather DE, Hayden MJ. 2009. Temperature switch PCR (TSP): robust assay design for reliable amplification and genotyping of SNPs. BMC Genomics 10:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao XL, et al. 2009. Simultaneous detection of human enterovirus 71 and coxsackievirus A16 in clinical specimens by multiplex real-time PCR with an internal amplification control. Arch. Virol. 154:121–125 [DOI] [PubMed] [Google Scholar]
- 27. Xiao XL, et al. 2009. Simultaneous detection of enterovirus 70 and coxsackievirus A24 variant by multiplex real-time RT-PCR using an internal control. J. Virol. Methods 159:23–28 [DOI] [PubMed] [Google Scholar]
- 28. Yang CF, et al. 1992. Genotype-specific in vitro amplification of sequences of the wild type 3 polioviruses from Mexico and Guatemala. Virus Res. 24:277–296 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y, et al. 2009. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 44:262–267 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y, et al. 2010. Molecular evidence of persistent epidemic and evolution of subgenotype B1 coxsackievirus A16-associated hand, foot, and mouth disease in China. J. Clin. Microbiol. 48:619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou F, et al. 2009. Identification of 20 common human enterovirus serotypes by use of a reverse transcription-PCR-based reverse line blot hybridization assay. J. Clin. Microbiol. 47:2737–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu Z, et al. 2007. Molecular epidemiological analysis of echovirus 19 isolated from an outbreak associated with hand, foot, and mouth disease (HFMD) in Shandong Province of China. Biomed. Environ. Sci. 20:321–328 [PubMed] [Google Scholar]